Abstract
Human African trypanosomiasis, caused by the Trypanosoma brucei protozoan parasite, is fatal when left untreated. Current therapies are antiquated, and there is a need for new pharmacologic agents against T. brucei targets that have no human ortholog. Trypanosomes have a single mitochondrion with a unique mitochondrial DNA, known as kinetoplast DNA (kDNA), a topologically complex network that contains thousands of interlocking circular DNAs, termed minicircles (∼1 kb) and maxicircles (∼23 kb). Replication of kDNA depends on topoisomerases, enzymes that catalyze reactions that change DNA topology. T. brucei has an unusual type IA topoisomerase that is dedicated to kDNA metabolism. This enzyme has no ortholog in humans, and RNA interference (RNAi) studies have shown that it is essential for parasite survival, making it an ideal drug target. In a large chemical library screen, two compounds were recently identified as poisons of bacterial topoisomerase IA. We found that these compounds are trypanocidal in the low micromolar range and that they promote the formation of linearized minicircles covalently bound to protein on the 5′ end, consistent with the poisoning of mitochondrial topoisomerase IA. Surprisingly, however, band depletion studies showed that it is topoisomerase IImt, and not topoisomerase IAmt, that is trapped. Both compounds are planar aromatic polycyclic structures that intercalate into and unwind DNA. These findings reinforce the utility of topoisomerase IImt as a target for development of new drugs for African sleeping sickness.
Human African trypanosomiasis, also known as African sleeping sickness, is a protozoal infection caused by Trypanosoma brucei that is fatal without treatment (3). Parasites are transmitted to humans via the tsetse fly, which is found exclusively in sub-Saharan Africa and thus limits the geographic distribution of the disease. Currently, there are an estimated 50,000 to 70,000 cases (25). Despite the lethality of sleeping sickness, the drugs currently available for its treatment are toxic and they require parenteral administration (10). In addition to these obstacles, resistance has been a growing concern, and availability and cost are major problems for the population affected. In light of these issues, new drugs for African trypanosomiasis are in demand. The recently completed T. brucei genome sequence has made possible the identification of potential drug targets which are essential to trypanosomes and have no human ortholog (2).
Trypanosomes have a single mitochondrion with an unusual mitochondrial DNA, known as kinetoplast DNA (kDNA). The kinetoplast is a massive and topologically complex structure, comprised of thousands of interlocked ∼1-kb DNA minicircles and a few dozen ∼23-kb maxicircles (structure and replication of kDNA are reviewed in reference 16). This intricate network is in striking contrast with the monomeric circular DNA molecule found in several copies in each human mitochondrion. Replication of kDNA requires the release of individual covalently closed minicircles from the network into a pool of “free” minicircles, DNA synthesis via theta structure intermediates, and reattachment to the network of progeny minicircles, which retain nicks or gaps until all circles have been replicated. Every step of kDNA replication depends on the function of topoisomerases. These enzymes catalyze reactions that alter the topological state of DNA by cleaving the phosphodiester backbone through the formation of a covalent phosphotyrosine link, transferring segments of DNA through the break, and then ligating the DNA together (topoisomerases are reviewed in references 7, 9, and 28). They are classified into two types, based on the number of DNA strands cleaved: type I enzymes cleave one strand while type II enzymes cleave both strands in reactions requiring ATP. Type I enzymes are further classified into A and B subfamilies based on structural and mechanistic differences. Topoisomerase IA enzymes catalyze the relaxation of negatively supercoiled DNA through an “enzyme bridging mechanism.” After cleavage of a single strand, topoisomerase IA remains covalently bound to the 5′ end as it processively unwinds the DNA. If the enzyme acts across from a preexisting nick, it can pass a double helix through the resulting double-stranded break. The type II enzymes also utilize a 5′-end linkage during catalysis, in contrast to IB topoisomerases, which create a covalent link at the 3′ end of the cleavage site.
T. brucei has at least six catalytically active topoisomerases, and two of these appear to function exclusively in the mitochondrion: topoisomerase IAmt and topoisomerase IImt (15, 21). These mitochondrial enzymes are especially attractive drug targets since they are essential according to RNA interference (RNAi) and drug studies (21), and their depletion leads to akinetoplasty (the loss of kDNA), which in turn results in death (13).
Compounds that target topoisomerases can be classified as poisons or nonpoisoning inhibitors (17). Poisons bind to the Michaelis complex comprised of DNA and enzyme, forming a ternary DNA-protein-inhibitor “cleavable complex.” In situ, the presence of a poison prevents strand ligation, and thus, the topoisomerase remains in an inert complex that is fixed on the DNA helix. Covalent adducts of topoisomerase and DNA can be forged by rapid denaturation of the topoisomerase with detergents such as sodium dodecyl sulfate (SDS). Nonspecific inhibitors also decrease topoisomerase activity but do not promote the formation of cleavable complexes. They may include agents that deform the DNA substrate, bind proteins with high affinity, or compete for binding at active sites, such as DNA and ATP analogs. In the cell, stationary cleavable complexes prove lethal when replication machinery collides with them, producing breaks in the DNA (11, 17). Therapeutically useful topoisomerase-targeting agents are all poisons: fluoroquinolones and etoposide are topoisomerase II poisons for antibacterial and antitumor therapy, respectively, while camptothecin is an antitumor agent that poisons topoisomerase IB. For T. brucei, poisons against topoisomerase IAmt would be ideal since this enzyme has no human ortholog and is found exclusively in the mitochondrion with kDNA (21).
Recently, a luciferase reporter system that detects the accumulation of covalent DNA complexes with recombinant Yersinia pestis topoisomerase IA in Escherichia coli was developed (8). The system was used in a high-throughput assay to screen for compounds that poisoned Y. pestis topoisomerase IA. Three compounds were found to promote intracellular cleavable complex formation by the recombinant topoisomerase IA and to inhibit relaxation of DNA by the isolated enzyme. These compounds also inhibited the growth of bacterial cells.
Lead compounds 1 and 2 identified in the Y. pestis model (Fig. 1), hereafter referred to as 1895 and 0020, respectively, were tested against T. brucei to assess cytotoxic activity and ability to poison topoisomerase IAmt. These compounds were found to be trypanocidal at micromolar concentrations, they appear to intercalate into DNA, and, unexpectedly, they promote linearization of free minicircles by poisoning intracellular topoisomerase IImt.
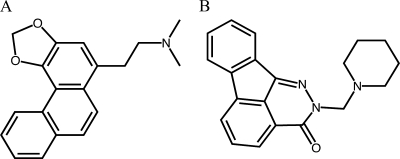
FIG. 1.
Molecular structure of 1895 (A), which is N,N-dimethyl-2-(phenanthro[3,4-d][1,3]dioxol-5-yl)ethanamine, and 0020 (B), which is 2-(piperidin-1-ylmethyl)indeno[1,2,3-de]phthalazin-3-(2H)-one. Of interest, 1895 contains a methylenedioxy motif seen also in etoposide and some semisynthetic derivatives of camptothecin, which are topoisomerase poisons.
MATERIALS AND METHODS
All experiments were performed with bloodstream-form Trypanosoma brucei brucei (MiTat 1.2 strain 427; doubling time, 6 to 8 h), containing a c-Myc-tagged topoisomerase IA allele (21), grown at 37°C in HMI-9 containing 10% fetal bovine serum, 10% Serum Plus, and 0.8 μg/ml G418 (Gibco BRL Life Technologies, Inc.). Compounds 1895 and 0020 were purchased from ChemDiv (San Diego, CA; catalog numbers 4333-1895 and 4711-0020, respectively), dissolved in dimethyl sulfoxide (DMSO), aliquoted, and stored at −20°C.
Cytotoxicity assay.
Compounds 1895 and 0020 were assayed for antitrypanosomal activity using an acid phosphatase-based 96-well plate method (4). Briefly, 100 μl of cells (2 × 105/ml) was incubated with 100 μl of medium containing solvent or 2× serially diluted compound (20 to 24 h, 37°C). Each concentration was assayed in quadruplicate. Final DMSO concentrations did not exceed 1%. Lethality was confirmed by microscopic examination for motility. Acid phosphatase activity from surviving cells was measured by adding 20 μl of buffer containing 20 mg/ml p-nitrophenyl phosphate in 1 M sodium acetate, pH 5.5, 1% Triton X-100 (5 h, 37°C). Absorbance was measured at 405 nm. Each compound was assayed three times. Curve fitting and 50% effective concentration (EC50) determinations were obtained with the Emax model (12) and DeltaGraph Pro 3.5. Based on Chauvenet's criterion (26), values from three (of 576 total) wells were identified as outliers and were dropped from analysis.
Free minicircle profiles.
For each concentration, 12 μl of 10× compound was added to 1.2 × 107 cells in 108 μl of medium (30 min, 37°C). Just before lysis, a portion was taken for Western blot analysis (see below). The remaining cells were lysed with an equal volume of 10 mM Tris HCl, pH 8.0, 75 mM Na2-EDTA, 1% SDS, with or without 2 mg/ml proteinase K (2 h, 55°C), and then treated with 100 μg/ml RNase A (30 min, 37°C). Samples were separated by electrophoresis in 1.5% agarose gels containing 89 mM Tris base, 89 mM boric acid, 2 mM EDTA with 1 μg/ml ethidium bromide (35 V, 24 h). DNA was transferred to a Hybond XL membrane (GE Healthcare), washed in 750 mM NaCl and 75 mM sodium citrate, UV cross-linked (Stratagene UV Cross-linker 1800) to the membranes, and hybridized in Church-Gilbert solution with a 32P-labeled probe that recognizes the 124-bp conserved minicircle sequence (overnight, 45°C) (21). Membranes were exposed to a PhosphorImager plate that was scanned (Fuji BAS 2500 PhosphorImager) and visualized with Image Gauge version 3.45 (Fuji Photo Films Company). Unless indicated otherwise, Adobe Photoshop was used to adjust contrast and brightness only.
Exonuclease treatment.
Trypanosomes were treated with 25.5 μM 1895 or 63 μM 0020 (1 h, 37°C) and then lysed and digested with proteinase K and RNase A, as described above. DNA was extracted with buffer-saturated phenol and then with phenol-chloroform-isoamyl alcohol (25:24:1), followed by back extraction and ethanol precipitation. Samples were redissolved in 100 μl of 10 mM Tris HCl, pH 8.0, 1 mM Na2-EDTA; applied to a QiaAmp DNA microkit (Qiagen) for cleanup; eluted in 30 μl distilled water; and treated with 10 U λ exonuclease in buffer containing 67 mM glycine-KOH, pH 9.4, 2.5 mM MgCl2, and 50 μg/ml bovine serum albumin (BSA) or with 10 U exonuclease III in buffer containing 10 mM Bis-Tris-propane-HCl, pH 7.0, 10 mM MgCl2, 1 mM dithiothreitol in a final volume of 40 μl (30 min, 37°C). HindIII-linearized pBluescript (80 ng) was added to each treatment as an internal control. Minicircle DNA was separated and visualized as described above.
Western blot band depletion.
The 8 × 106 cells reserved above were lysed with an equal volume of 117 mM Tris HCl, pH 6.8, 3.4% SDS, 10% glycerol, 0.2 M dithiothreitol, 0.004% bromophenol blue. Proteins were separated by SDS-PAGE and transferred to polyvinyl difluoride membranes (Millipore). Blots were blocked with 3% bovine serum albumin in 25 mM Tris HCl, pH 7.4, 150 mM NaCl, 0.05% Tween 20, and probed with the following primary and then secondary antibodies: anti-c-Myc (1:1,000; Santa Cruz Biotechnologies)-anti-rabbit-horseradish peroxidase (anti-rabbit-HRP; 1:20,000; Santa Cruz Biotechnologies) and anti-T. brucei-topoisomerase IImt (1:5,000 [15])-anti-guinea pig-HRP (1:40,000; Sigma). Loading control was provided by anti-human enolase (1:200; Santa Cruz Biotechnologies)-anti-rabbit-HRP (1:30,000). Quantitation was done with Image J (1).
Precipitation of topoisomerase-DNA complexes.
The ability of compounds 1895 and 0020 to promote intracellular cleavable complex formation with nuclear DNA was determined by a potassium-SDS (KSDS) assay (5, 27). Trypanosomes (107 cells per ml) in thymidine-free medium were radiolabeled with 333 μCi/ml [3H]thymidine (20 Ci/mmol; Perkin-Elmer; 1 h, 37°C), washed three times, resuspended at 107 cells/ml, and treated with DMSO, 500 μM etoposide, 50 μM 1895, or 50 μM 0020 (30 min, 37°C). Samples were assayed in triplicate. An equal volume of lysis buffer (1.25% SDS, 5 mM EDTA, 0.4 mg/ml calf thymus DNA) was added, and lysates were incubated (10 min, 65°C). Covalent DNA-protein complexes were precipitated by the addition of ice-cold 65 mM KCl and washed three times with 10 mM Tris HCl, pH 8.0, 100 mM KCl, 1 mM EDTA, 0.1 mg/ml calf thymus DNA, and radioactivity was counted by liquid scintillation. Total incorporation of radioactivity by control cells was measured in triplicate by aliquoting 5 × 105 untreated labeled cells onto glass filter paper and precipitating DNA with 5% ice-cold trichloroacetic acid. To calculate the percentage of DNA trapped in cleavable complexes, the averaged counts per minute detected in KSDS precipitates were divided by the averaged counts per minute in trichloroacetic acid precipitates.
DNA unwinding.
DNA unwinding by 1895 and 0020 was assayed as described previously (24). In brief, 250 ng of supercoiled pBluescript or of pBluescript relaxed by Escherichia coli topoisomerase IA was treated with various concentrations of 1895 and 0020 in 50 mM Tris HCl, pH 7.5, 50 mM NaCl, and 5 mM MgCl2, after which 1 pM vaccinia virus topoisomerase IB (gift from James Stivers) was added (1 h, 37°C). Reactions were quenched by addition of 10× DNA loading dye containing 0.625% SDS, 0.125% xylene cyanol, 0.125% bromophenol blue, 62.5% glycerol. DNA was separated in 1% agarose in 89 mM Tris base, 89 mM boric acid, 2 mM EDTA and visualized by ethidium bromide fluorescence.
RESULTS
Trypanocidal activity in vitro.
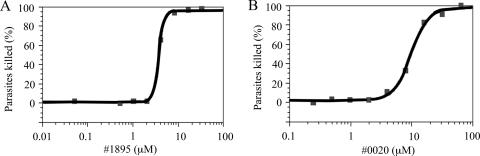
Increasing concentrations of 1895 or 0020 were tested against bloodstream T. brucei to assess their killing activity (Fig. 2). In three determinations, the EC50 for 1895 is 6.3 ± 4.3 μM and that for 0020 is 16.9 ± 8.8 μM. For perspective, these potencies against trypanosomes are comparable to those of known topoisomerase poisons (camptothecin, 1.5 μM [4]; ciprofloxacin, 52 μM [18]) and are low compared to some clinically used agents for trypanosomiasis (pentamidine, 14 nM [4]; berenil, 94 nM [4]), but not difluoromethylornithine (>147 μM [29]).
FIG. 2.
Killing activity against T. brucei in vitro for 1895 (A) and 0020 (B). For each concentration tested (assayed in quadruplicate), the coefficients of variation were less than 10%. Data were fitted to the equation for the Emax model to obtain EC50s; R2 values are 0.99 for both graphs.
Compounds 1895 and 0020 promote minicircle linearization.
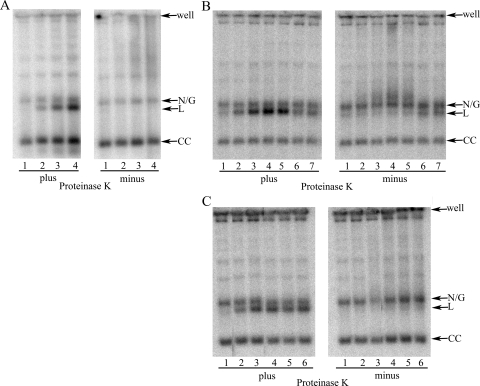
To determine whether antitrypanosomal activity is related to topoisomerase IAmt, free minicircle replication intermediates from treated cells were analyzed by Southern hybridization (Fig. 3). When electrophoresed in the presence of ethidium bromide, free minicircles are separated into nicked/gapped daughter circles, linear forms, and covalently closed template circles. As expected, etoposide, a known poison of T. brucei topoisomerase IImt (23), increased linearized minicircles in a concentration-dependent fashion (Fig. 3A, left panel). These linear forms are substantially reduced in samples not treated with proteinase K, consistent with their covalent linkage to protein (Fig. 3A, right panel).
FIG. 3.
Free minicircle profiles. T. brucei was treated as indicated and lysed with SDS buffer. Lysates were digested (or not) with proteinase K, as indicated, and DNA was separated by electrophoresis prior to Southern blotting for minicircle DNA. (A) Lanes 1 to 4, 0, 4, 20, and 100 μM etoposide, respectively (5 × 106 cells/lane). (B) Lanes 1 to 7, 0, 10, 20, 40, 60, 80, and 100 μM 1895, respectively (1.7 × 106 cells/lane). (C) Lanes 1 to 6, 0, 20, 40, 80, 100, and 120 μM 0020, respectively (1.7 × 106 cells/lane). Samples treated with 10 and 60 μM had incomplete proteolysis and are not shown. The position of two lanes in this blot have been switched to present results in increasing concentrations. N/G, nicked/gapped; L, linear; CC, covalently closed.
Compounds 1895 and 0020 also give rise to an increase in protease-sensitive linear forms (Fig. 3B and 3C). For 1895, at concentrations between 0 and 40 μM, there is a steady increase in linear forms, but above 60 μM, the population decreases (Fig. 3B). Based on proteinase K sensitivity, at 80 and 100 μM, the linear forms are not protein bound. For 0020, the pattern of protein-bound linear forms is more complicated (Fig. 3C). In the presence of proteinase K, linear forms increase steadily between 0 and 40 μM and level off at 80 μM. At 100 μM there appears to be a decrease in linear forms, which then rebound at 120 μM. This wavering pattern has been observed reproducibly in several experiments.
Linear forms would not be the expected product if 1895 and 0020 were poisoning topoisomerase IAmt. Since type I enzymes make a single-stranded break, their poisoning should lead to an increase of nicked, not linearized, minicircles. However, the population of nicked/gapped minicircles from treated cells is comparable to that of controls (Fig. 3B and 3C). Perhaps if topoisomerase IAmt works across from a preexisting nick in a minicircle or if two molecules of topoisomerase IAmt work adjacent to each other on opposite strands, linear forms could be generated by treatment with compounds 1895 and 0020.
Linearized minicircles are protected from exonuclease digestion.
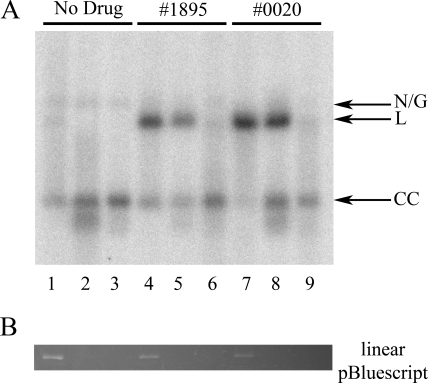
To determine whether the protein is covalently bound to the 5′ or 3′ end of cleavage sites in the linear forms, minicircles extracted from cells treated with either 1895 or 0020 were subjected to λ exonuclease, which degrades linear DNA from the 5′ end, or to exonuclease III, which degrades from the 3′ end (Fig. 4). Internal control linear pBluescript was completely digested in all samples containing exonuclease, verifying enzymatic activity (Fig. 4B). Minicircle linear forms from cells treated with either compound were protected from degradation by λ exonuclease but not from that by exonuclease III, indicating that the DNA is blocked by protein on the 5′ end (Fig. 4A). This finding rules out a topoisomerase IB mechanism (which requires a 3′-phosphotyrosine link) and is most consistent with a topoisomerase II, in which protein is covalently bound to the 5′ end of both strands. However, type IA enzymes also bind to the 5′ end of their cleavage site. If the linear forms were produced by a topoisomerase IAmt molecule which acted across from a preexisting nick, only a single strand of DNA would be protected from digestion by λ exonuclease. A new population of minicircle DNA that runs faster than covalently closed circles does appear in samples digested with λ exonuclease, but this is also present in the control sample treated with λ exonuclease. If two topoisomerase IAmt molecules worked in close proximity and on opposite strands, linearized forms with both strands protected at the 5′ end could be produced.
FIG. 4.
Exonuclease susceptibility of minicircle DNA from treated T. brucei. Cells were treated with no drug, 25.5 μM 1895, or 63 μM 0020 as indicated and lysed with SDS. Internal control HindIII-linearized pBluescript was added to each sample prior to exonuclease treatment. (A) Southern blot of minicircle DNA. Lanes 1, 4, and 7, no exonuclease. Lanes 2, 5, and 8, λ exonuclease. Lanes 3, 6, and 9, exonuclease III. There were 2.45 × 107 cells/lane. (B) Reaction internal control pBluescript DNA visualized by ethidium bromide fluorescence prior to DNA transfer for minicircle hybridization.
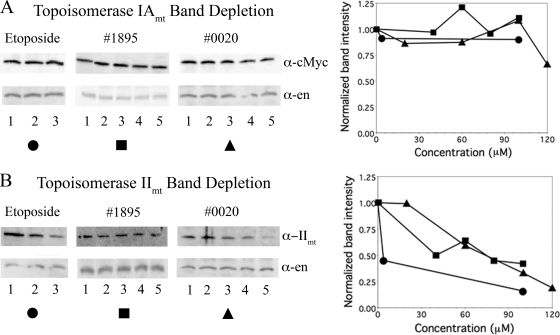
Depletion of free topoisomerase in cell lysates.
To determine which mitochondrial topoisomerase is covalently linked to linearized minicircles, lysates of treated cells were analyzed by immunoblotting. In this method, a topoisomerase enzyme covalently bound to DNA is unable to enter the gel matrix and only free topoisomerase molecules are detected (14). As expected, etoposide treatment depletes topoisomerase IImt and has no effect on topoisomerase IAmt (Fig. 5A and 5B, left panels). Interestingly, in cells treated with 1895 or 0020, there was no decrease in topoisomerase IAmt, indicating that these compounds do not promote cleavable complex formation with this enzyme in trypanosomes (Fig. 5A, central and right panels). However, they do lead to a significant decrease in free topoisomerase IImt, indicating that this is the protein responsible for minicircle linearization (Fig. 5B, central and right panels). The depletion of free topoisomerase IImt by 1895 and 0020 has reproducibly been obtained in five and three independent experiments, respectively. There is no effect on the catalytic subunit of topoisomerase IB (data not shown), an enzyme that acts in both the nucleus and mitochondrion of trypanosomes (5).
FIG. 5.
Western blots of T. brucei proteins after treatment as indicated and SDS lysis. (A) Free topoisomerase IAmt signals with enolase controls. (Left) Lanes 1 to 3, 0, 4, and 100 μM etoposide, respectively (1 × 106 cells/lane). (Center) Lanes 1 to 5, 0, 40, 60, 80, and 100 μM 1895, respectively (8.33 × 105 cells/lane). (Right) Lanes 1 to 5, 0, 20, 60, 100, and 120 μM 0020, respectively (8.33 × 105 cells/lane). In the far-right panel these results were quantitated by first correcting each topoisomerase signal with its cognate enolase control and then normalizing treated samples to untreated controls. Etoposide, filled circles; 1895, filled squares; 0020, filled triangles. (B) Free topoisomerase IImt signals with enolase controls; concentrations as in panel A. Membranes were hybridized with antibodies to c-Myc (α-cMyc) to visualize c-Myc-tagged topoisomerase IAmt, to topoisomerase IImt (α-IImt), or to enolase (α-en). In the far-right panel, quantitation and symbols are as given above. The quantitative data for all three panels were verified in an analysis by an independent investigator.
Nuclear topoisomerase II is not trapped in cleavable complexes.
To assess further the possible effect on nuclear topoisomerases, we utilized the KSDS assay. Since 95% of DNA in trypanosomes is nuclear (6), this assay monitors cleavable complex formation in the nucleus. In this experiment, the total incorporation of [3H]thymidine was 3.7 × 105 cpm/107 cells. In the presence of DMSO, 2.2% of total DNA was covalently bound to protein and precipitated, attributable to naturally occurring topoisomerase catalytic complexes in situ (Table 1). As expected, etoposide increased this signal more than fourfold. In trypanosomes, the trapping of nuclear topoisomerase II is apparent at etoposide concentrations as low as 1 μM and is maximal at ∼100 μM (data not shown). For Table 1, the 500 μM concentration was utilized to ensure a robust positive control. Interestingly, and in contrast to etoposide, both 1895 and 0020 reduced cleavable complex formation with nuclear DNA, at concentrations that maximize the linearization of minicircles (Fig. 3 and Table 1).
TABLE 1.
In situ formation of topoisomerase-DNA cleavable complexes
| Compounda (μM) | Cleavable complexes |
|
|---|---|---|
| % of total DNAb | Ratio to DMSO | |
| DMSO | 2.2 ± 0.05 | 1.0 |
| Etoposide (500) | 9.0 ± 0.81 | 4.2 |
| 1895 (50) | 1.9 ± 0.09 | 0.86 |
| 0020 (50) | 1.7 ± 0.13 | 0.79 |
Each compound was assayed in triplicate.
KSDS-precipitable radioactivity in treated cells, as a percentage of trichloroacetic acid-precipitable radioactivity in untreated cells. Values are means ± standard deviations.
Compounds 1895 and 0020 unwind DNA.
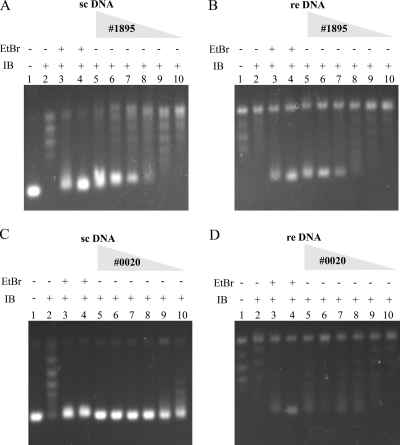
The polycyclic, aromatic, planar structures of 1895 and 0020 suggest that these compounds may be DNA intercalators. To assess this possibility, plasmid DNA was incubated with various concentrations of 1895 or 0020 and treated with vaccinia virus topoisomerase IB, prior to electrophoresis. The binding of intercalators such as ethidium bromide causes a DNA helix to unwind and form positive supertwists. In the presence of a topoisomerase these are relaxed to equilibrium, and when the topoisomerase and intercalator are subsequently removed, the DNA becomes hypernegatively supercoiled. In the absence of added compound, vaccinia virus topoisomerase IB relaxes the DNA substrates (Fig. 6A to D, lanes 2). However, when relaxation occurs in the presence of ethidium bromide, the isolated products are hypernegatively supercoiled (Fig. 6A to D, lanes 3 and 4). This unwinding effect was also seen, in a concentration-dependent fashion, for 1895 and 0020 and with DNA substrates that were negatively supercoiled or relaxed (Fig. 6A to D, lanes 5 to 10).
FIG. 6.
DNA unwinding assay. Lanes 1 to 4, supercoiled or relaxed plasmid DNA (lanes 1) was treated with topoisomerase IB alone (lanes 2) or with topoisomerase IB in the presence of 1.0 or 2.5 μg/ml ethidium bromide (lanes 3 and 4, respectively). Lanes 5 to 10, topoisomerase IB treatment in the presence of 500, 250, 125, 67.5, 33.8, or 16.9 μM 1895, respectively (A and B), or 310, 155, 77.5, 38.8, 19.4, or 9.70 μM 0020, respectively (C and D). There was 100 ng plasmid DNA/lane. EtBr, ethidium bromide; IB, vaccinia virus topoisomerase IB; sc DNA, supercoiled pBluescript plasmid substrate; re DNA, relaxed pBluescript plasmid substrate.
DISCUSSION
With the discovery of an essential mitochondrial topoisomerase IAmt in T. brucei that has no ortholog in humans (21), it was of great interest to study the effects on African trypanosomes of newly described antibacterial topoisomerase IA poisons (8). Two of these compounds had trypanocidal activity at low micromolar concentrations. Furthermore, they promoted the formation of covalent protein adducts with minicircle DNA, as would be expected for topoisomerase poisons operating in the mitochondrion of trypanosomes. Protein was attached at the 5′ end of minicircle DNA, consistent with poisoning of a type IA enzyme. However, band depletion studies unexpectedly revealed that the enzyme being captured was the trypanosome's mitochondrial type II, not type IA, topoisomerase. Trapping of a mitochondrial type II enzyme is also consistent with protection of the 5′ end of minicircle DNA, and it provides a satisfying explanation for the formation of protein-bound linearized, rather than nicked, minicircles upon SDS lysis of treated cells. Interestingly, although T. brucei has a second type II topoisomerase that operates in the nucleus, this enzyme was not poisoned by 1895 or 0020 as shown by the KSDS assay.
In trypanosomes, the selective poisoning of mitochondrial, but not nuclear, topoisomerase II was seen previously with a number of structurally unrelated DNA-binding antitrypanosomal drugs (22). This finding is in contrast to that seen with etoposide, which does not bind directly to DNA but traps both nuclear and mitochondrial topoisomerase II (23). The planar aromatic structure of 1895 and 0020 suggested that they might be acting as intercalators in T. brucei and perhaps lead to differential poisoning in the mitochondrion and nucleus. DNA unwinding assays indeed confirmed 1895 and 0020 to be DNA intercalators. The exact basis for selective poisoning of mitochondrial but not nuclear type II topoisomerases by DNA-binding agents is unknown, but it may relate to intrinsic differences in the two enzymes, which are phylogenetically quite distinct (15), as well as to marked structural differences between chromosomal DNA and minicircle DNA. It is also possible that these compounds are selectively concentrated in the mitochondrion.
Ability to intercalate into DNA may also explain the variation seen reproducibly in the accumulation of linearized minicircles as a function of concentration of 1895 or 0020 (Fig. 3B and 3C). Intercalators that insert between DNA bases and unwind DNA can have mixed effects on topoisomerases. Some intercalators inhibit topoisomerases without poisoning them, as seen with 4′-(9-acridinylamino)-methanesulfon-o-anisidide (o-AMSA). Others, such as the closely related isomer m-AMSA, poison topoisomerase II, promote the formation of double-stranded breaks, and are clinically useful antitumor agents (17, 19, 20). 2-Methyl-9-hydroxyellipticinium (2-Me-9-OH-E+), another intercalator, has an effect similar to what we observe for 1895 and 0020: low concentrations stimulate cleavable complex formation by topoisomerase II, but at higher concentrations the trapping of topoisomerase II is reduced (19, 20). These effects may result from distortion of the substrate DNA induced by strong intercalators present in high ratios. The intercalation effect may account not only for a decrease in linearization of minicircles at high concentrations but also for the reduction in SOS response seen when high concentrations of 1895 were used in the Y. pestis model (8). In keeping with an intercalation effect, in the unwinding assay 0020 appears to be a stronger intercalator than 1895, consistent with its greater inhibition of cleavable complex formation with nuclear DNA (Table 1).
Compounds 1895 and 0020 do not poison the mitochondrial type IA topoisomerase in T. brucei, despite their ability to poison the cognate enzyme in bacteria. Instead, these compounds act against the mitochondrial, but not nuclear, topoisomerase II in trypanosomes. Though unexpected, these findings do corroborate the mitochondrial topoisomerase II as a suitable target for the development of much-needed new antitrypanosomal drugs.
Acknowledgments
This work was supported by NIH R01AI028855, the National Institutes of Health Medical Scientist Training Grant T32GM07309, and the PhRMA Foundation's Paul Calabresi Medical Student Research Fellowship. The work received the Excellence in Medical Student Research award from the Johns Hopkins University School of Medicine.
We thank Rahul Bakshi and Jane Scocca for their assistance in methodology, helpful discussions, and suggestions for the manuscript. We are grateful to Paul Englund and Robert Jensen for their suggestions, James Stivers for providing vaccinia virus topoisomerase IB, and Jeannette Fanning for administrative support.
Footnotes
Published ahead of print on 14 December 2009.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with Image J. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Aguero, F., B. Al-Lazikani, M. Aslett, M. Berriman, F. S. Buckner, R. K. Campbell, S. Carmona, I. M. Carruthers, A. W. Chan, F. Chen, G. J. Crowther, M. A. Doyle, C. Hertz-Fowler, A. L. Hopkins, G. McAllister, S. Nwaka, J. P. Overington, A. Pain, G. V. Paolini, U. Pieper, S. A. Ralph, A. Riechers, D. S. Roos, A. Sali, D. Shanmugam, T. Suzuki, W. C. Van Voorhis, and C. L. Verlinde. 2008. Genomic-scale prioritization of drug targets: the TDR Targets database. Nat. Rev. Drug Discov. 7:900-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett, M. P., R. J. Burchmore, A. Stich, J. O. Lazzari, A. C. Frasch, J. J. Cazzulo, and S. Krishna. 2003. The trypanosomiases. Lancet 362:1469-1480. [DOI] [PubMed] [Google Scholar]
- 4.Bodley, A. L., M. W. McGarry, and T. A. Shapiro. 1995. Drug cytotoxicity assay for African trypanosomes and Leishmania species. J. Infect. Dis. 172:1157-1159. [DOI] [PubMed] [Google Scholar]
- 5.Bodley, A. L., and T. A. Shapiro. 1995. Molecular and cytotoxic effects of camptothecin, a topoisomerase I inhibitor, on trypanosomes and Leishmania. Proc. Natl. Acad. Sci. U. S. A. 92:3726-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borst, P., M. van der Ploeg, J. F. van Hoek, J. Tas, and J. James. 1982. On the DNA content and ploidy of trypanosomes. Mol. Biochem. Parasitol. 6:13-23. [DOI] [PubMed] [Google Scholar]
- 7.Champoux, J. J. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70:369-413. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, B., I. F. Liu, and Y. C. Tse-Dinh. 2007. Compounds with antibacterial activity that enhance DNA cleavage by bacterial DNA topoisomerase I. J. Antimicrob. Chemother. 59:640-645. [DOI] [PubMed] [Google Scholar]
- 9.Corbett, K. D., and J. M. Berger. 2004. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 33:95-118. [DOI] [PubMed] [Google Scholar]
- 10.Fairlamb, A. H. 2003. Chemotherapy of human African trypanosomiasis: current and future prospects. Trends Parasitol. 19:488-494. [DOI] [PubMed] [Google Scholar]
- 11.Froelich-Ammon, S. J., and N. Osheroff. 1995. Topoisomerase poisons: harnessing the dark side of enzyme mechanism. J. Biol. Chem. 270:21429-21432. [DOI] [PubMed] [Google Scholar]
- 12.Holford, N. H., and L. B. Sheiner. 1981. Understanding the dose-effect relationship: clinical application of pharmacokinetic-pharmacodynamic models. Clin. Pharmacokinet. 6:429-453. [DOI] [PubMed] [Google Scholar]
- 13.Jensen, R. E., L. Simpson, and P. T. Englund. 2008. What happens when Trypanosoma brucei leaves Africa. Trends Parasitol. 24:428-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaufmann, S. H., and P. Svingen. 1999. Immunoblot analysis and band depletion assays, p. 253-268. In M. Bjornsti and N. Osheroff (ed.), DNA topoisomerase protocols, vol. 1. DNA topology and enzymes. Humana Press Inc., Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 15.Kulikowicz, T., and T. A. Shapiro. 2006. Distinct genes encode type II topoisomerases for the nucleus and mitochondrion in the protozoan parasite Trypanosoma brucei. J. Biol. Chem. 281:3048-3056. [DOI] [PubMed] [Google Scholar]
- 16.Liu, B., Y. Liu, S. A. Motyka, E. E. Agbo, and P. T. Englund. 2005. Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol. 21:363-369. [DOI] [PubMed] [Google Scholar]
- 17.Liu, L. F. 1989. DNA topoisomerase poisons as antitumor drugs. Annu. Rev. Biochem. 58:351-375. [DOI] [PubMed] [Google Scholar]
- 18.Nenortas, E., C. Burri, and T. A. Shapiro. 1999. Antitrypanosomal activity of fluoroquinolones. Antimicrob. Agents Chemother. 43:2066-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pommier, Y., J. K. Minford, R. E. Schwartz, L. A. Zwelling, and K. W. Kohn. 1985. Effects of the DNA intercalators 4′-(9-acridinylamino)methanesulfon-m-anisidide and 2-methyl-9-hydroxyellipticinium on topoisomerase II mediated DNA strand cleavage and strand passage. Biochemistry 24:6410-6416. [DOI] [PubMed] [Google Scholar]
- 20.Pommier, Y., R. E. Schwartz, L. A. Zwelling, and K. W. Kohn. 1985. Effects of DNA intercalating agents on topoisomerase II induced DNA strand cleavage in isolated mammalian cell nuclei. Biochemistry 24:6406-6410. [DOI] [PubMed] [Google Scholar]
- 21.Scocca, J. R., and T. A. Shapiro. 2008. A mitochondrial topoisomerase IA essential for late theta structure resolution in African trypanosomes. Mol. Microbiol. 67:820-829. [DOI] [PubMed] [Google Scholar]
- 22.Shapiro, T. A., and P. T. Englund. 1990. Selective cleavage of kinetoplast DNA minicircles promoted by antitrypanosomal drugs. Proc. Natl. Acad. Sci. U. S. A. 87:950-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shapiro, T. A., V. A. Klein, and P. T. Englund. 1989. Drug-promoted cleavage of kinetoplast DNA minicircles. Evidence for type II topoisomerase activity in trypanosome mitochondria. J. Biol. Chem. 264:4173-4178. [PubMed] [Google Scholar]
- 24.Shen, L. L. 2001. DNA-unwinding test using eukaryotic DNA topoisomerase I, p. 149-160. In N. Osheroff and M. Bjornsti (ed.), DNA topoisomerase protocols, part II. Enzymology & drugs. Humana Press Inc., Totowa, NJ. [DOI] [PubMed]
- 25.Steverding, D. 2008. The history of African trypanosomiasis. Parasit. Vectors 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor, J. R. 1997. Rejection of data, p. 165-172. In A. McGuire (ed.), An introduction to error analysis: the study of uncertainties in physical measurements. University Science Books, Sausalito, CA.
- 27.Trask, D. K., J. A. DiDonato, and M. T. Muller. 1984. Rapid detection and isolation of covalent DNA/protein complexes: application to topoisomerase I and II. EMBO J. 3:671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, J. C. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3:430-440. [DOI] [PubMed] [Google Scholar]
- 29.Zweygarth, E., R. Kaminsky, P. D. Sayer, and S. van Nieuwenhove. 1990. Synergistic activity of 5-substituted 2-nitroimidazoles (Ro 15-0216 and benznidazole) and DL-alpha-difluoromethylornithine on Trypanosoma brucei brucei. Ann. Soc. Belg. Med. Trop. 70:269-279. [PubMed] [Google Scholar]