Abstract
Large multidrug resistance plasmids of the A/C incompatibility complex (IncA/C) have been found in a diverse group of Gram-negative commensal and pathogenic bacteria. We present three completed sequences from IncA/C plasmids that originated from Escherichia coli (cattle) and Salmonella enterica serovar Newport (human) and that carry the cephamycinase gene blaCMY-2. These large plasmids (148 to 166 kbp) share extensive sequence identity and synteny. The most divergent plasmid, peH4H, has lost several conjugation-related genes and has gained a kanamycin resistance region. Two of the plasmids (pAM04528 and peH4H) harbor two copies of blaCMY-2, while the third plasmid (pAR060302) harbors a single copy of the gene. The majority of single-nucleotide polymorphisms comprise nonsynonymous mutations in floR. A comparative analysis of these plasmids with five other published IncA/C plasmids showed that the blaCMY-2 plasmids from E. coli and S. enterica are genetically distinct from those originating from Yersinia pestis and Photobacterium damselae and distal to one originating from Yersinia ruckeri. While the overall similarity of these plasmids supports the likelihood of recent movements among E. coli and S. enterica hosts, their greater divergence from Y. pestis or Y. ruckeri suggests less recent plasmid transfer among these pathogen groups.
Plasmids are extrachromosomal genetic elements that play key roles in the maintenance and dissemination of novel genetic traits in bacterial populations. Antibiotic resistance is one of the most important medicine-related traits that can be encoded by plasmids. Over the past decade, increasing attention has been focused on plasmids that harbor the antibiotic resistance gene blaCMY-2 (hereafter called blaCMY-2 plasmids). blaCMY-2 encodes an AmpC-type beta-lactamase that hydrolyzes third-generation cephalosporins (4, 36). This gene, which probably originated from Citrobacter freundii (2, 27, 35), has been disseminated horizontally within and between several bacterial species from the family Enterobacteriaceae, including important enteric pathogens (9, 14, 18, 24, 34, 35).
Plasmids that harbor blaCMY-2 are typically large (ca. 140-plus kbp), and they encode multiple drug resistance traits (7, 8, 10, 11, 34). Kang et al. (15) examined a collection of blaCMY-2-positive strains and verified that the blaCMY-2 gene was carried on a large plasmid and that the plasmids frequently harbored two copies of the gene as part of a repeated region. Welch et al. (33) described a complete sequence from a blaCMY-2-positive plasmid that originated from a strain of Salmonella enterica serovar Newport (strain SL254). This plasmid also carried two copies of blaCMY-2 and was from the A/C incompatibility plasmid group (IncA/C) (9, 21). Welch et al. (33) also reported the full sequences of IncA/C plasmids from Yersinia pestis (strain IP275) and from the fish pathogen Yersinia ruckeri (strain YR71). These three plasmids shared a plasmid backbone with a high degree of sequence similarity and conserved gene synteny, although only pSN254 harbored blaCMY-2. The authors concluded that the plasmids shared a recent common ancestor. They also reported the previously unappreciated fact that IncA/C plasmids can be recovered from a range of meat products throughout the United States. The implication of this finding is that agricultural reservoirs have the potential to share these multidrug resistance plasmids with zoonotic agents of significant public health concern, including Y. pestis.
The objective of the current study was to closely examine several additional blaCMY-2 plasmids to better understand how these plasmids are changing over short periods of time. We report complete plasmid sequences for the blaCMY-2 plasmid from a strain of S. enterica serovar Newport and from two geographically separated Escherichia coli strains recovered from cattle. Additional IncA/C plasmid sequences are now available (17), and this permitted us to assess the genetic relatedness between IncA/C plasmids from a diverse collection of Gram-negative bacteria. We found that most changes in blaCMY-2 plasmids involved insertions and deletions of sequences related to antibiotic resistance. Analysis of eight IncA/C plasmids showed that blaCMY-2 plasmids harbored by E. coli and S. enterica are relatively distinct from IncA/C plasmids described from other taxa.
MATERIALS AND METHODS
Strains used in this study.
E. coli strain H4H (the source of plasmid peH4H) was originally isolated from a dairy cow in Washington State by the Washington State University College of Veterinary Medicine Field Disease Investigation Unit (Pullman, WA) (Table 1). E. coli strain AR060302 (the source of plasmid pAR060302) was originally isolated from a dairy calf in Illinois as part of a project investigating antibiotic resistance in production environments (R. S. Singer, unpublished data). S. enterica serovar Newport strain AM04528 (the source of pAM04528) was provided by the Centers for Disease Control and Prevention (Atlanta, GA) and originated from a case of human salmonellosis in Kansas. E. coli DH10B and DH5α were used as host strains for plasmid isolation and preparation for sequencing. Plasmids were categorized by the restriction fragment length polymorphism (RFLP) classification system described by Giles et al. (13) and with additional restriction digests. Antibiotic susceptibility was assessed using agar diffusion according to Clinical and Laboratory Standards Institute guidelines (3, 23), and MICs for a range of antibiotics were determined using an automated Sensititer assay (Trek Diagnostic Systems, Cleveland, OH) at the Washington Animal Disease Diagnostic Laboratory (Pullman, WA). To estimate conjugation efficiency, plasmids were first transferred to E. coli strain DH10B by electroporation (29). Single colonies were grown overnight (3 ml LB broth at 37°C; 200-rpm shaker), and 10 μl was then mixed with 10 μl overnight culture of a recipient strain (E. coli strain DH5α; nalidixic acid resistant [Nalr]). This mixture was spread onto a nitrocellulose membrane overlaid onto LB agar without antibiotics. After overnight incubation (37°C), the culture mat was diluted with 0.5 ml phosphate-buffered saline (PBS) and spread onto LB agar plates with florfenicol (50 μg/ml) and nalidixic acid (20 μg/ml) and incubated overnight (37°C). The conjugation efficiency was expressed as the number of transconjugants divided by the number of donor bacteria. A subset of putative transconjugants was verified for antibiotic resistance phenotype as described above and for plasmid transfer by plasmid profiling (10).
TABLE 1.
Basic characteristics of three sequenced blaCMY-2plasmids (this study) and the reference sequence, pSN254
| Characteristic | pAR060302 (FJ621588) | peH4H (FJ621586) | pAM04528 (FJ621587) | pSN254a (NC_009140) |
|---|---|---|---|---|
| Mammalian host | Cow | Cow | Human | NA |
| Bacterial host | E. coli | E. coli | S. Newport | S. Newport |
| Strain | AR060302 | H4H | AM04528 | SN254 |
| Year of isolation | 2002 | 2002 | 1998 | NA |
| Antibiotic resistance phenotypeb | AM-AMC-C-CAZ-GM-S-SUL-TE | AM-AMC-C-CAZ-GM-K-S-SUL-TE | AM-AMC-C-CAZ- S- SUL-TE | AM-AMC-C-CAZ-GM-S-SUL-TEc |
| Size (bp) | 166,531 | 148,106 | 158,195 | 176,473 |
| %GC | 53.1 | 48.3 | 51.9 | 52.8 |
| Conjugation efficiencyd | 1.3 × 10−3 | 0.0 | 0.0 | NA |
| Giles typee | A | C | C | Cf |
| No. of copies of blaCMY-2 | 1 | 2 | 2 | 2 |
| floR regiong | Yes | Yes | Yes | Yes |
| aadA regiong | Yes | Yes | No | Yes |
| kan regiong | No | Yes | No | No |
| merA regiong | Yes | Yes | Yes | Yes |
The pSN254 sequence was published by Welch et al. (33). NA, not applicable.
AM, ampicillin; AMC, amoxicillin-clavulanic acid; C, chloremphenicol; CAZ, ceftazidime; GM, gentamicin; K, kanamycin; S, streptomycin; SUL, sulfasoxazole; TE, tetracycline.
The antibiotic resistance phenotype is inferred from reference 33 and the plasmid sequence.
Conguation efficiencies for plasmids that were first transferred to E. coli DH10B by electroporation.
Giles type based on plasmid RFLP after PstI digestion (13).
The Giles type for pSN254 is presumed to be group C based on sequence similarity with pAM04528.
The floR, aadA, kan, and merA regions are shown in Fig. 1.
Plasmid sequencing and annotation.
Plasmids were shotgun sequenced from purified plasmid preparations (30). Plasmid pAR060302 was transconjugated to E. coli DH5α in broth culture, and one transconjugant was submitted to Agencourt Bioscience Corp. (Beverly, MA) for plasmid purification, library construction, and sequencing. Plasmids pAM04528 and peH4H were transformed into E. coli DH10B and purified by cesium chloride gradient (30). After the cloning, the resulting plasmid libraries were sequenced either in house or through services provided by Amplicon Express (Pullman, WA). Plasmid sequences were assembled using phrap and Sequencher (Gene Codes Corp., Ann Arbor, MI); direct plasmid sequencing and PCR were used to close gaps. A combination of PCR and RFLP experiments were used to test ambiguous assemblies. Preliminary annotation was done using an annotation pipeline that included Glimmer 2 (12) for gene detection and BLAST and PFam database comparisons for initial gene identification. In addition, the pipeline employed RBS Finder (31), which was used in comparisons with database information to verify coding region start sites. Curation of the annotation was completed manually using Artemis ver. 10 (28), and pSN254 (GenBank NC_009140) was used as the reference sequence for nomenclature and repeat identification. Descriptive information about each plasmid was summarized with the aid of Vector NTI (Invitrogen Corp., Carlsbad, CA).
Phylogenetic analysis.
Our phylogenetic analysis considered the three new sequences presented in our study compared with pSN254 (originating from S. enterica serovar Newport strain SN254; GenBank NC_009140 [33]) with an additional comparison of plasmids from Yersinia pestis (pIP1202; GenBank CP000603 [33]), Yersinia ruckeri (pYR1; GenBank CP000602 [33]), and Photobacterium damselae subsp. piscicida (pP91278 and pP99-018; GenBank AB277724 and AB277723 [17]). Because plasmids are typically a mosaic of sequences from multiple, potentially divergent sources, a sequence-based phylogenetic analysis must necessarily focus on gene sequences that are “conserved” between plasmids. The challenges are to identify these sequences in an efficient manner and to devise a reasonable definition for “conserved,” because 100% conservation would produce no differentiation and genes that are “too divergent” can generate exaggerated differences. As detailed below, our approach to this challenge was to identify all the genes common to every plasmid, choose a threshold value to select conserved genes, concatenate these genes in the same order for each plasmid, and perform an alignment of the resulting concatenated sequences.
For a set S of n plasmids S = (p1, p2,…, pn), a conserved gene among these plasmids can be represented by a tuple Gi = (gi1, gi2,…, gin), where gij denotes a specific gene i from the jth plasmid. Assuming there are mj genes for the jth plasmid, then there are m1m2m3… mn possible tuples. We used BLAST (1) to obtain similarity scores for each pair of genes in a tuple where the score was calculated as follows: (length of matching sequence) × (BLAST identity score)/(length of reference gene + length of matching sequence gene), which gives a maximum score of 0.5. If all scores in a tuple were larger than a chosen threshold value, we considered the gene represented by the tuple Gi to be conserved. We then concatenated the respective genes gij for each plasmid. For K conserved genes G1 = (g11, g12,…, g1n), G2 = (g21, g22,…, g2n),…, GK = (gK1, gK2,…, gKn), we have P1 = [g11g21… gK1], P2 = [g12g22… gK2], and…, Pn = [g1ng2n… gKn], where Pj is the concatenation of all conserved genes for the jth plasmid. P1 to Pn were aligned and analyzed using MEGA4 (32) with a maximum composite likelihood method underlying a neighbor-joining algorithm. This method assumes equality of nucleotide substitution patterns and mutation rates among lineages; however, some substitution sites are likely to be under selective pressure while others probably represent neutral mutations. Relationships within distinct clusters of the tree potentially can be made more accurate by repeating the procedure described above for the subgroups of plasmids in a cluster, and this process can be repeated until no subgroups remain.
Obtaining BLAST scores for all combinations of gene pairs is computationally impractical for more than a few plasmids. Instead, one plasmid can be selected and BLAST scores can be obtained for the genes of all the other plasmids relative to the selected reference. We have shown empirically that for a given set of plasmids the choice of reference plasmid does not alter the outcome of the analysis. Finally, when one gene from the reference plasmid had two or more similar genes in another plasmid or when two or more genes from the reference plasmid had a similar gene in another plasmid, these genes were considered duplicates and were removed from the list of conserved genes.
Nucleotide sequence accession numbers.
The plasmid sequences were archived at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) under accession numbers FJ621586, FJ621587, and FJ621588.
RESULTS AND DISCUSSION
Basic characteristics of blaCMY-2 plasmids.
Plasmids pAM04528, peH4H, pAR060302, and pSN254 (33) ranged in size between ca. 148 kbp and 176 kbp and encoded up to 11 antibiotic resistance genes, in addition to a mercury resistance operon (Table 1 and Fig. 1). Disc diffusion tests verified that all three plasmids conveyed multidrug resistance phenotypes, with pAR060302 conveying additional resistance to gentamicin compared to pAM04528 and peH4H conveying additional resistance to both gentamicin and kanamycin compared to pAM04528. MICs demonstrated consistent resistance phenotypes (data not shown). The H4H strain was also resistant to trimethoprim due to the presence of a resistance gene that was harbored by a coresident plasmid, and strain AR060302 was also resistant to kanamycin, but this trait appeared to be encoded on the chromosome (data not shown).
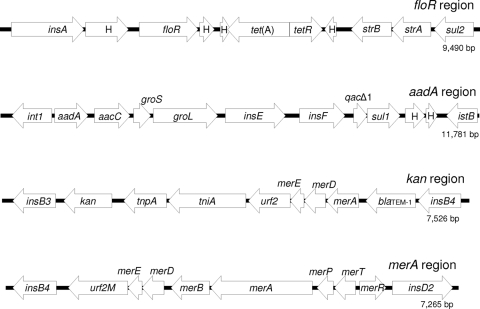
FIG. 1.
Major accessory gene elements found in IncA/C plasmids peH4H, pAM04528, pAR060302, and pSN254. Shown are the floR resistance region (found in all four plasmids), the aadA resistance region (found in peH4H, pAR060302, and pSN254), the kan resistance region (found in peH4H only), and the merA resistance region (found in all four plasmids). H, hypothetical open reading frames.
Restriction digests and Southern blots demonstrated that pAR060302 most closely resembles Giles type A (13) while the remaining plasmids most closely resemble Giles type C (data not shown). Sequence analysis demonstrated that antimicrobial resistance genes were grouped into four main regions (Fig. 1) in addition to the region that encompasses blaCMY-2. Each region includes multiple genes and potentially more than one insertion sequence, integrase, or tranposase. The “floR region” includes floR, tet(A), strA, strB, and sul2. The “aadA region” includes aadA, aacC, and two heat-shock chaperones (groS and groL), followed by a 3-prime conserved region of a class I integron composed of qacΔ, sul1, and two hypothetical open reading frames. The “kan region” includes kan and a truncated mercury resistance operon plus blaTEM-1. All four blaCMY-2 plasmids carry a Tn21 mercury resistance operon (19) and the floR region, whereas the aadA and kan regions were not present in all cases (Fig. 2).
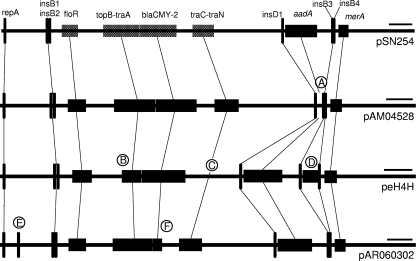
FIG. 2.
Schematic diagram of full-length, IncA/C, and blaCMY-2 plasmids. (A) The aadA region is absent from pAM04528. (B) The traD, traL, traE, and traK genes are absent from peH4H. (C) A 22-kbp region downstream from the blaCMY-2 region is absent from peH4H. (D) The kan region is present in peH4H between insB3 and insB4. (E) IS1294 is found only in pAR060302. (F) Only a single copy of blaCMY-2 is present in pAR060302. The portions of the sequence that are not highlighted are identical in these plasmids, except for minor differences noted in Table 2. Bars, 10,000 bp.
Aside from the absence of the aadA region from pAM04528, the only other differences between this plasmid and the previously published pSN254 were a single-nucleotide polymorphism (SNP) found in a noncoding region of the plasmid (Table 2) and an inverted tnpA (see below). Plasmid peH4H differed the most from pSN254, with two major deletions associated with tra genes (Fig. 2), addition of the kan region, 9 SNPs, an 842-bp alteration of the aadA coding region, and insertion of a tandem repeat (Table 2). Three of six nonsynonymous changes occurred in the floR sequence, which is clearly a nonrandom distribution relative to the entire peH4H sequence. floR encodes an efflux pump that exports both florfenicol and chloramphicol (6). The concentration of nonsynonymous SNPs at this locus is consistent with selection for functional changes for this trait. Plasmid pAR060302 had an additional transposase insertion (Fig. 2) plus 4 SNPs, including 3 nonsynonymous SNPs in the floR sequence, 1 of which was different from the peH4H SNPs at this locus (Table 2).
TABLE 2.
Summary of single-nucleotide and small-scale polymorphisms relative to the pSN254 sequence
| Position (bp) | Name | DNA | Amino acid | Position in: |
Comments | ||
|---|---|---|---|---|---|---|---|
| pAM04528 | pAR060302 | peH4H | |||||
| 14859 | Hyp. proteina | G→A | P→L | NAb | 16547 | NA | |
| 25054 | Hyp. protein | C→T | A→V | NA | NA | 25054 | |
| 25968 | floR | G→A | V→M | 27656 | 25968 | ||
| 26148 | floR | C→T | P→S | NA | NA | 26148 | |
| 26212 | floR | T→A | F→Y | NA | 27900 | 26212 | |
| 26235 | floR | C→A | P→T | NA | 27923 | NA | |
| 126062-126419 | aadA | Multiple | ∼50% divergence | NA | NA | 91634-92476 | 5′ region, aadA sequence, spanning 842 bp |
| 127040 | Repeat region | A→C | Noncoding | NA | NA | 92621 | aadA flanking region |
| 132430 | insF | G→A | Synonymous | NA | NA | 98011 | |
| 160, 25 | Putative exonuclease | T→C | S→P | NA | NA | 131846 | |
| 161189 | Repeat sequence | Insertion | Noncoding | NA | NA | 132811-132820 | TTTTTCTCTCT |
| 171923 | Noncoding | A→T | Noncoding | 153,663 | NA | NA | |
| 174236 | Hyp. protein | “A” insertion | Frameshift | NA | 164292 | 145867 | |
| 176058 | stbA | A→G | Synonymous | NA | NA | 147690 | |
Hyp. protein, hypothetical protein or conserved hypothetical protein.
NA, not applicable.
Comparison of blaCMY-2 regions.
Plasmid pAR060302 is also unique because it harbors a single copy of blaCMY-2, whereas the other three plasmids harbor two copies (Fig. 3), as verified by a double restriction enzyme digest (XbaI and EcoRI) followed by Southern blotting for the blaCMY-2 sequence (data not shown). There was no apparent difference in the resistance phenotype for one (pAR060302) or two (pAM04528 and peH4H) copies of blaCMY-2, as judged by zone size differences from disc diffusion tests (ceftazidime) or by results from MIC tests where the ceftiofur MIC for pAM04528 was 8 μg/ml and the MICs for the other two plasmids were >8 μg/ml.
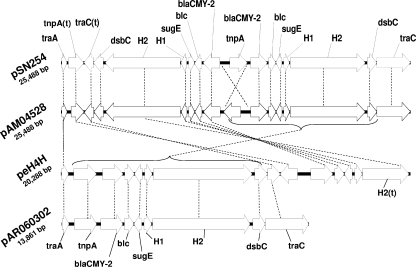
FIG. 3.
Schematic diagram showing different blaCMY-2 regions. The primary element consists of eight genes downstream of traA, as shown for pAR060302. A second copy of this segment (inverted) is inserted immediately upstream of the original segment in pSN254 and pAM04528. Note that the proposed reversal in directionality of tnpA is suspect due to the sequence symmetry in this region (see Discussion). A duplicated blaCMY-2 region is also present in peH4H as a tandem repeat downstream of the original element. Gene names with a (t) suffix are truncated relative to the full length shown for pAR060302. H1 and H2 refer to hypothetical open reading frames.
As observed previously (15, 33), the blaCMY-2 region is composed of that gene, in addition to tnpA, blc, sugE, dsbC, and two hypothetical open reading frames (Fig. 3). This region is identical in pSN254 and pAM04528, except that our assembly showed tnpA as inverted relative to pSN254. The tnpA found in the blaCMY-2 region is an ISEcp1 insertion sequence that has been associated with several other plasmid-encoded, expanded-spectrum β-lactamases (16, 20, 26). The size (>13,000 bp) of the blaCMY-2 region, combined with a high degree of sequence symmetry, makes it very difficult to confirm the directionality of tnpA for pSN254 and pAM04528. While the repeated blaCMY-2 regions in pSN254 and pAM04528 are tandem and encode in opposite directions, the tandem repeat in peH4H encodes in the same direction, but the 5-prime repeated region is truncated (Fig. 3).
The addition of tandem repeats of the blaCMY-2 region is likely due to transposition rather than homologous exchange because for all duplicated blaCMY-2 regions, the first copy of the blaCMY-2 region is flanked by a short sequence at the 3-prime terminus (ATTTCCCTA), and this short sequence is nearly identical at the 5-prime terminus of the single (pAR060402) or repeated blaCMY-2 region (ATTTCCTTA). The latter short sequence is also found at the termini of duplicated blaCMY-2 regions in pSN254, pAM04528, and peH4H. All four blaCMY-2-negative plasmids included in our study have the ATTTCCTTA sequence located adjacent to traA, and the proximal sequence located “downstream” from this region is also conserved across all of the plasmids, indicating that this short flanking sequence is a probably a transposon insertion site that is conserved in the IncA/C plasmid backbone. There are no deletions of existing plasmid sequence associated with tandem duplication of the blaCMY-2 region, and therefore it is unlikely that homologous exchange produced the tandem repeat.
Theoretical models indicate that plasmids will only be retained over the short term if they convey fitness advantages on the host bacterium or if the rate of horizontal transmission exceeds the rate of segregation or competitive loss (5). Clearly, antibiotic selection pressure could select for expansion and dissemination of these plasmids in human and animal populations, although further work is needed to identify the primary variables driving this process. In our hands, plasmid pAR060302 self-conjugated with relatively high efficiency (1.3 × 10−3 conjugations per donor cell) (Table 1). The failure of self-conjugation for peH4H is probably explained by the loss of multiple tra genes (Fig. 2), but a comparison of tra genes between pAR060302 and pAM04528 showed no loss of tra genes or base mutations in tra genes that might explain this difference in function. The only difference involving tra genes was partial duplication of traC associated with the duplication of the blaCMY-2 region (Fig. 3). Given the sequence conservation between pAM04528 and pSN254, we speculate that the latter also lacks the ability to self-conjugate.
Genetic relationships between plasmids.
The high degree of sequence similarity and synteny makes it almost certain that the four blaCMY-2 plasmids examined in this study were derived from a common ancestor, although it is not possible to know with certainty which of the blaCMY-2 plasmids most closely resembles the ancestral state. Plasmid pAR060302 has a single copy of the blaCMY-2 region and retains the aadA region, suggesting that pAR060302 is representative of the most ancestral sequence, but there are multiple scenarios that could be derived depending on how the large antibiotic-resistance-encoding sequences were lost or gained.
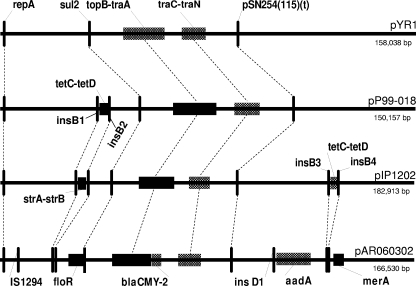
There are a number of differences between the blaCMY-2 plasmids and other IncA/C plasmids, and we have highlighted some of the more interesting differences in Fig. 4. Plasmid pYR1 from Y. ruckeri appears to be the most dissimilar, with only a single antibiotic resistance gene (sul2), and one common reference point is present in truncated form in the pYR1 sequence [pSN254(115)]. Plasmid pP99-018 from Photobacterium damselae subsp. piscicida acquired tet(C) and tet(D), flanked by transposase genes insB1 and insB2. These transposase genes appear to have been duplicated in pIP1202 from Y. pestis with tet(C) and tet(D). strA and strB appeared between transposase genes insB1 and insB2. The pAR060302 sequence shows that insertions have vacated the region between insB1 and insB2, and in addition to insertion of the blaCMY-2, region there has been the addition of the aadA and merA regions (Fig. 4).
FIG. 4.
Schematic diagram highlighting several conserved segments and variable regions relative to antimicrobial resistance for pAR060302 and other IncA/C plasmids from Yersinia ruckeri (pYR1) (33), Photobacterium damselae (pP99-018) (17), and Y. pestis (pIP1202) (33). Relative to pYR1, plasmid pP99-018 (also see pP91278 [17]) has the addition of insB1 and insB2 sequences associated with tet(C) and tet(D). The tet(C) and tet(D) genes are relocated in pIP1202 (between insB3and insB4), while strA and strB are found between insB1 and insB2. In contrast, the blaCMY-2 plasmids (from pSN254, peH4H, pAM04528, and pAR060302) harbor no inserts between insB1 and insB2 (Fig. 2); if present, the region between insB3 and insB4 is occupied by the kan region. In addition, these plasmids include a floR region, a merA region, and, with the exception of pAM04528, an aadA region (associated with insD1), as shown for pAR060302. Locus pSN254(115) is shown here as a reference point, and this sequence is identical for all plasmids considered in this study except for a 3-prime truncation in pYR-1 and a 5-prime truncation in peH4H.
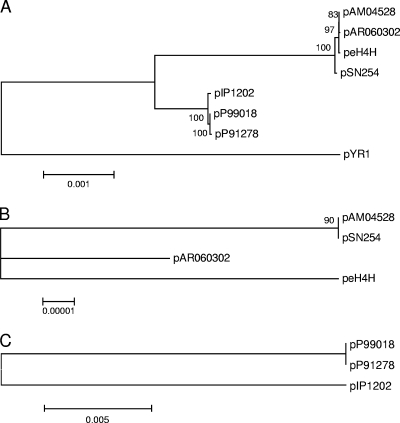
We used a sequence conservation-based method to build a tree for the eight IncA/C plasmids, and we conducted multiple numerical experiments to ensure that tree construction was robust. There are three distinct subgroups within the main tree, with one subgroup represented by plasmids from E. coli and S. enterica and a second subgroup represented by plasmids from Y. pestis and P. damselae (Fig. 5A). The Y. ruckeri plasmid was distal to these two subgroups. Conserved genes were identified within the two plasmid subgroups (see Tables S2 and S3 in the supplemental material), and sequence analysis confirmed that pAM04528 and pSN254 were virtually identical while peH4H was the most divergent member of one subgroup (Fig. 5B). Plasmid pIP1202 from Y. pestis was the most divergent member of the second subgroup (Fig. 5C). The Y. pestis plasmid (pIP1202) is most closely related to the IncA/C plasmids from Photobacterium damselae. This analysis supports the likelihood of recent dissemination of blaCMY-2 plasmids among E. coli and S. enterica hosts, but their greater divergence from Y. pestis or Y. ruckeri suggests less recent plasmid transfer among these pathogen groups. The last is a marine fish pathogen with no obvious ecological overlap with Y. pestis. At present, it is not possible to determine whether this plasmid clade originated from marine or terrestrial systems. The pIP-1202-like plasmids are disseminating in marine systems, judging by a recent report of Vibrio cholerae strains harboring these plasmids (25). blaCMY-2 plasmids have been detected in Aeromonas salmonicida isolated from Atlantic salmon (Salmo salar) (22). While the full plasmid sequence was not reported, PCR and some sequence data suggest that the blaCMY-2 plasmids detected in A. salmonicida closely match the pSN254 sequence (33). If this is the case, then it appears that there has been successful dissemination of blaCMY-2 plasmids first documented in mammalian enteric flora to an aquatic ecosystem.
FIG. 5.
(A) Bootstrap consensus tree for a neighbor-joining algorithm showing relative genetic relatedness between eight IncA/C plasmids. The tree was based on sequences from 91 conserved genes (BLAST score threshold = 0.3) (see Table S1 in the supplemental material; see Materials and Methods for details), with pIP1202 arbitrarily chosen as the reference plasmid. (B) Subtree for blaCMY-2 plasmids based on 25 conserved genes (BLAST score threshold = 0.3) (see Table S2 in the supplemental material; see Materials and Methods for details), with pSN254 used as the reference sequence. (C) Subtree for three additional plasmids based on 49 conserved genes (BLAST score threshold = 0.3; see Table S3 in the supplemental material; see Materials and Methods for details), with pIP1202 used as the reference sequence. Neighbor-joining consensus trees were calculated from 500 bootstrap iterations, with numbers at nodes representing percent correspondence. The scale bars represent the number of base substitutions per site (32).
Supplementary Material
Acknowledgments
Kelly Brayton, Deborah Duricka, Patrick Friel, Murugan Subbiah, and Yubei Zhang provided technical assistance. Jean Whichard (CDC, Atlanta, Georgia) provided strain AM04528.
This project has been funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. NO1-AI-30055 (D.R.C., T.E.B., and S.L.B.); the USDA-ARS Animal Disease Research Unit, Pullman WA; and the Agricultural Animal Health Program, College of Veterinary Medicine, Washington State University, Pullman, WA, and in part by USDA National Research Initiative Competitive Grants 2000-35212-9398 and 2003-35212-13853 (R.S.S.) and the Biomedical Genomics Center at the University of Minnesota (R.S.S.).
Footnotes
Published ahead of print on 30 November 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barlow, M., and B. G. Hall. 2002. Origin and evolution of the AmpC beta-lactamases of Citrobacter freundii. Antimicrob. Agents Chemother. 46:1190-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer, A. W., W. M. Kirby, J. C. Sherris, and M. Turck. 1966. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45:493-496. [PubMed] [Google Scholar]
- 4.Bauernfeind, A., I. Stemplinger, R. Jungwirth, and H. Giamarellou. 1996. Characterization of the plasmidic beta-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob. Agents Chemother. 40:221-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergstrom, C. T., M. Lipsitch, and B. R. Levin. 2000. Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics 155:1505-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braibant, M., J. Chevalier, E. Chaslus-Dancla, J. M. Pages, and A. Cloeckaert. 2005. Structural and functional study of the phenicol-specific efflux pump FloR belonging to the major facilitator superfamily. Antimicrob. Agents Chemother. 49:2965-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carattoli, A. 2003. Plasmid-mediated antimicrobial resistance in Salmonella enterica. Curr. Issues Mol. Biol. 5:113-122. [PubMed] [Google Scholar]
- 8.Carattoli, A., V. Miriagou, A. Bertini, A. Loli, C. Colinon, L. Villa, J. M. Whichard, and G. M. Rossolini. 2006. Replicon typing of plasmids encoding resistance to newer beta-lactams. Emerg. Infect. Dis. 12:1145-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carattoli, A., F. Tosini, W. P. Giles, M. E. Rupp, S. H. Hinrichs, F. J. Angulo, T. J. Barrett, and P. D. Fey. 2002. Characterization of plasmids carrying CMY-2 from expanded-spectrum cephalosporin-resistant Salmonella strains isolated in the United States between 1996 and 1998. Antimicrob. Agents Chemother. 46:1269-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniels, J. B., D. R. Call, and T. E. Besser. 2007. Molecular epidemiology of blaCMY-2 plasmids carried by Salmonella enterica and Escherichia coli isolates from cattle in the Pacific Northwest. Appl. Environ. Microbiol. 73:8005-8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, M. A., D. D. Hancock, T. E. Besser, J. B. Daniels, K. N. Baker, and D. R. Call. 2007. Antimicrobial resistance in Salmonella enterica serovar Dublin isolates from beef and dairy sources. Vet. Microbiol. 119:221-230. [DOI] [PubMed] [Google Scholar]
- 12.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giles, W. P., A. K. Benson, M. E. Olson, R. W. Hutkins, J. M. Whichard, P. L. Winokur, and P. D. Fey. 2004. DNA sequence analysis of regions surrounding blaCMY-2 from multiple Salmonella plasmid backbones. Antimicrob. Agents Chemother. 48:2845-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, I. F., C. H. Chiu, M. H. Wang, C. Y. Wu, K. S. Hsieh, and C. C. Chiou. 2005. Outbreak of dysentery associated with ceftriaxone-resistant Shigella sonnei: first report of plasmid-mediated CMY-2-type AmpC beta-lactamase resistance in S. sonnei. J. Clin. Microbiol. 43:2608-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang, M. S., T. E. Besser, and D. R. Call. 2006. Variability in the region downstream of the blaCMY-2 beta-lactamase gene in Escherichia coli and Salmonella enterica plasmids. Antimicrob. Agents Chemother. 50:1590-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karim, A., L. Poirel, S. Nagarajan, and P. Nordmann. 2001. Plasmid-mediated extended-spectrum beta-lactamase (CTX-M-3 like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201:237-241. [DOI] [PubMed] [Google Scholar]
- 17.Kim, M. J., I. Hirono, K. Kurokawa, T. Maki, J. Hawke, H. Kondo, M. D. Santos, and T. Aoki. 2008. Complete DNA sequence and analysis of the transferable multiple-drug resistance plasmids (R plasmids) from Photobacterium damselae subsp. piscicida isolates collected in Japan and the United States. Antimicrob. Agents Chemother. 52:606-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koeck, J. L., G. Arlet, A. Philippon, S. Basmaciogullari, H. V. Thien, Y. Buisson, and J. D. Cavallo. 1997. A plasmid-mediated CMY-2 beta-lactamase from an Algerian clinical isolate of Salmonella senftenberg. FEMS Microbiol. Lett. 152:255-260. [DOI] [PubMed] [Google Scholar]
- 19.Liebert, C. A., R. M. Hall, and A. O. Summers. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Literacka, E., B. Bedenic, A. Baraniak, J. Fiett, M. Tonkic, I. Jajic-Bencic, and M. Gniadkowski. 2009. blaCTX-M genes in Escherichia coli strains from Croatian hospitals are located in new (blaCTX-M-3a) and widely spread (blaCTX-M-3a and blaCTX-M-15) genetic structures. Antimicrob. Agents Chemother. 53:1630-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Llanes, C., P. Gabant, M. Couturier, and Y. Michel-Briand. 1994. Cloning and characterization of the Inc A/C plasmid RA1 replicon. J. Bacteriol. 176:3403-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIntosh, D., M. Cunningham, B. Ji, F. A. Fekete, E. M. Parry, S. E. Clark, Z. B. Zalinger, I. C. Gilg, G. R. Danner, K. A. Johnson, M. Beattie, and R. Ritchie. 2008. Transferable, multiple antibiotic and mercury resistance in Atlantic Canadian isolates of Aeromonas salmonicida subsp. salmonicida is associated with carriage of an IncA/C plasmid similar to the Salmonella enterica plasmid pSN254. J. Antimicrob. Chemother. 61:1221-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.NCCLS. 2003. Performance standards for antimicrobial susceptibility testing, 13th informational supplement, 13th ed. Approved standard M100-S13. NCCLS, Wayne, PA.
- 24.Odeh, R., S. Kelkar, A. M. Hujer, R. A. Bonomo, P. C. Schreckenberger, and J. P. Quinn. 2002. Broad resistance due to plasmid-mediated AmpC beta-lactamases in clinical isolates of Escherichia coli. Clin. Infect. Dis. 35:140-145. [DOI] [PubMed] [Google Scholar]
- 25.Pan, J. C., R. Ye, H. Q. Wang, H. Q. Xiang, W. Zhang, X. F. Yu, D. M. Meng, and Z. S. He. 2008. Vibrio cholerae O139 multiple-drug resistance mediated by Yersinia pestis pIP1202-like conjugative plasmids. Antimicrob. Agents Chemother. 52:3829-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park, Y. J., S. Y. Kim, J. K. Yu, S. I. Kim, Y. Uh, S. G. Hong, L. Jongwook, and H. S. Kwak. 2009. Spread of Serratia marcescens coharboring aac(6′)-Ib-cr, bla(CTX-M), armA, and bla(OXA-1) carried by conjugative IncL/M type plasmid in Korean hospitals. Microb. Drug Resist. 15:97-102. [DOI] [PubMed] [Google Scholar]
- 27.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-determined AmpC-type beta-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning, a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Suzek, B. E., M. D. Ermolaeva, M. Schreiber, and S. L. Salzberg. 2001. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics 17:1123-1130. [DOI] [PubMed] [Google Scholar]
- 32.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 33.Welch, T. J., W. F. Fricke, P. F. McDermott, D. G. White, M. L. Rosso, D. A. Rasko, M. K. Mammel, M. Eppinger, M. J. Rosovitz, D. Wagner, L. Rahalison, J. E. Leclerc, J. M. Hinshaw, L. E. Lindler, T. A. Cebula, E. Carniel, and J. Ravel. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winokur, P. L., A. Brueggemann, D. L. DeSalvo, L. Hoffmann, M. D. Apley, E. K. Uhlenhopp, M. A. Pfaller, and G. V. Doern. 2000. Animal and human multidrug-resistant, cephalosporin-resistant salmonella isolates expressing a plasmid-mediated CMY-2 AmpC beta-lactamase. Antimicrob. Agents Chemother. 44:2777-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, S. W., K. Dornbusch, G. Kronvall, and M. Norgren. 1999. Characterization and nucleotide sequence of a Klebsiella oxytoca cryptic plasmid encoding a CMY-type beta-lactamase: confirmation that the plasmid-mediated cephamycinase originated from the Citrobacter freundii AmpC beta-lactamase. Antimicrob. Agents Chemother. 43:1350-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao, S., D. G. White, P. F. McDermott, S. Friedman, L. English, S. Ayers, J. Meng, J. J. Maurer, R. Holland, and R. D. Walker. 2001. Identification and expression of cephamycinase bla(CMY) genes in Escherichia coli and Salmonella isolates from food animals and ground meat. Antimicrob. Agents Chemother. 45:3647-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.