Abstract
Triclosan, a very widely used biocide, specifically inhibits fatty acid synthesis by inhibition of enoyl-acyl carrier protein (ACP) reductase. Escherichia coli FabI is the prototypical triclosan-sensitive enoyl-ACP reductase, and E. coli is extremely sensitive to the biocide. However, other bacteria are resistant to triclosan, because they encode triclosan-resistant enoyl-ACP reductase isozymes. In contrast, the triclosan resistance of Pseudomonas aeruginosa PAO1 has been attributed to active efflux of the compound (R. Chuanchuen, R. R. Karkhoff-Schweizer, and H. P. Schweizer, Am. J. Infect. Control 31:124-127, 2003). We report that P. aeruginosa contains two enoyl-ACP reductase isozymes, the previously characterized FabI homologue plus a homologue of FabV, a triclosan-resistant enoyl-ACP reductase recently demonstrated in Vibrio cholerae. By deletion of the genes encoding P. aeruginosa FabI and FabV, we demonstrated that FabV confers triclosan resistance on P. aeruginosa. Upon deletion of the fabV gene, the mutant strain became extremely sensitive to triclosan (>2,000-fold more sensitive than the wild-type strain), whereas the mutant strain lacking FabI remained completely resistant to the biocide.
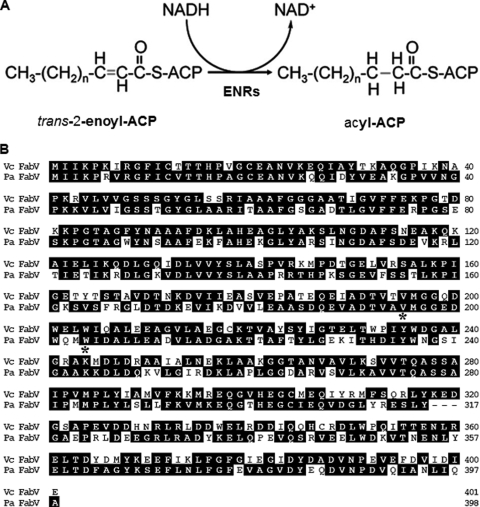
Fatty acids are major components of cell membrane as well as metabolic intermediates in bacteria (9, 20, 25). The bacterial fatty acid synthesis system (FAS II), which is carried out by a series of discrete enzymes, differs significantly from the mammalian and fungal system (FAS I), which uses a large complex multifunctional enzymes (25, 43). The differences between the FAS I and FAS II systems make the FAS II enzymes good targets for antibacterial inhibitors (19, 20, 32, 45). The enoyl-acyl carrier protein (ACP) reductase (ENR) components of the type II system catalyze the last step of the elongation cycle in the synthesis of fatty acids (28). ENRs catalyze the reduction of trans-2-acyl-ACPs (an enoyl-ACP) to the fully saturated acyl-ACP species (Fig. 1A). In bacteria four ENR isozymes have been reported. These are FabI (3, 39), FabL (18), FabV (29), and FabK (17, 27). FabI, FabL, and FabV are members of the short-chain dehydrogenase/reductase (SDR) superfamily, whereas FabK, a TIM barrel flavoprotein, is unrelated to this family (28). Most bacteria contain an easily identifiable fabI gene in their chromosomes; the expressed proteins are about 40% identical to Escherichia coli FabI and contain a conserved Tyr-156-(Xaa)6-Lys-163 (E. coli numbering) catalytic dyad (19, 29). In E. coli, FabI has been shown to be the site of action of triclosan (30), a biocide used in hand soaps and a large variety of other everyday products. Since FabI is the only E. coli ENR, it acts in each step of the fatty acid elongation cycle and thus is essential for cell growth and survival (2, 3, 15). Soon after the identification of FabI as the triclosan target, the existence of a number of bacterial species naturally resistant to triclosan was recognized. FabL was identified in Bacillus subtilis by the presence of the Tyr-(Xaa)6-Lys dyad, and although it shares a degree of sequence similarity with FabI proteins, it is not a member of the FabI family (18). FabL is moderately resistant to triclosan and uses NADPH as the coenzyme (18). More recently, FabV, the third SDR family ENR, was discovered in Vibrio cholerae. FabV is unlike the other SDR ENRs in that it is completely refractory to triclosan inhibition, is 60% larger than the typical SDR family member (which are generally about 250 residues long), and has an eight-residue space between the active-site tyrosine and lysine residues (Tyr-X8-Lys) (29). This spacing has two more residues than those in the FabI and FabL active sites and one more than the maximum reported for other SDR proteins (33). However, like FabI, FabV has the classical Rossman fold motif (29). The only non-SDR-family ENR reported to date is Streptococcus pneumoniae FabK which is as refractory to triclosan as FabV (17, 27). The crystal structure of FabK has been reported recently (36). Unlike the SDR enzymes, FabK has a TIM barrel (α8-β8) structure and is an FMN-dependent oxidoreductase of the NAD(P)H-dependent flavin oxidoreductase family (28, 36).
FIG. 1.
The ENR reaction and alignment of P. aeruginosa FabV with V. cholerae FabV. (A) The ENR reaction. Depending on the enzyme, the reductant can be NADPH or FMNH2 (coupled to NADH oxidation) rather than NADH. (B) The V. cholerae FabV (top line) and P. aeruginosa FabV sequences have 58% identity and 79% similarity. The active-site tyrosine and lysine residues are asterisked.
The important opportunistic pathogen Pseudomonas aeruginosa contains a fatty acid synthase system very similar to that of E. coli (21, 22, 24). A fabI gene was identified in the P. aeruginosa genome and disrupted (21). The fabI-null mutant strain grew normally, and thus, it was concluded that unlike E. coli, P. aeruginosa must contain another ENR (21). Indeed, assay of extracts of the fabI-null mutant strain showed that it retained 62% of the activity seen in wild-type extracts and that this activity was resistant to triclosan (21). Searches of the P. aeruginosa genome database revealed at least 18 possible FabI paralogs (averaging 25% identity and 40% similarity), but all lacked the signature motifs found in FabI proteins (21). Hence, Heath and Rock (17) suggested that the second triclosan-resistant ENR of P. aeruginosa was a FabK homologue, three of which are encoded in the P. aeruginosa genome. However, we have expressed all three putative FabK homologues and found that none possessed ENR activity (data not shown). The discovery of Vibrio cholerae FabV suggested another possibility for the highly triclosan resistant ENR of P. aeruginosa. In this report we describe the characterization of the P. aeruginosa fabV gene and its product, the second ENR of this bacterium.
MATERIALS AND METHODS
The supply sources were as follows: malonyl coenzyme A (malonyl-CoA), acetyl-CoA, fatty acids, triclosan, NADH, NADPH, and antibiotics were from Sigma; Takara Biotechnology Co. provided molecular biology reagents; Novagen provided pET vectors; American Radiolabeled Chemicals, Inc., provided sodium [1-14C]acetate (specific activity, 50 mCi/mM); Invitrogen provided the Ni2-agarose column; and Bio-Rad provided the UNOsphere Q strong anion-exchange media and the Quick Start Bradford dye reagent. All other reagents were of the highest available quality.
Bacterial strains, plasmids, and growth media.
The E. coli K-12 strains, P. aeruginosa PAO1 strains, and plasmids used in this study are listed in Table 1. Luria-Bertani (LB) medium (44) was used as the rich medium for E. coli and P. aeruginosa growth. ME medium (31) supplemented with 0.1% yeast extract was used to screen P. aeruginosa mutants, and if needed, 5% or 10% sucrose was added. Antibiotics were used at the following concentrations (in micrograms per milliliter): sodium ampicillin, 100; kanamycin sulfate, 30; chloramphenicol, 30; and gentamicin, 10 (for E. coli) or 100 (for P. aeruginosa). l-Arabinose was used at a final concentration of 0.01%. Isopropyl-β-d-thiogalactoside (IPTG) was used at a final concentration of 1 mM, and 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) was used at a final concentration of 20 μg/ml.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant characteristic(s) | Source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F φ80ΔlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 | Laboratory collection |
| W3110 | Wild-type strain | Laboratory collection |
| MG1655 | Wild-type strain | Laboratory collection |
| BL21(DE3) | E. coli B; F−ompT rB− mB− (λDE3) | Laboratory collection |
| S17-1 | F−thi pro hsdR [RP4-2 Tc::Mu Km::Tn7 (Tp Sm)] | Laboratory collection |
| JP1111 | Hfr galE45 fabI392(Ts) relA1 spoT1 | 3 |
| P. aeruginosa | ||
| PAO1 | PAO1 | Laboratory collection |
| PAO170 | PAO1 fabV::Gmr | This work |
| PAO272 | PAO1 fabI::Gmr | This work |
| Plasmids | ||
| pMD19 | T-vector; Ampr | Takara |
| pBAD24M | Ampr; NcoI site of expression vector pBAD24 changed to an NdeI site | Reference 14 and this work |
| pET-28b | Kmr, expression vector | Novagen |
| pBluescript SK(+) | Ampr; clone vector | Laboratory collection |
| p34s-Gm | Ampr; Gm resistance cassette-carrying vector | 7 |
| pK18mobsacB | Kmr; sacB-based gene replacement vector | 37 |
| pFX7 | Kmr; EcfabI (ligation of E. coli fabI between the NdeI and HindIII sites of pET28b) | Laboratory collection |
| pCD1 | Kmr; PaacpP (ligation of P. aeruginosa acpP between the NcoI and HindIII sites of pET28b) | Laboratory collection |
| pZL1 | Ampr; PafabV in pMD19 (800-bp PafabV PCR fragment) | This work |
| pZL2 | Ampr; PafabI in pMD19 (1,200-bp PafabI PCR fragment) | This work |
| pZL6 | Ampr; PafabV in pBAD24 M (ligation of the NdeI-HindIII fragment from pZL1 into the same sites of pBAD24M) | This work |
| pZL7 | Ampr; PafabI in pBAD24M (ligation of the NdeI-HindIII fragment from pZL2 into the same sites of pBAD24M) | This work |
| pZL11 | Kmr; PafabV in pET-28b (ligation of the NdeI-HindIII fragment from pZL1 into the same sites of pET28b) | This work |
| pZL12 | Kmr; PafabI in pET-28b (ligation of the NdeI-HindIII fragment from pZL2 into the same sites of pET28b) | This work |
| pZL16 | Ampr; PafabV in pBluescript SK(+) [ligation of the XbaI-HindIII fragment of pZL11 into pBluescript SK(+)] | This work |
| pZL17 | Ampr; PafabI [XbaI-HindIII fragment of pZL12 in pBluescript SK(+)] | This work |
| pZL18 | Ampr; EcfabI in pBAD24M (NdeI-HindIII fragment of pFX7 inserted into the same sites of pBAD24M) | This work |
| pZL19 | Ampr; EcfabI in pBluescript SK(+) [ligation of the XbaI-HindIII fragment of pFX7 into pBluescript SK(+)] | This work |
| pZL20 | Ampr; PafabV Y235F mutant in pBAD24M | This work |
| pZL21 | Ampr; PafabV K244M mutant in pBAD24M | This work |
| pZL22 | PafabV Y235F K244M double mutant in pBAD24M | This work |
| pZL23 | Kmr; PafabV Y235F mutant in pET-28b | This work |
| pZL24 | Kmr; PafabV K244M mutant in pET-28b | This work |
| pZL25 | Kmr; PafabV Y235F K244M double mutant in pET-28b | This work |
| pZL26 | Kmr Gmr; a 1,500-bp PCR fragment containing the PafabV::Gmr cassette in pK18mobsacB | This work |
| pZL27 | Kmr, Gmr; a 1,900-bp fragment containing the PafabI::Gmr cassette in pK18mobsacB | This work |
Recombinant DNA techniques and construction of plasmids.
To clone the P. aeruginosa fabV and fabI genes, genomic DNA was extracted from P. aeruginosa strain PAO1 using the Takara DNA extraction kit. The PCR products were amplified from strain PAO1 genomic DNA using Pfu DNA polymerase and the primers listed in Table 2 and were inserted into T-vector plasmid pMD19 to produce plasmids pZL1 (PafabV) and pZL2 (PafabI). The fab gene sequences were confirmed by sequencing by Shanghai Sangon, Inc.
TABLE 2.
Sequences of the PCR primers used
| Primer name | Primer sequence (5′→3′)a |
|---|---|
| PafabI NdeI | CGGCATATGGGATTTCTCACAGGAA |
| PafabI HindIII | GGGAAGCTTAGTCGTCGTCCAGCGG |
| PafabV NdeI | ACCGAGGTTCATATGATCATCAAACCGCGC |
| PafabV HindIII | AGGCAAGCTTGTCGCTGAAAACGCGAAC |
| FabV 235Y to F 1 | GATCACCCACGACATCTTCTGGAACGGTTCCATCGGC |
| FabV 235Y to F 2 | GCCGATGGAACCGTTCCAGAAGATGTCGTGGGTGATC |
| FabV 244K to M 1 | GTTCCATCGGCGCAGCCATGAAGGATCTCGACCAGAAG |
| FabV 244K to M 2 | CTTCTGGTCGAGATCCTTCATGGCTGCGCCGATGGAAC |
| Gmdown | TTAGGTGGCGGTACTTGG |
| Gmup | CCTGTTCGGTTCGTAAAC |
| FabVdown | CCAAGTACCGCCACCTAATCTCGGTGCTCAAGGCGGTG |
| FabVup | GTTTACGAACCGAACAGGTCGACCTTGCCGAGATCG |
| PaI up | GCAGGTGGATCCCCAGGACCCGTATGGCAG |
| PaI down | CACAACAAGCTTAAGGAGCAACCGCTGGAG |
| GmNcoI1 | AAGCTTCCATGGGAATTGACATAAGCCTGTTCGGTTCG |
| GmNcoI2 | AAGCTTCCATGGGAATTGGCCGCGGCGTTGTGACAATTTAC |
| P1 | CCGATTCCAAGCCGACAAGAC |
| P2 | AAACCGCGCAGCACTACCAG |
| P9 | GGCAAGCGCGGAAAACTGAGTAGCAGAC |
| P10 | CATTTTTCATGGGCGGGGTGATCAGTCG |
Restriction sites are underlined.
To produce plasmids pZL6 (PafabV) and pZL7 (PafabI), or pZL11 (PafabV) and pZL12 (PafabI), the T-vector pMD19 fab gene plasmids were digested with NdeI and a second restriction enzyme, the site for which had been designed into the downstream primers listed in Table 2. The fragment was gel purified and ligated into pBAD24M or pET-28b digested with the same enzymes. The XbaI-HindIII fragments of pZL11 (PafabV) and pZL12 (PafabI) were ligated into pBluescript SK(+) cut with the same enzymes to yield pZL16 (PafabV) and pZL17 (PafabI), respectively. To obtain pZL18 (EcfabI), plasmid pFX7 was digested with NdeI and HindIII, and the EcfabI fragment was inserted into the same sites of pBAD24M, whereas the XbaI-HindIII fragment of plasmid pFX7 was ligated to pBluescript SK(+) cut with the same enzymes to produce pZL19 (EcfabI). Plasmids pZL20 and pZL21, carrying a single mutation within the PafabV gene coding sequence, were constructed by site-directed mutagenesis using the primers listed in Table 2 with pZL6 as the PCR template. Plasmid pZL22 carrying the double mutant PafabV was made using pZL20 as the template. The mutant PafabV gene of plasmid pZL20, pZL21, or pZL22 were cloned into vector pET-28b to generate plasmid pZL23, pZL24, or pZL25, respectively. All plasmids were mobilized into E. coli strains by CaCl2-mediated transformation.
Analysis of phospholipid compositions.
The cultures were grown aerobically at 37°C in LB medium overnight. Cells were harvested from 10-ml cultures and washed three times with LB medium at room temperature. The phospholipids were extracted by a previously published procedure (41). Fatty acid methyl esters were synthesized and extracted as described previously (11, 40). Briefly, the phospholipids were dissolved in 1.2 ml dry methanol, and 0.2 ml of 25% (vol/vol) sodium methoxide was added. After the solution was allowed to stand for 15 min at room temperature, 1.2 ml of 2 M HCl was added, and the fatty acid methyl esters were obtained by three extractions each with 1.2 ml of petroleum ether. The solvent was removed under a stream of nitrogen, and the residue was analyzed by gas chromatography-mass spectrometry (GC-MS).
The ability of PafabV or PafabI to restore fatty acid synthesis in vivo was tested by transforming the E. coli fabI(Ts) mutant strain, JP1111, with plasmids carrying genes encoding a putative PafabV or PafabI homologue in the arabinose-inducible vector pBAD24M. The cultures were grown at a permissive temperature, induced with arabinose, and shifted to 42°C, and then [1-14C]acetate was added as described previously (13). Labeled phospholipids were extracted, analyzed by thin-layer chromatography (TLC) (29), and quantitated by phosphorimaging.
Expression and purification of plasmid-encoded proteins.
The pET28b-dervied plasmids pZL11, pZL12, pZL23, pZL24, and pZL25 were introduced into E. coli strain BL21(DE3), and the respective proteins, PaFabV, PaFabI, PaFabV mutant (Y to F), PaFabV mutant (K to M), and PaFabV double mutant (Y to F and K to M) were expressed at high levels and purified as described previously (16, 42). The enzymes were homogeneous as judged by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (data not shown). The E. coli FabD, FabH, FabG, FabZ, and FabI and Vibrio harveyi AasS proteins were purified by their hexahistidine tags as described previously (16, 23). To purify P. aeruginosa holo-ACP, the PCR product of PaacpP (NP251656) was cloned into the NcoI and HindIII sites of pET-28b to yield plasmid pET-ACP. This plasmid was introduced into E. coli BL21(DE3), and the holo-ACP was expressed at high levels and purified as described previously (38).
Acyl-ACP preparations.
We synthesized crotonyl-ACP, trans-2-hexenoyl-ACP, trans-2-octenoyl-ACP, trans-2-decenoyl-ACP, trans-2-dodecenoyl-ACP, trans-2-tetradecanoyl-ACP, and trans-2-hexadecadienoyl-ACP as described by Jiang and coworkers (23, 46). Briefly, the 1-ml reaction mixture contained 10 mM ATP, 20 mM MgSO4, 0.1 M Tris-HCl (pH 7.8), 1 mM dithiothreitol, 0.66 mg of P. aeruginosa holo-ACP, 40 μg of His-tagged V. harveyi acyl-ACP synthetase purified from E. coli, and 0.5 mM fatty acid. For trans-2-enoyl-ACP with chain lengths between 12 and 16, the reaction mixture contained 30 μg E. coli FabZ (EcFabZ) plus a 3-hydroxy-fatty acid. The reaction mixtures were incubated at 37°C for 4 h. Two volumes of acetone were then added to each sample, and the mixtures were allowed to precipitate overnight at −20°C. The precipitates were pelleted at 20,000 × g for 20 min at 4°C and were then washed twice with 3 volumes of acetone. The pellet was air dried and resuspended in 20 mM Tris-HCl (pH 7.4) at an ACP concentration of 3.3 mg/ml. Completion of the acyl-ACP synthesis reaction was determined by the gel shift assay (35).
Assay of PaFabV and PaFabI activities in vitro.
The abilities of PaFabV and PaFabI to function in the first cycle of fatty acid synthesis were assessed in reaction mixtures containing 0.1 M sodium phosphate (pH 7.0); 0.1 μg each of EcFabD, EcFabH, EcFabG, and EcFabZ; 50 μM NADH; 50 μM NADPH; 1 mM β-mercaptoethanol; 100 μM acetyl-CoA; 100 μM malonyl-CoA; and 50 μM holo-ACP in a final volume of 40 μl. In the PaFabV- or PaFabI-catalyzed reduction of trans-2-enoyl-ACPs with chain lengths between 6 and 10 carbons, the reaction mixtures contained 0.1 M sodium phosphate (pH 7.0); 1 mM β-mercaptoethanol; and 50 μM acyl-ACP (trans-2-hexenoyl-ACP, trans-2-octenoyl-ACP, or trans-2-decenoyl-ACP). For trans-2-enoyl-ACPs with chain lengths between 12 and 16 carbons, the reaction mixtures contained EcFabZ and 3-hydroxydodecanoyl-ACP, 3-hydroxytetradecanoyl-ACP, or 3-hydroxyhexadecanoyl-ACP as the substrate. The reactions were initiated by addition of PaFabV or PaFabI, followed by incubation for 1 h. The reaction products were resolved by conformationally sensitive gel electrophoresis on 15% polyacrylamide gels containing a concentration of urea optimized for the separation (34). The gel was stained with Coomassie brilliant blue R250. A stock solution of triclosan was prepared in 95% ethanol. When needed in order to assess the possible inhibitory effects of triclosan on enzyme activities, the appropriate volumes of triclosan solution were added to the assay tubes, and the triclosan was dried before the addition of the assay ingredients.
NADH oxidation assay.
ENR activity was monitored spectrophotometrically by the decrease in the absorbance at 340 nm using an NADH extinction coefficient of 6,220 M−1 (29). Each reaction (reaction mixture volume, 100 μl) was performed in UV-transparent microcuvettes. The reaction mixtures for activity assays of PaFabV or PaFabI for enoyl-ACP contained 150 μM NADH, 10 ng of the purified native PaFabV or 100 ng of PaFabI, 100 μM enoyl-ACP, and 0.1 M LiCl in 0.1 M sodium phosphate buffer (pH 7.0). Kinetic constants were determined using GraphPad Prism software, version 4.
Essentiality testing of the PafabV gene and disruption of the fabV gene.
To disrupt P. aeruginosa fabV, a suicide plasmid was constructed as follows. The 400-bp regions upstream and downstream of fabV (called UpfabV and DownfabV, respectively) were amplified with Pfu DNA polymerase using plasmid pZL1 as the template and either PafabVNdeI and FabVup (for UpfabV) or PafabVHindIII and FabVdown (for DownfabV) as the primers (Table 2). A 750-bp gentamicin resistance cassette was also amplified from plasmid p34s-Gm (8) with Gmdown plus Gmup (Table 2) as the primers. The products of these PCRs were purified, and overlapping PCR was carried out using PafabVNdeI and PafabVHindIII as the primers. The 1,500-bp ΔPafabV::Gm PCR fragment was ligated into the T-vector plasmid pMD19 to yield pMD-ΔPafabV::Gm. The BamHI-HindIII fragment of the ΔPafabV::Gm PCR fragment was cloned into the same sites of pK18mobsacB to yield pZL26. Following mating of a derivative of E. coli strain S17-1 carrying plasmid pZL26 with P. aeruginosa PAO1 on LB plates for 12 h at 37°C, the cells were suspended in LB medium and appropriate dilutions were spread on LB plates containing chloramphenicol (to select against the donor strain) plus gentamicin to select for integration of the nonreplicating plasmid into the chromosome of the recipient. Several colonies were inoculated into LB medium, and the cultures were incubated at 37°C overnight, after which appropriate dilutions were spread onto ME plates containing 10% sucrose. Several of the resulting colonies were inoculated onto ME plates containing gentamicin using sterile toothpicks. Colonies resistant to gentamicin were screened by colony PCR utilizing the primers listed in Table 2. The fabI mutant was isolated by the same procedure as that used for the fabV mutant.
RESULTS
Identification of a new P. aeruginosa ENR.
Since a P. aeruginosa fabI insertion mutant was viable (21) and was resistant to inhibition by triclosan, Heath and Rock (17) suggested that P. aeruginosa must contain a FabK-type ENR. This seemed a reasonable hypothesis, and thus, we searched the P. aeruginosa PAO1 genome database against the Streptococcus pneumoniae FabK sequence using BLAST. Three candidates were obtained: NP249715 (also called PA1024, annotated as a 2-nitropropane dioxygenase), NP252891 (also called PA4202, annotated as a hypothetical protein), and NP249351 (also called PA0660, annotated as a hypothetical protein), which exhibited 31%, 34%, and 31% identity and 51%, 48%, and 51% similarity to S. pneumoniae FabK protein, respectively. Each of these proteins was tested for the ability to restore growth to an E. coli fabI mutant strain and for ENR activity as was done with FabV (as described below), and none of the three showed activity in either assay (data not shown). In a second approach, the P. aeruginosa PAO1 genome database was searched for a homologue to V. cholerae FabV, a recently identified SDR-family ENR (29). A single strong homologue was found, NP251640 (also called PA2950), which had 58% identity and 79% similarity to V. cholerae FabV (Fig. 1B). Moreover, the P. aeruginosa FabV homologue contained the Tyr-(Xaa)8-Lys motif first seen in V. cholerae FabV, a further indication that the protein might have ENR activity.
P. aeruginosa fabV functionally replaces E. coli fabI in vivo and renders E. coli resistant to triclosan.
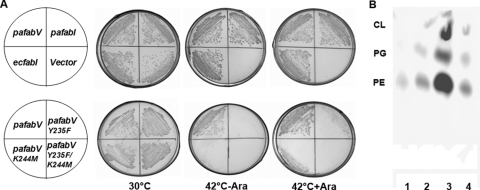
To test the function of the PafabV homologue, this gene and the PafabI gene were inserted into the arabinose-inducible vector pBAD24M, a derivative of pBAD24 in which the NcoI site was converted to an NdeI site by site-directed mutagenesis. The resulting plasmids were then introduced into the E. coli fabI(Ts) strain JP1111. At the nonpermissive temperature, this mutant strain lacks ENR activity and thus is unable to grow (2, 3). Derivatives of strain JP1111 carrying plasmids encoding PafabV, PafabI, or EcfabI (pZL6, pZL7, and pZL18, respectively) grew at 42°C in the presence or absence of arabinose, whereas strains carrying the vector plasmid or a plasmid encoding one of the putative FabK homologues failed to grow on either medium (Fig. 2A). Thus, PafabV complemented the E. coli fabI mutation, showing that P. aeruginosa FabV catalyzed enoyl-ACP reduction. Alignment of P. aeruginosa FabV with V. cholerae FabV showed that PaFabV contains the Tyr-(Xaa)8-Lys motif first seen in V. cholerae FabV. We mutated PaFabV Tyr-235 to Phe and Lys-244 to Met by site-directed mutagenesis to give three plasmids: pZL20 (PaFabV Y235F), pZL21 (PaFabV K244M), and pZL22 (the Y235F K244M double mutant). The growth of E. coli strain JP1111 carrying the plasmids encoding the mutant proteins was tested at the nonpermissive temperature (Fig. 2A). The strains expressing the Y235F and K244M proteins grew weakly in the presence of arabinose but failed to grow in the absence of arabinose, whereas strain JP1111 expressing the double mutant Y235F K244M protein, failed to grow either in the presence or in the absence of arabinose. These results demonstrated that Tyr-235 and Lys-244 are required for PaFabV activity. The function of P. aeruginosa FabV in E. coli fatty acid synthesis was also assayed by measuring de novo fatty acid synthesis by [1-14C]acetate incorporation into membrane phospholipids (Fig. 2B). Although E. coli JP1111 failed to incorporate [1-14C]acetate into membrane phospholipids at the nonpermissive temperature (Fig. 2B) because of the lack of ENR activity at that temperature, derivatives of strain JP1111 carrying plasmid pZL18 (EcfabI), pZL6 (PafabV), or pZL7 (PafabI) incorporated high levels of [1-14C]acetate into membrane phospholipids (Fig. 2B). Strain JP1111 expressing pZL6 (PaFabV) incorporated 5.5-fold more label into phospholipids than did the strains expressing either EcfabI or PafabI, suggesting that the strain expressing PaFabV encoded a higher level of ENR activity.
FIG. 2.
Expression of P. aeruginosa fabV restores growth and phospholipid synthesis in an E. coli fabI mutant strain. (A) Transformants of strain JP1111 (a temperature-sensitive E. coli fabI mutant) were grown at 30°C or 42°C on RB medium. At 42°C, growth was tested in either the presence or the absence of arabinose (strain JP1111 grows at 30°C but not at 42°C). The strains carried plasmid pZL6, pZL7, pZL18, pZL20, pZL21,or pZL22 encoding PafabV, PafabI, EcfabI, or the Y235F, K244M, or Y235F K244M PafabV mutant, respectively, or the vector plasmid, pBAD24M. (B) Cultures of the E. coli fabI(Ts) strain JP1111 carrying either the pBAD24M, pZL6 (PafabV), pZL7 (PafabI), or pZL18 (EcfabI) plasmid were labeled with [1-14C]acetate at the nonpermissive temperature, followed by extraction of the cellular phospholipids and analysis by TLC. An autoradiogram of the TLC plate is shown. Lane 1, phospholipids from JP1111 carrying vector plasmid pBAD24M; lanes 2 to 4, phospholipids from JP1111 carrying plasmid pZL18, pZL6, or pZL7, respectively. The levels of radioactivity incorporated relative to that in the strain carrying plasmid pZL18 (EcfabI) were as follows: 0.25 for plasmid pBAD24M, 6.48 for plasmid pZL6 (PafabV), and 1.05 for plasmid pZL7 (PafabI). The phospholipids are phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL).
We also inserted the PafabV, PafabI, and EcfabI genes into plasmid pBluescript SK(+) (see Materials and Methods) and introduced these plasmids into the wild-type E. coli strains W3110 and MG1655. Strains W3110 and MG1655 were sensitive to triclosan, and expression of the E. coli and P. aeruginosa FabI proteins from the pBluescript SK(+)-derived plasmids increased triclosan resistance; the MIC was shifted from 0.2 μg/ml to 3 to 4 μg/ml (Table 3). In contrast, introduction of the plasmid carrying PafabV into strains W3110 and MG1655 shifted the MIC of triclosan to a concentration greater than the solubility of triclosan in the growth medium (2,000 μg/ml) (Table 3), indicating that P. aeruginosa FabV, like V. cholerae FabV, is not inhibited by triclosan.
TABLE 3.
Triclosan resistance of E. coli and P. aeruginosa strains
| Strain | Clone | Triclosan MIC (μg/ml)a |
|---|---|---|
| E. coli | ||
| W3110/pBluescript | Empty vector | 0.2 |
| W3110/pZL19 | EcfabI | 3 |
| W3110/pZL17 | PafabI | 4 |
| W3110/pZL16 | PafabV | >2,000 |
| MG1655/pBluescript | Empty vector | 0.2 |
| MG1655/pZL19 | EcfabI | 3 |
| MG1655/pZL17 | PafabI | 4 |
| MG1655/pZL16 | PafabV | >2,000 |
| P. aeruginosa | ||
| PAO1 | None | >2,000 |
| PAO170 (fabV::Gm) | None | 1 |
| PAO272 (fabI::Gm) | None | >2,000 |
Triclosan-containing LB agar plates were prepared with serial dilutions of triclosan. Cells were plated, and the plates were incubated at 37°C. The MIC is defined as the concentration of triclosan in the first plate on which there was no growth.
In vitro characterization of the P. aeruginosa ENRs.
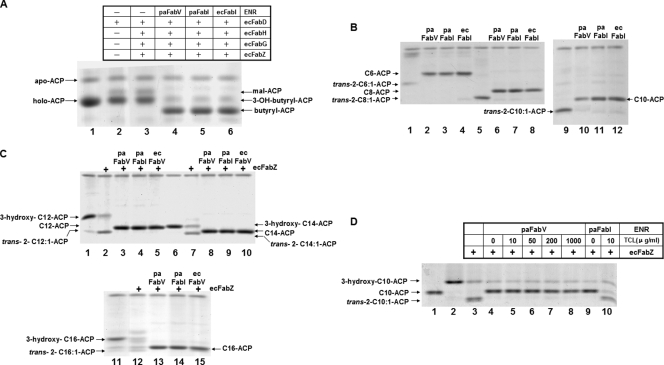
In order to facilitate the purification of FabV and FabI so as to allow direct in vitro assays of their ENR activities, the proteins were expressed in E. coli. SDS-gel electrophoresis showed that all of these proteins were expressed well and had molecular weights in good agreement with those expected for the His6-tagged proteins (data not shown). The N-terminally His6 tagged versions of P. aeruginosa FabV and FabI were purified by nickel chelate chromatography to preparations that each gave a single band upon SDS-gel electrophoresis (data not shown). The N-terminally His6 tagged versions of the E. coli fatty acid biosynthetic proteins FabD, FabG, FabZ, and FabI, plus the V. harveyi acyl-ACP synthetase (AasS), were also purified by nickel chelate chromatography (data not shown). The P. aeruginosa holo-ACP protein was also expressed in E. coli and purified (see Materials and Methods). To test the functions of PaFabV and PaFabI, the initiation steps of the fatty acid synthesis reaction were reconstituted by the sequential addition of purified components, followed by analysis by conformationally sensitive gel electrophoresis. The addition of malonyl-CoA:ACP transacylase (EcFabD) plus malonyl-CoA led to the formation of malonyl-ACP (Fig. 3A). The subsequent additions of 3-ketoacyl-ACP synthase III (EcFabH), 3-ketoacyl-ACP reductase (EcFabG), and 3-hydroxyacyl-ACP dehydrase (EcFabZ) should result in the accumulation of 3-hydroxybutyryl-ACP, since the EcFabZ reaction rapidly reaches an equilibrium in favor of the 3-hydroxyacyl-ACP species (1, 16) (Fig. 3A), such that crotonyl-ACP was not seen. The addition of PaFabV, PaFabI, or EcFabI plus NADH to the reaction mixture yielded butyryl-ACP (Fig. 3A). These data showed that PaFabV, as well as PaFabI and EcFabI, could pull the elongation cycle such that 3-hydroxybutyryl-ACP was converted to butyryl-ACP.
FIG. 3.
Enzymatic characterization of P. aeruginosa FabV. (A) The first cycle of fatty acid biosynthesis was reconstructed in vitro by the sequential addition of each purified enzyme to a reaction mixture containing NADPH, NADH, and PaACP as cofactors plus acetyl-CoA and malonyl-CoA as substrates as described in Materials and Methods. (B) The reaction mixture for assays of the reduction of trans-2-enoyl-ACPs with acyl chain lengths between 6 and 10 carbons by PaFabV or PaFabI contained 0.1 M sodium phosphate (pH 7.0), 1 mM β-mercaptoethanol, and 50 μM acyl-ACP (trans-2-hexenoyl-ACP [lane 1], trans-2-octenoyl-ACP [lane 5], or trans-2-decenoyl-ACP [lane 9]). (C) For assays of the reduction of various trans-2-enoyl-ACPs (lane 2, trans-2-C12:1-ACP; lane 7, trans-2-C14:1-ACP; lane 12, trans-2-C16:1-ACP) with chain lengths between 12 and 16 carbons, the reaction mixtures contained EcFabZ and the appropriate 3-hydroxyacyl-ACP (lane 1, 3-hydroxy-C12-ACP; lane 6, 3-hydroxy-C14-ACP; lane 11, 3-hydroxy-C16-ACP). Acyl-ACPs were synthesized as described in Materials and Methods. (D) Effect of triclosan on the activities of FabV or FabI. The appropriate amounts of triclosan solution were added to 1.5-ml tubes, and the solvent was evaporated. The reaction mixture contained EcFabZ to convert 3-hydroxydecanoyl-ACP to trans-2-decenoyl-ACP. The reactions were initiated by the addition of PaFabV or PaFabI. The reaction mixtures were incubated for an additional 1 h. The reaction products were resolved by conformationally sensitive gel electrophoresis on 17% polyacrylamide gels containing concentrations of urea optimized to effect the separation (34).
We also directly investigated the reduction of long-chain enoyl-ACPs by PaFabV. First, trans-2-hexenoyl-ACP, trans-2-octenoyl-ACP, and trans-2-decenoyl-ACP were synthesized from the acids and P. aeruginosa holo-ACP using V. harveyi acyl-ACP synthetase; then PaFabV and NADH were added to the reaction mixtures. The synthesis of hexanoyl-ACP, octanoyl-ACP, and decanoyl-ACP products was demonstrated by conformationally sensitive gel electrophoresis in the presence of various concentrations of urea (Fig. 3B). For the C12 to C16 enoyl-ACPs, the 2-enoic acids were not available, so we used the 3-hydroxyacids to generate the 3-hydroxyacyl-ACPs. E. coli FabZ-catalyzed dehydration of the 3-hydroxyacyl-ACPs generated the trans-2-enoyl-ACP derivatives, which were used as PaFabV substrates. As expected from the in vivo results, PaFabV quantitatively converted each of the trans-2-enoyl-ACPs to the corresponding acyl-ACP (Fig. 3C). The diversity of acyl-ACP substrates that could be generated allowed us to examine the acyl chain length specificity of both P. aeruginosa ENRs (data not shown). Both ENRs were active on all substrates tested (C4 to C16). FabV was about 10-fold more active than FabI. FabV activity increased with the chain length but differed only about 3-fold over the range of chain lengths tested, with a maximum at C12, whereas FabI did not show this dropoff in activity at the longer chain lengths. We also assayed the activities of the F235T, M244K, and double mutant FabV proteins. The Y235F, K244M, or double mutant (Y235F K244M) FabV protein retained 17%, 1.7%, or 0.9%, respectively, of the activity of wild-type FabV with trans-2-decenoyl-ACP as the substrate, results consistent with the in vivo results.
In vivo experiments indicated that PaFabV was insensitive to triclosan (Table 3). To test this directly, 3-hydroxydecanoyl-ACP was generated and then converted to trans-2-decenoyl-ACP by incubation with EcFabZ. The reaction mixture was then divided into tubes containing various amounts of triclosan, and after the addition of PaFabV, the reaction products were assayed by conformationally sensitive gel electrophoresis in the presence of 2.5 M urea. In agreement with the in vivo results, PaFabV remained active in the conversion of trans-2-decenoyl-ACP to decanoyl-ACP in the presence of 1,000 μg/ml triclosan, whereas 10 μg/ml triclosan almost completely eliminated PaFabI activity (Fig. 3D).
The kinetic properties of FabV and FabI were also examined (Table 4). The experimentally observed maximal velocities of FabV with crotonyl-CoA and trans-2-decenoyl-ACP were higher than the maximal velocities of FabI with the same substrates, although FabI had lower Km values for these substrates than FabV (Table 4).
TABLE 4.
Kinetic parameters of P. aeruginosa FabV and FabI
| Substrate |
Km (μM)a |
Vmax (nmol min−1 mg−1) |
||
|---|---|---|---|---|
| FabV | FabI | FabV | FabI | |
| Crotonyl-CoA | 704.3 | 4.7 | 7.9 | 1.5 |
| trans-2-Decenoyl-ACP | 691.0 | 20.6 | 243.3 | 3.1 |
Determined at an NADH concentration of 100 μM. The concentrations of the hexahistidine-tagged enzymes were 3.57 pM for FabV and 71.4 pM for FabI.
Properties of fabV and fabI deletion mutants.
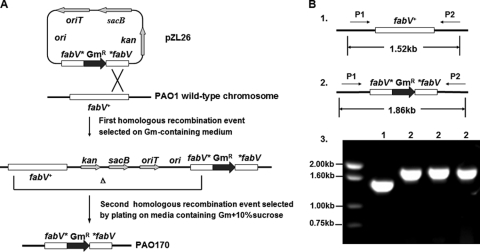
The results described above indicated that P. aeruginosa FabV was refractory toward triclosan. Moreover, expression of PaFabV in E. coli rendered the host bacterium resistant to high levels of triclosan. The remaining question was whether or not P. aeruginosa FabV was responsible for the triclosan resistance of the parent organism. This is particularly important because active efflux of the compound has been reported to be responsible for the triclosan resistance of P. aeruginosa PAO1 (see Discussion). The physiological functions of the two P. aeruginosa ENRs and their roles in triclosan resistance were examined by deleting each of the genes and determining the effects of the gene disruptions on cell growth and triclosan sensitivity. Strains carrying insertion/deletion mutants of either fabV or fabI were constructed by replacement of most the coding sequence of the gene by a gentamicin resistance cassette (Fig. 4A). The disruption alleles were assembled by overlapping PCR, followed by insertion into the small mobilizable vector pK18mobsacB (37). These plasmids were then transformed into E. coli S17-1 (which encodes the mobilization functions required for RP4-mediated conjugation), and the plasmid was transferred into P. aeruginosa PAO1 by conjugation. Single-crossover integrants into the strain PAO1 chromosome were selected by gentamicin resistance. Cultures grown from the integrant colonies were then plated onto a medium containing sucrose and gentamicin in order to select for loss of the vector sequences. Successful construction of the designed mutations was assayed by colony PCR analysis using the primers listed in Table 2. As expected for the fabV mutant, the primers (P1 and P2) amplified a 1.52-kb fabV-containing fragment from wild-type PAO1 DNA. In strain PAO170, this fragment was increased to 1.86 kb by insertion of the Gmr cassette plus deletion of fabV sequences (Fig. 4B). The strains were further verified by sequencing of the 1.86-kb fragment, which demonstrated the expected construction. A ΔfabI strain was made and verified by exactly the same approaches (data not shown).
FIG. 4.
Construction and characterization of the ΔfabV mutation. (A) Recombination effects required to generate the mutant strain by sacB counterselection gene replacement. (B) Characterization of the mutation by PCR. Part 1 shows the map of the wild-type allele, whereas part 2 shows the map of the fabV::Gmr insertion mutant and part 3 shows the PCR analysis of genomic DNA from the two strains. The PCRs were primed with primers P1 and P2. Abbreviations: kan, kanamycin-resistant gene; Gm, Gmr marker; ori, pMB1 origin of replication; oriT, origin of transfer; sacB, levansucrase-encoding gene.
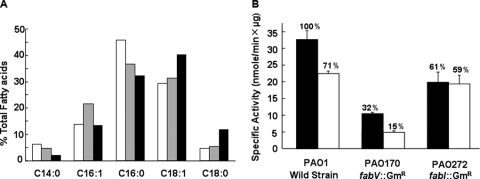
First, the effects of deletion of fabV or fabI on the fatty acid composition of P. aeruginosa were determined by GC-MS (see Materials and Methods) (Fig. 5A). The fabV and fabI deletion strains, PAO170 and PAO272, produced significantly more unsaturated fatty acids than wild-type strain PAO1. The ratio of unsaturated fatty acids to saturated fatty acids in strain PAO1 was 0.76, whereas in the fabV or fabI deletion strain this ratio was increased to 1.12 or 1.16, respectively. However, in the fabV deletion strain, the major unsaturated fatty acid was the C16:1 species, whereas the C18:1 species was the major unsaturated fatty acid in the fabI deletion strain. These findings indicate that the deletion of fabV or fabI affected P. aeruginosa fatty acid biosynthesis.
FIG. 5.
Phospholipid fatty acid compositions and ENR activities of wild-type PAO1, the fabV mutant PAO170, and the fabI mutant PAO272. (A) The fatty acid compositions were obtained from cultures grown in LB medium at 37°C to an optical density at 600 nm of 0.8. The total lipids were extracted and transesterified to yield fatty acid methyl esters (see Materials and Methods), which were analyzed by gas chromatography-mass spectrometry. The methyl esters were C14:0 (tetradecanoic), C16:1 (hexadecenoic), C16:0 (hexadecanoic), C18:1 (octadecenoic), and C18:0 (octadecanoic). Open bars, PAO1; shaded bars, PAO170; filled bars, PAO272. (B) The 70-μl ENR reaction mixtures contained 0.1 M sodium phosphate buffer (pH 7.5), 0.1 mM NADH, and 2 μg of cell extract for determination of NADH-dependent ENR activities. The reactions were initiated by addition of 100 μM trans-2-decenoyl-ACP. Filled bars, no addition of triclosan; open bars, reaction mixtures containing 15 μg/ml triclosan. The results shown are means plus standard deviations for three experiments.
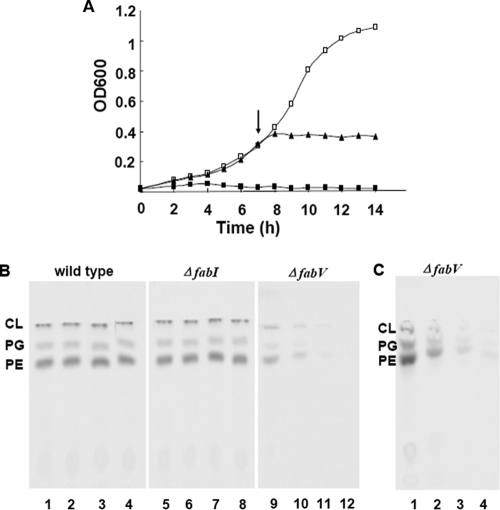
Hoang and Schweizer (21) reported that the major ENR of P. aeruginosa PAO1 was not FabI but rather a triclosan-resistant species encoded by an unknown gene. Determination of ENR activities in cell extracts of the wild type and two mutant strains indicated that FabV was the major enoyl-ACP reductase of P. aeruginosa and that FabI constituted only 32% of the total activity (Fig. 5B), results consistent with the data of Hoang and Schweizer (21). When 15 μg/ml triclosan was added to the reaction mixture, the total ENR activity of the extract of the wild-type strain PAO1 decreased by 29%, whereas the residual reductase activity of the ΔfabV extract decreased to 15% of the untreated wild-type levels upon triclosan addition. In contrast, the ENR activity of the ΔfabI extract was not affected by triclosan. In defined minimal medium, the ΔfabV mutant was viable at 37°C, although its generation time (∼118 min) was significantly longer than that of the wild-type strain (∼40 min) (Fig. 6A). The generation time of the ΔfabI mutant was about 44 min, only marginally less than that of the wild-type strain. We next examined the sensitivity of the two strains to triclosan. The MIC of triclosan for the ΔfabI strain was the same (above 2,000 μg/ml) as that for the wild-type strain PAO1, and [1-14C]acetate incorporation into the phospholipids of these two strains was essentially unaffected by the presence of triclosan (Fig. 6B). In contrast, the ΔfabV strain was extremely sensitive to triclosan, with a MIC of 1 μg/ml, and triclosan efficiently and rapidly blocked [1-14C]acetate incorporation into its phospholipid (Fig. 6C). These data demonstrate that FabV is responsible for the resistance of P. aeruginosa to triclosan.
FIG. 6.
Inhibition of growth and phospholipid synthesis of P. aeruginosa mutants by triclosan. (A) Immediate growth inhibition of the P. aeruginosa fabV mutant PAO170 by triclosan. Cultures were grown in LB medium overnight at 37°C and were subcultured 1:100 into fresh medium at 37°C. The optical density at 600 nm (OD600) was measured at intervals, and triclosan was added to 5 μg/ml at the time indicated by the arrow. Symbols: □, no triclosan added; ▴, triclosan added when the OD600 of cells reached 0.4; ▪, triclosan added at time zero. (B) Effects of triclosan on the phospholipid synthesis of P. aeruginosa strains. Cultures of the P. aeruginosa wild-type strain PAO1, the ΔfabI mutant strain PAO272, and the ΔfabV mutant strain PAO170 were labeled with [1-14C]acetate, followed by extraction of the cellular phospholipids and analysis by TLC. Lanes 1 to 4, phospholipids extracted from the wild-type strain PAO1; lane 5 to 8, phospholipids extracted from the ΔfabI strain PAO272; lanes 9 to 12, phospholipids extracted from the ΔfabV strain PAO170. In each set of lanes, the first lane contains the culture without triclosan addition; the second contains the culture labeled immediately following triclosan addition; the third, the culture labeled after exposure to triclosan for 30 min; the last, the culture labeled after exposure to triclosan for 1 h. The triclosan concentration was 10 μg/ml. The wild-type and ΔfabI strains incorporated [1-14C]acetate at the same rate in the first three lanes and at 86 to 88% of the original level in the last lanes of their respective panels, whereas the untreated ΔfabV strain incorporated only 49.7% of the radioactivity incorporated by the untreated wild-type strain PAO1, and this level progressively declined with the duration of triclosan treatment to 2.5% of the level in the untreated wild-type strain after 1 h of treatment. (C) Inhibition of the incorporation of [1-14C]acetate into the phospholipids of the P. aeruginosa ΔfabV strain by different concentrations of triclosan after exposure to the compound for 30 min. Lane 1, no triclosan; lane 2, 1 μg/ml triclosan; lane 3, 2 μg/ml triclosan, lane 4, 3 μg/ml triclosan. The labeling period was 1 h, and the experiments were initiated when the OD600 of the cultures reached 0.4. In lanes 2, 3, and 4, the levels of [1-14C]acetate incorporated into phospholipids were 43%, 15%, and 6%, respectively, of the level for the untreated control (lane 1).
DISCUSSION
Hoang and Schweizer (21) showed that a P. aeruginosa PAO1 strain carrying a fabI deletion mutation grew normally and retained about 60% of the normal ENR activity in cell extracts. They therefore concluded that the P. aeruginosa PAO1 genome encodes two ENR isoforms. Moreover, unlike that of FabI, the residual activity present in the extracts of the ΔfabI strain was insensitive to triclosan. Following the discovery of FabK in S. pneumoniae (17, 27), Heath and Rock (17) suggested that the non-FabI ENR activity might be encoded by one of the FabK homologues found in the P. aeruginosa PAO1 genome. We have shown that this is not the case. None of the three proteins with similarity to FabK had ENR activity as assayed both in vivo and in vitro (data not shown). Instead, we have shown that the triclosan-insensitive activity of Hoang and Schweizer (21) is due to an ENR that very closely resembles those of V. cholerae FabV, a recently discovered ENR of the SDR family (29). The ENR activity of P. aeruginosa FabV and its resistance to triclosan were demonstrated both in vivo and in vitro.
A ΔfabV derivative of P. aeruginosa PAO1 was constructed, and for purposes of comparison, a ΔfabI strain similar to that made by Hoang and Schweizer (21) was also constructed. The ΔfabI strain grew almost normally and retained about 60% of the wild type ENR activity in cell extracts, and all of the residual ENR activity was insensitive to triclosan inhibition, in excellent agreement with the prior report (21). Moreover, the ΔfabI strain was as resistant to triclosan as the wild-type strain. In contrast, the ΔfabV derivative grew about 3-fold more slowly than the wild-type strain and was exquisitely (1 μg/ml) sensitive to triclosan. Indeed, the ΔfabV strain was more than 2,000-fold more sensitive than the wild-type strain and displayed the lowest triclosan MIC of any genetically defined P. aeruginosa PAO1 mutant strain. These data indicate that FabV plays the major role in the inherent triclosan resistance of this bacterium.
Other mechanisms have been proposed to be responsible for the triclosan resistance of P. aeruginosa PAO1. It has been reported that inactivation of the MexAB-OprM multidrug efflux pump results in a decrease of P. aeruginosa PAO1 triclosan resistance from 1,000 μg/ml to 16 μg/ml (4, 5). However, other investigators have reported that inactivation of the MexAB-OprM efflux pump had either no effect (4, 10) or a minimal effect (10) on triclosan resistance unless the outer membrane was permeabilized by chemical agents (10). The differences among the MexAB-OprM reports may be due to the fact that the workers who reported the dramatic effects of MexAB-OprM inactivation (5) solubilized triclosan with 2-methoxyethanol, and this solvent may have permeabilized the outer membrane. However, even given efflux pump activation and a permeabilized outer membrane (10), the triclosan MIC was still 8-fold higher than that seen for the ΔfabV strain. Moreover, the effects on permeabilized cells may be off-target effects of triclosan, since the compound binds to phospholipids (13), and this binding could nonspecifically exacerbate the effects of permeabilization agents.
The increased unsaturated fatty acid contents seen in the ΔfabV and ΔfabI strains were unexpected but may reflect competition for trans-2-decenoyl-ACP between the ENRs and the FabA 3-hydroxydecanoyl-ACP dehydratase/isomerase. FabA dehydrates 3-hydroxydecanoyl-ACP to trans-2-decenoyl-ACP and then goes on to isomerize this product to cis-3-decenoyl-ACP, the key intermediate of unsaturated fatty acid synthesis. However, FabA does not bind trans-2-decenoyl-ACP tightly (6, 12), and thus, a portion of the FabA dehydration product could dissociate, be captured by FabI or FabV, and thereby become shunted off to the saturated fatty acid synthetic branch. In this scenario, the decreased ENR levels of the mutant strains would increase the level of unsaturated fatty acid synthesis by giving FabA a better chance to rebind trans-2-decenoyl-ACP and isomerize it to cis-3-decenoyl-ACP.
It should be noted that our inability to demonstrate ENR activity for the putative P. aeruginosa PAO1 FabK homologues is not unprecedented. Several proteins that aligned well with S. pneumoniae FabK also lack ENR activity (18, 27) (H. Wang and J. E. Cronan, unpublished data). Hence, simple alignments are not effective at detecting FabK homologues. Other criteria, such as genomic context, are needed. Indeed, S. pneumoniae FabK was discovered by its location within a cluster of fatty acid synthetic genes plus the lack of a FabI ENR encoded in the genome (17, 26).
Acknowledgments
This work was supported by the President Foundation of the South China Agricultural University and by grants from The National Natural Science Foundation of China (30870036/C010201) and the National Institute of Allergy and Infectious Diseases (AI15650).
Footnotes
Published ahead of print on 23 November 2009.
REFERENCES
- 1.Baker, M. E. 1995. Enoyl-acyl-carrier-protein reductase and Mycobacterium tuberculosis InhA do not conserve the Tyr-Xaa-Xaa-Xaa-Lys motif in mammalian 11 b-and 17 b-hydroxysteroid dehydrogenases and Drosophila alcohol dehydrogenase. Biochem. J. 309:1029-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergler, H., G. Hogenauer, and F. Turnowsky. 1992. Sequences of the envM gene and of two mutated alleles in Escherichia coli. J. Gen. Microbiol. 138:2093-2100. [DOI] [PubMed] [Google Scholar]
- 3.Bergler, H., P. Wallner, A. Ebeling, B. Leitinger, S. Fuchsbichler, H. Aschauer, G. Kollenz, G. Hogenauer, and F. Turnowsky. 1994. Protein EnvM is the NADH-dependent enoyl-ACP reductase (FabI) of Escherichia coli. J. Biol. Chem. 269:5493-5496. [PubMed] [Google Scholar]
- 4.Champlin, F. R., M. L. Ellison, J. W. Bullard, and R. S. Conrad. 2005. Effect of outer membrane permeabilisation on intrinsic resistance to low triclosan levels in Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 26:159-164. [DOI] [PubMed] [Google Scholar]
- 5.Chuanchuen, R., R. R. Karkhoff-Schweizer, and H. P. Schweizer. 2003. High-level triclosan resistance in Pseudomonas aeruginosa is solely a result of efflux. Am. J. Infect. Control 31:124-127. [DOI] [PubMed] [Google Scholar]
- 6.Clark, D. P., D. DeMendoza, M. L. Polacco, and J. E. Cronan, Jr. 1983. β-Hydroxydecanoyl thio ester dehydrase does not catalyze a rate-limiting step in Escherichia coli unsaturated fatty acid synthesis. Biochemistry 22:5897-5902. [DOI] [PubMed] [Google Scholar]
- 7.Dennis, J. J., and G. J. Zylstra. 1998. Improved antibiotic-resistance cassettes through restriction site elimination using Pfu DNA polymerase PCR. Biotechniques 25:772-774. [DOI] [PubMed] [Google Scholar]
- 8.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, Y. H., L. Y. Wang, and L. H. Zhang. 2007. Quorum-quenching microbial infections: mechanisms and implications. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362:1201-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellison, M. L., A. L. Roberts, and F. R. Champlin. 2007. Susceptibility of compound 48/80-sensitized Pseudomonas aeruginosa to the hydrophobic biocide triclosan. FEMS Microbiol. Lett. 269:295-300. [DOI] [PubMed] [Google Scholar]
- 11.Grogan, D. W., and J. E. Cronan, Jr. 1997. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol. Mol. Biol. Rev. 61:429-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerra, D. J., and J. A. Browse. 1990. Escherichia coli β-hydroxydecanoyl thioester dehydrase reacts with native C10 acyl-acyl-carrier proteins of plant and bacterial origin. Arch. Biochem. Biophys. 280:336-345. [DOI] [PubMed] [Google Scholar]
- 13.Guillén, J., A. Bernabeu, S. Shapiro, and J. Villalain. 2004. Location and orientation of triclosan in phospholipid model membranes. Eur. Biophys. J. 33:448-453. [DOI] [PubMed] [Google Scholar]
- 14.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heath, R. J., and C. O. Rock. 1995. Enoyl-acyl carrier protein reductase (fabI) plays a determinant role in completing cycles of fatty acid elongation in Escherichia coli. J. Biol. Chem. 270:26538-26542. [DOI] [PubMed] [Google Scholar]
- 16.Heath, R. J., and C. O. Rock. 1996. Roles of the FabA and FabZ β-hydroxyacyl-acyl carrier protein dehydratases in Escherichia coli fatty acid biosynthesis. J. Biol. Chem. 271:27795-27801. [DOI] [PubMed] [Google Scholar]
- 17.Heath, R. J., and C. O. Rock. 2000. A triclosan-resistant bacterial enzyme. Nature 406:145-146. [DOI] [PubMed] [Google Scholar]
- 18.Heath, R. J., N. Su, C. K. Murphy, and C. O. Rock. 2000. The enoyl-[acyl-carrier-protein] reductases FabI and FabL from Bacillus subtilis. J. Biol. Chem. 275:40128-40133. [DOI] [PubMed] [Google Scholar]
- 19.Heath, R. J., S. W. White, and C. O. Rock. 2002. Inhibitors of fatty acid synthesis as antimicrobial chemotherapeutics. Appl. Microbiol. Biotechnol. 58:695-703. [DOI] [PubMed] [Google Scholar]
- 20.Heath, R. J., S. W. White, and C. O. Rock. 2001. Lipid biosynthesis as a target for antibacterial agents. Prog. Lipid Res. 40:467-497. [DOI] [PubMed] [Google Scholar]
- 21.Hoang, T. T., and H. P. Schweizer. 1999. Characterization of Pseudomonas aeruginosa enoyl-acyl carrier protein reductase (FabI): a target for the antimicrobial triclosan and its role in acylated homoserine lactone synthesis. J. Bacteriol. 181:5489-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoang, T. T., and H. P. Schweizer. 1997. Fatty acid biosynthesis in Pseudomonas aeruginosa: cloning and characterization of the fabAB operon encoding β-hydroxyacyl-acyl carrier protein dehydratase (FabA) and β-ketoacyl-acyl carrier protein synthase I (FabB). J. Bacteriol. 179:5326-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang, Y., C. H. Chan, and J. E. Cronan. 2006. The soluble acyl-acyl carrier protein synthetase of Vibrio harveyi B392 is a member of the medium chain acyl-CoA synthetase family. Biochemistry 45:10008-10019. [DOI] [PubMed] [Google Scholar]
- 24.Kutchma, A. J., T. T. Hoang, and H. P. Schweizer. 1999. Characterization of a Pseudomonas aeruginosa fatty acid biosynthetic gene cluster: purification of acyl carrier protein (ACP) and malonyl-coenzyme A:ACP transacylase (FabD). J. Bacteriol. 181:5498-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu, Y. J., Y. M. Zhang, and C. O. Rock. 2004. Product diversity and regulation of type II fatty acid synthases. Biochem. Cell Biol. 82:145-155. [DOI] [PubMed] [Google Scholar]
- 26.Marrakchi, H., K. H. Choi, and C. O. Rock. 2002. A new mechanism for anaerobic unsaturated fatty acid formation in Streptococcus pneumoniae. J. Biol. Chem. 277:44809-44816. [DOI] [PubMed] [Google Scholar]
- 27.Marrakchi, H., W. E. Dewolf, Jr., C. Quinn, J. West, B. J. Polizzi, C. Y. So, D. J. Holmes, S. L. Reed, R. J. Heath, D. J. Payne, C. O. Rock, and N. G. Wallis. 2003. Characterization of Streptococcus pneumoniae enoyl-(acyl-carrier protein) reductase (FabK). Biochem. J. 370:1055-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Massengo-Tiassé, R. P., and J. E. Cronan. 2009. Diversity in enoyl-acyl carrier protein reductases. Cell. Mol. Life Sci. 66:1507-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massengo-Tiassé, R. P., and J. E. Cronan. 2008. Vibrio cholerae FabV defines a new class of enoyl-acyl carrier protein reductase. J. Biol. Chem. 283:1308-1316. [DOI] [PubMed] [Google Scholar]
- 30.McMurry, L. M., M. Oethinger, and S. B. Levy. 1998. Triclosan targets lipid synthesis. Nature 394:531-532. [DOI] [PubMed] [Google Scholar]
- 31.Morris, T. W., K. E. Reed, and J. E. Cronan, Jr. 1995. Lipoic acid metabolism in Escherichia coli: the lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein. J. Bacteriol. 177:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Payne, D. J., P. V. Warren, D. J. Holmes, Y. Ji, and J. T. Lonsdale. 2001. Bacterial fatty-acid biosynthesis: a genomics-driven target for antibacterial drug discovery. Drug Discov. Today 6:537-544. [DOI] [PubMed] [Google Scholar]
- 33.Persson, B., Y. Kallberg, U. Oppermann, and H. Jornvall. 2003. Coenzyme-based functional assignments of short-chain dehydrogenases/reductases (SDRs). Chem. Biol. Interact. 143-144:271-278. [DOI] [PubMed] [Google Scholar]
- 34.Post-Beittenmiller, D., J. G. Jaworski, and J. B. Ohlrogge. 1991. In vivo pools of free and acylated acyl carrier proteins in spinach. Evidence for sites of regulation of fatty acid biosynthesis. J. Biol. Chem. 266:1858-1865. [PubMed] [Google Scholar]
- 35.Rock, C. O., J. L. Garwin, and J. E. Cronan, Jr. 1981. Preparative enzymatic synthesis of acyl-acyl carrier protein. Methods Enzymol. 72:397-403. [DOI] [PubMed] [Google Scholar]
- 36.Saito, J., M. Yamada, T. Watanabe, M. Iida, H. Kitagawa, S. Takahata, T. Ozawa, Y. Takeuchi, and F. Ohsawa. 2008. Crystal structure of enoyl-acyl carrier protein reductase (FabK) from Streptococcus pneumoniae reveals the binding mode of an inhibitor. Protein Sci. 17:691-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schäfer, A., A. Tauch, W. Jager, J. Kalinowski, G. Thierbach, and A. Puhler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 38.Thomas, J., and J. E. Cronan. 2005. The enigmatic acyl carrier protein phosphodiesterase of Escherichia coli: genetic and enzymological characterization. J. Biol. Chem. 280:34675-34683. [DOI] [PubMed] [Google Scholar]
- 39.Turnowsky, F., K. Fuchs, C. Jeschek, and G. Hogenauer. 1989. envM genes of Salmonella typhimurium and Escherichia coli. J. Bacteriol. 171:6555-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulrich, A. K., D. de Mendoza, J. L. Garwin, and J. E. Cronan, Jr. 1983. Genetic and biochemical analyses of Escherichia coli mutants altered in the temperature-dependent regulation of membrane lipid composition. J. Bacteriol. 154:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, H., and J. E. Cronan. 2003. Haemophilus influenzae Rd lacks a stringently conserved fatty acid biosynthetic enzyme and thermal control of membrane lipid composition. J. Bacteriol. 185:4930-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, H., and J. E. Cronan. 2004. Only one of the two annotated Lactococcus lactis fabG genes encodes a functional β-ketoacyl-acyl carrier protein reductase. Biochemistry 43:11782-11789. [DOI] [PubMed] [Google Scholar]
- 43.White, S. W., J. Zheng, Y. M. Zhang, and C. O. Rock. 2005. The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 74:791-831. [DOI] [PubMed] [Google Scholar]
- 44.Wilkins, P. O. 1982. Effects of growth temperature on transport and membrane viscosity in Streptococcus faecalis. Arch. Microbiol. 132:211-215. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Y. M., S. W. White, and C. O. Rock. 2006. Inhibiting bacterial fatty acid synthesis. J. Biol. Chem. 281:17541-17544. [DOI] [PubMed] [Google Scholar]
- 46.Zhao, X., J. R. Miller, and J. E. Cronan. 2005. The reaction of LipB, the octanoyl-[acyl carrier protein]:protein N-octanoyltransferase of lipoic acid synthesis, proceeds through an acyl-enzyme intermediate. Biochemistry 44:16737-16746. [DOI] [PubMed] [Google Scholar]