Abstract
The selection of drug-resistant variants of human immunodeficiency virus type 1 (HIV-1) is an impediment to the efficiency of antiretroviral (ARV) therapy. We have developed an allele-specific real-time PCR assay to explore the presence of K65R minority species among treated HIV-1 subtype B and C infections. Thirty HIV-1 subtype C- and 26 subtype B-infected patients lacking K65R as determined by conventional sequencing methods were studied, and viral minority species were found in four HIV-1 subtype C samples.
Currently, there are nine recognized subtypes of human immunodeficiency virus type 1 (HIV-1) (A to D, F to H, J, and K) and 43 circulating recombinant forms (http://www.hiv.lanl.gov/). More than 50% of the worldwide epidemic is due to HIV-1 subtype C (23). Subtype C viruses display certain unique genetic characteristics that are associated with specific genotypic resistance pathways. For example, a V106M mutation (GTG → ATG) confers high-level resistance against multiple nonnucleoside reverse transcriptase inhibitors (NNRTIs), with the V106A substitution being more common in subtype B (1). Moreover, unique polymorphisms at reverse transcriptase (RT) codons 64 (AAG → AAA), 65 (AAA → AAG), and 66 (AAA → AAG) may cause the K65R nucleoside resistance mutation to be selected more rapidly by several members of the nucleoside RT inhibitor (NRTI) family of drugs in subtype C than by those in subtype B both in cell culture and in the clinic (2, 5).
Among the drugs that can be affected by the K65R mutation is tenofovir (TFV), a compound with a generally favorable resistance and safety profile as well as a long intracellular half-life. TFV-based therapies are recommended by current guidelines for the treatment of antiretroviral-naïve patients, and TFV is also a drug of choice in HIV prophylaxis drug-based strategies directed against the spread of HIV (8).
In developing-country settings, the use of stavudine (d4T) and didanosine (ddI) with nevirapine (NVP) has been associated with increased rates of selection of K65R. The incidences of K65R in genotyped persons for whom treatment failed were 7%, 9%, 14%, 23%, and 30% in clinical studies in Thailand, Senegal, South Africa, Malawi, and Botswana, respectively (5, 7, 9, 20, 21). Tissue culture studies have confirmed that d4T and ddI can select K65R with high efficiency in subtype C viruses (10).
It is important to develop sensitive techniques to detect minority K65R species in drug-naïve persons and cases of early virological failure. Primary infection with K65R may decrease the efficacy of initial therapy. In this regard, a study in Malawi showed that K65R could have been transmitted together with NVP resistance to 3 of 4 breast-feeding mothers treated postpartum with d4T/lamivudine (3TC)/NVP (13).
There is a need to explore the prevalence of certain mutations that are present as members of minority species in both drug-naïve and treatment-experienced patients (11, 22). Conventional genotyping may fail to detect K65R due to the poor replicative capacity of viruses containing this mutation as well as to K65R/M184V and K65R/thymidine analogue mutation (TAM) bidirectional mutational antagonism (14, 18, 19). In this study, subtype B- and C-specific allele-specific PCR (AS-PCR) assays were developed to analyze the presence of K65R in patients on antiretroviral therapy.
(This study was presented in part at the 15th International Conference on AIDS in Africa [ICASA], Dakar, Senegal, 3 to 7 December 2008 [25].)
Drug resistance genotyping.
A random sampling of 42 HIV-1 subtype C- and 26 subtype B-infected patients were studied as part of the Quebec provincial HIV genotyping program. Among individuals bearing subtype C infections, 30 had received therapy, and at least one drug regimen had failed for them, while 11 were drug naïve. All of the subtype B patients had at least once received therapy that failed for them. All patients possessed HIV viral loads of >1,000 copies of RNA/ml. All of the 30 subtype C patients for whom therapy had failed and 25 of the 26 subtype B patients for whom therapy had failed possessed mutations associated with resistance to at least one nucleoside reverse transcriptase inhibitor (NRTI). None of these individuals possessed the K65R mutation, as determined by bulk sequencing, which analyzed a region of the HIV pol gene spanning the entire protease (PR)-coding region and most of the RT-coding region (codons 1 to 400) (Virco; BVBA, Mechelen, Belgium) (International AIDS Society—USA [12]). Viral subtypes were determined using Mega 3 software.
Quantification of minority resistance species by AS-PCR.
The PCR products of HIV-1 subtype B and C reactions were further studied by K65R allele-specific PCR (AS-PCR) (3). Purified PCR products amplified from plasma RNA, as described for the genotypic resistance analysis, were then submitted for nested real-time PCR enabling the specific amplification of wild-type or mutated sequences or whole populations for codon 65. The PCRs were performed using SYTO9 (Invitrogen), an intercalating dye, and HIV-1 subtype B and C 3′ lock nucleic acid (LNA)-specific primers. The forward primers used for subtype B were B65R-fw (5′-CTC CAG TAT TTG CCA TAA AGA+G-3′), B65wt-fw (5′-CTC CAG TAT TTG CCA TAA AGA+A-3′), and B65tot-fw (5′-CTC CAG TAT TTG CCA TAA AGA-3′) for K65R, wild-type K65, and the total population, respectively. The equivalent forward primers for subtype C were C65R-fw (5′-CTC CAG TAT TTG CCA TAA AAA+G-3′), C65wt-fw (5′-CTC CAG TAT TTG CCA TAA AAA+A-3′), and C65tot-fw (5′-CTC CAG TAT TTG CCA TAA AAA-3′), respectively. The same reverse primer, Rev65m (5′-TAT TCC TAA TTG AAC TTC CCA-3′), was used in all reactions. The real-time PCRs were performed with a Rotor-Gene 6000 real-time rotary analyzer (Corbett Research, Sydney, Australia) with a Platinum Taq DNA polymerase kit (Invitrogen). The final concentration of each primer was 0.3 μM, and the final SYTO9 concentration was 1.5 μM.
As a control, K65R was introduced by site-directed mutagenesis (SDM) into HIV-1 subtype B pNL4-3 and subtype C MJ4 wild-type plasmids by using a QuikChange SDM kit as specified by the manufacturer (Stratagene, La Jolla, CA). The introduction of the K65R mutation into the plasmid was confirmed by sequencing, and the plasmid was ultimately transformed into DH5α (Invitrogen) for high-yield plasmid production. The wild-type and mutated standards were tested in duplicate for each experiment and were prepared by serial dilution from 109 to 106 copies/μl. The viral PCR products were tested at 107 copies/μl. The absence of the K65R mutation in the corresponding patient PCR products was confirmed by sequencing. For quantification of various viral populations, DNA standards were prepared and run in parallel with each sample reaction.
Real-time PCRs were executed with 50 cycles of melting at 94°C for 10 s, annealing at 55°C for 15 s, and extension at 72°C for 30 s. This was followed by two steps, the first at 72°C for 1 min and the second at 50°C for 30 s. The assays were ended with a high-resolution melt (HRM), with a ramp from 65°C to 90°C. The specificity of detection of the amplified product was verified by analysis of the melting curve for each reaction.
To evaluate the discriminatory ability of each assay, mutant and wild-type DNA standards were amplified using the corresponding and noncorresponding AS primers. For the HIV-1 subtype C MJ4 standard, the discriminatory thresholds for amplification of K65R mutant and wild-type sequences were 8,192 (213) and 1,024 (210) copies/μl, respectively, and these thresholds were less than those for amplification of the incorrect target. For HIV-1 subtype B pNL4-3, the discriminatory thresholds for amplification of the K65R mutant and the wild type were 512 (29) copies/μl for both K65R and wild-type AS-PCR. In addition, our assay was controlled by adding 107 copies of wild-type plasmid into equal volumes of serial dilutions of samples of K65R-containing plasmid (107 to 103 copies/μl). The sensitivity for detection of minority species was 1%. The supplemental material includes details of the AS-PCR assay for detection of the K65R mutation in HIV-1 as well as information related to the discriminatory ability of the assay.
Detection of minority resistance variants.
The results in regard to the prevalence of different drug resistance mutations in this study are summarized in Table 1 and show that the K65R mutation was not found in any of the samples by bulk sequencing. Minority K65R species were found in 4 of the 30 treated subtype C patients but not in any of the 26 subtype B patients (Fig. 1). Viruses from the first of these four patients (Table 2) also contained mutations M41L, D67N, L210W, T215Y, K101E, and K103N; the patient was receiving ddI, d4T, and efavirenz (EFV) at the time of study and had received zidovudine (ZDV) in the past. The second patient exhibited mutations L74V, M184V, and K103N. This patient had received lamivudine (3TC), abacavir (ABC), and nevirapine (NVP). The third sample contained mutations T215Y, K103N, and Y181C, reflecting exposure to d4T and NVP. The fourth patient developed K65R on a TDF/FTC/EFV regimen; in this case, minority K65R species were later also detected by bulk sequencing. No major protease inhibitor (PI) drug resistance mutations were found in any of the samples, although minor PR substitutions were observed at positions 10, 33, 36, 63, 70, 71, 77, and 93. HIV-1 RNA quantification yielded results of 19,363, 1,377 and 32,000 copies/ml for patients 1, 2, and 4, respectively. This confirms that HIV viral load has a low impact on the sensitivity of our assay.
TABLE 1.
Resistance mutational profiles of HIV-1 of subtype B and subtype C origin as determined by bulk sequencing
| Subtype | % Patients with mutation(s) of indicated type |
|||||
|---|---|---|---|---|---|---|
| TAMsa | Other NRTIb | K65R | NNRTIc | Major PRd | Minor PRe | |
| B (n = 26) | 69 | 62 | 0 | 69 | 50 | 50 |
| C (n = 30) | 66 | 62 | 0 | 66 | 17 | 83 |
TAMs, thymidine analogue mutations (M41L, D67N, K70R, L210W, T215Y, and K219Q).
Other nucleoside reverse transcriptase inhibitor mutations as listed by the International AIDS Society—USA panel (http://www.iasusa.org/ [December 2008 update]).
Nonnucleoside reverse transcriptase inhibitor mutations (A98S, K101E, K103N, V106M, Y181C, Y188C, G190A, and P225H).
Major protease inhibitor mutations included D30N, M46I, V82A, I84V, N88D, and L90M.
Other minor protease mutations can be found in a report from the International AIDS Society—USA (http://www.iasusa.org/ [December 2008 update]).
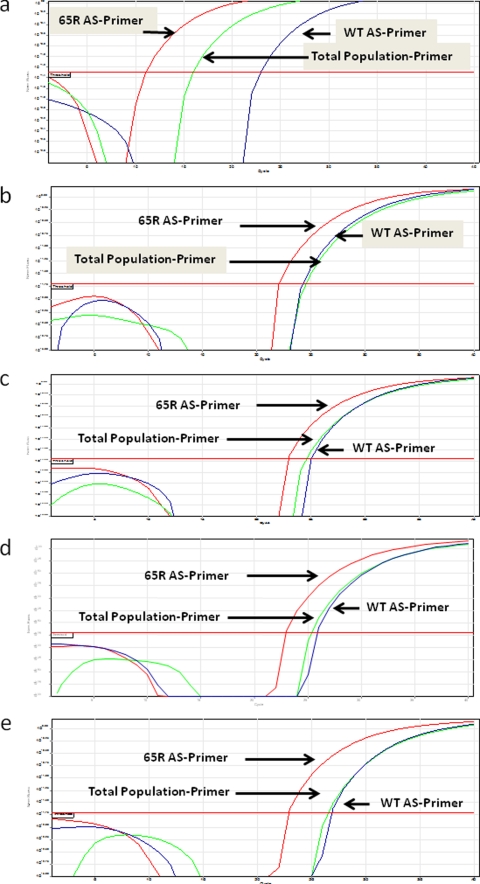
FIG. 1.
Presence and absence of K65R as a minority species. We tested HIV-1 samples and standards for the presence of the K65R mutation as a minority species by using our novel AS-PCR assay as described in the text. (a) The amplification plots show that K65R was present in subtype C MJ4 K65R plasmid (107 copies/μl). The K65R curve crosses the threshold level earlier than that of the total virus population and of the wild type, indicating the presence of K65R only in the former. Positive results were obtained for each of the four patient samples tested under the same conditions, as shown in panels b, c, d, and e.
TABLE 2.
Drug resistance mutations in subtype C patients testing positive by AS-PCR for K65R
| Patient | Drugs received | Mutations in reverse transcriptase |
|---|---|---|
| 1a | ZDV, ddI, d4T, EFV | M41L, D67N, L210W, T215Y, K101E, K103N |
| 2 | d4T, 3TC, EFV | L74V, M184V, K103N |
| 3 | d4T, 3TC, NVP | T215Y, K103N, Y181C |
| 4 | TDF, FTC, EFV | K103K/N, M184V, G190C/G |
This patient had been on ZDV as part of a previous regimen.
AS-PCR has proven to be more sensitive than bulk sequencing for revealing the presence of minority species with drug resistance mutations in HIV-infected individuals. Recent findings suggest bidirectional antagonisms between K65R and thymidine analogue mutations (TAMs) in subtype B treatment-experienced patients (18, 19). In our study, two patients with K65R minority species also possessed TAMs. The presence of K65R can decrease susceptibility to TFV, ddI, ABC, and d4T (6). It is not yet known whether the antagonism between K65R and TAMs may exist to the same extent in subtype C as in subtype B viruses, but this antagonism may also contribute to the difficulty in detecting K65R by bulk sequencing.
Recent clinical studies have shown the preferential emergence of K65R in subtype C-infected patients in Botswana for whom d4T/ddI-based regimens failed (30%) and in subtype C-infected patients in Malawi and South Africa for whom d4T/3TC-based regimens failed (7 to 20%) (5, 17). In our study, no patient with subtype B viruses for whom therapy that included TDF, ddI, or d4T failed possessed K65R minority variants that were detectable by AS-PCR. In contrast, four subtype C patients developed K65R, confirming the tendency of subtype C viruses to preferentially develop this mutation (2, 5, 9). This underlines the utility of ultrasensitive assays.
The K65R mutation in RT has been demonstrated to compromise the replicative capacity of HIV-1 in tissue culture and is also known to compromise the catalytic efficiency of the RT enzyme (26), characteristics that are shared with each of the M184V and L74V mutations that also confer resistance to select NRTIs (4, 6, 26). An important issue will now be whether subtype C viruses containing K65R will be compromised in regard to replicative capacity to the same extent as occurs in subtype B. A related question will involve the extent to which viruses that contain the K65R mutation are likely to be sexually transmitted. Several studies of transmitted resistance with subtype B viruses have shown that the M184V mutation is vastly underrepresented in newly infected persons harboring drug resistance mutations in comparison to its presence among treated patients for whom antiviral chemotherapy has failed. The most likely explanation for this discrepancy, as shown by M184V AS-PCR, is reversion of M184V to the wild type, followed by the more rapid growth of wild-type viruses (15, 16, 24).
Until recently, there have been relatively few cases of treatment failure in which the K65R mutation has arisen, making it difficult to assess whether transmission of K65R-containing viruses may be likely to occur. Now that it is clear that K65R will be actively selected in subtype C viruses by regimens containing d4T, the importance of documenting the sexual transmission of this mutation is obvious. The availability of an AS-PCR test for the detection of K65R will assist in this process.
Supplementary Material
Acknowledgments
This work was sponsored by the Canadian Institutes of Health Research (CIHR). Thomas A. Toni was the recipient of a CIHR postdoctoral fellowship. Michel Ntemgwa was the recipient of a CIHR doctoral fellowship award.
Footnotes
Published ahead of print on 7 December 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Brenner, B., D. Turner, M. Oliveira, D. Moisi, M. Detorio, M. Carobene, R. G. Marlink, J. Schapiro, M. Roger, and M. A. Wainberg. 2003. A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. AIDS 17:F1-F5. [DOI] [PubMed] [Google Scholar]
- 2.Coutsinos, D., C. F. Invernizzi, H. Xu, D. Moisi, M. Oliveira, B. G. Brenner, and M. A. Wainberg. 2009. Template usage is responsible for the preferential acquisition of the K65R reverse transcriptase mutation in subtype C variants of human immunodeficiency virus type 1. J. Virol. 83:2029-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delaunay, C., F. Brun-Vezinet, R. Landman, G. Collin, G. Peytavin, A. Trylesinski, P. Flandre, M. Miller, and D. Descamps. 2005. Comparative selection of the K65R and M184V/I mutations in human immunodeficiency virus type 1-infected patients enrolled in a trial of first-line triple-nucleoside analog therapy (Tonus IMEA 021). J. Virol. 79:9572-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diallo, K., B. Marchand, X. Wei, L. Cellai, M. Gotte, and M. A. Wainberg. 2003. Diminished RNA primer usage associated with the L74V and M184V mutations in the reverse transcriptase of human immunodeficiency virus type 1 provides a possible mechanism for diminished viral replication capacity. J. Virol. 77:8621-8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doualla-Bell, F., A. Avalos, B. Brenner, T. Gaolathe, M. Mine, S. Gaseitsiwe, M. Oliveira, D. Moisi, N. Ndwapi, H. Moffat, M. Essex, and M. A. Wainberg. 2006. High prevalence of the K65R mutation in human immunodeficiency virus type 1 subtype C isolates from infected patients in Botswana treated with didanosine-based regimens. Antimicrob. Agents Chemother. 50:4182-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frankel, F. A., B. Marchand, D. Turner, M. Gotte, and M. A. Wainberg. 2005. Impaired rescue of chain-terminated DNA synthesis associated with the L74V mutation in human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 49:2657-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottlieb, G. S., N. M. Badiane, S. E. Hawes, L. Fortes, M. Toure, C. T. Ndour, A. K. Starling, F. Traore, F. Sall, K. G. Wong, S. L. Cherne, D. J. Anderson, S. A. Dye, R. A. Smith, J. I. Mullins, N. B. Kiviat, and P. S. Sow. 2009. Emergence of multiclass drug-resistance in HIV-2 in antiretroviral-treated individuals in Senegal: implications for HIV-2 treatment in resouce-limited West Africa. Clin. Infect. Dis. 48:476-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant, R. M., and M. A. Wainberg. 2006. Chemoprophylaxis of HIV infection: moving forward with caution. J. Infect. Dis. 194:874-876. [DOI] [PubMed] [Google Scholar]
- 9.Hosseinipour, M. C., J. J. van Oosterhout, R. Weigel, S. Phiri, D. Kamwendo, N. Parkin, S. A. Fiscus, J. A. Nelson, J. J. Eron, and J. Kumwenda. 2009. The public health approach to identify antiretroviral therapy failure: high-level nucleoside reverse transcriptase inhibitor resistance among Malawians failing first-line antiretroviral therapy. AIDS 23:1127-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Invernizzi, C. F., D. Coutsinos, M. Oliveira, D. Moisi, B. G. Brenner, and M. A. Wainberg. 2009. Signature nucleotide polymorphisms at positions 64 and 65 in reverse transcriptase favor the selection of the K65R resistance mutation in HIV-1 subtype C. J. Infect. Dis. 200:1202-1206. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, J. A., J. F. Li, X. Wei, J. Lipscomb, D. Irlbeck, C. Craig, A. Smith, D. E. Bennett, M. Monsour, P. Sandstrom, E. R. Lanier, and W. Heneine. 2008. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 5:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, V. A., F. Brun-Vezinet, B. Clotet, H. F. Gunthard, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2007. Update of the drug resistance mutations in HIV-1: 2007. Top. HIV Med. 15:119-125. [PubMed] [Google Scholar]
- 13.Lidstrom, J., N. Kumwnda, G. Kafulafula, D. Hoover, N. Kumwenda, L. Mofenson, M. Fowler, M. Thigpen, T. Taha, and S. Eshleman. 2009. Antiretroviral treatment of HIV-infected women can induce multiclass drug resistance in their breastfeeding infants. Antivir. Ther. 14(Suppl. 1):A158. [Google Scholar]
- 14.Margot, N. A., J. M. Waters, and M. D. Miller. 2006. In vitro human immunodeficiency virus type 1 resistance selections with combinations of tenofovir and emtricitabine or abacavir and lamivudine. Antimicrob. Agents Chemother. 50:4087-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metzner, K. J., S. G. Giulieri, S. A. Knoepfel, P. Rauch, P. Burgisser, S. Yerly, H. F. Gunthard, and M. Cavassini. 2009. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients. Clin. Infect. Dis. 48:239-247. [DOI] [PubMed] [Google Scholar]
- 16.Metzner, K. J., P. Rauch, V. Von Wyl, H. Kuster, H. J. Stellbrink, J. Böni, H. Trkola, R. Weber, and H. F. Günthard. 2007. Prevalence of minority quasispecies of drug-resistant HIV-1 in patients with primary HIV-1 infection in Zurich in the years 2002-2006. Antivir. Ther. 12:S47. [Google Scholar]
- 17.Orrell, C., R. P. Walensky, E. Losina, J. Pitt, K. A. Freedberg, and R. Wood. 2009. HIV type-1 clade C resistance genotypes in treatment-naive patients and after first virological failure in a large community antiretroviral therapy programme. Antivir. Ther. 14:523-531. [PMC free article] [PubMed] [Google Scholar]
- 18.Parikh, U. M., D. C. Barnas, H. Faruki, and J. W. Mellors. 2006. Antagonism between the HIV-1 reverse-transcriptase mutation K65R and thymidine-analogue mutations at the genomic level. J. Infect. Dis. 194:651-660. [DOI] [PubMed] [Google Scholar]
- 19.Parikh, U. M., S. Zelina, N. Sluis-Cremer, and J. W. Mellors. 2007. Molecular mechanisms of bidirectional antagonism between K65R and thymidine analog mutations in HIV-1 reverse transcriptase. AIDS 21:1405-1414. [DOI] [PubMed] [Google Scholar]
- 20.Pillay, V., C. Pillay, R. Kantor, F. Venter, L. Levin, and L. Morris. 2008. HIV type 1 subtype C drug resistance among pediatric and adult South African patients failing antiretroviral therapy. AIDS Res. Hum. Retrovir. 24:1449-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sungkanuparph, S., W. Manosuthi, S. Kiertiburanakul, N. Saekang, W. Pairoj, and W. Chantratita. 2008. Prevalence and risk factors for developing K65R mutations among HIV-1 infected patients who fail an initial regimen of fixed-dose combination of stavudine, lamivudine, and nevirapine. J. Clin. Virol. 41:310-313. [DOI] [PubMed] [Google Scholar]
- 22.Svarovskaia, E. S., N. A. Margot, A. S. Bae, J. M. Waters, D. Goodman, L. Zhong, K. Borroto-Esoda, and M. D. Miller. 2007. Low-level K65R mutation in HIV-1 reverse transcriptase of treatment-experienced patients exposed to abacavir or didanosine. J. Acquir Immune Defic. Syndr. 46:174-180. [DOI] [PubMed] [Google Scholar]
- 23.Thomson, M. M., and R. Najera. 2005. Molecular epidemiology of HIV-1 variants in the global AIDS pandemic: an update. AIDS Rev. 7:210-224. [PubMed] [Google Scholar]
- 24.Toni, T. A., E. L. Asahchop, D. Moisi, M. Ntemgwa, M. Oliveira, B. Masquelier, B. G. Brenner, and M. A. Wainberg. 2009. Detection of human immunodeficiency virus (HIV) type 1 M184V and K103N minority variants in patients with primary HIV infection. Antimicrob. Agents Chemother. 53:1670-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toni, T. A., et al. 2008. Abstr. 15th Int. Conf. AIDS Africa (ICASA), Dakar, Senegal, abstr. 357.
- 26.White, K. L., N. A. Margot, J. K. Ly, J. M. Chen, A. S. Ray, M. Pavelko, R. Wang, M. McDermott, S. Swaminathan, and M. D. Miller. 2005. A combination of decreased NRTI incorporation and decreased excision determines the resistance profile of HIV-1 K65R RT. AIDS 19:1751-1760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.