Abstract
The mechanism by which entecavir resistance (ETVr) substitutions of hepatitis B virus (HBV) can induce breakthrough (BT) during ETV therapy is largely unknown. We conducted a cross-sectional study of 49 lamivudine (LVD)-refractory patients and 59 naïve patients with chronic hepatitis B. BT was observed in 26.8% of the LVD-refractory group during weeks 60 to 144 of ETV therapy. A line probe assay revealed ETVr substitutions only in the LVD-refractory group, i.e., in 4.9% of patients at baseline, increasing to 14.6%, 24.4%, and 44.8% at weeks 48, 96, and 144, respectively. Multivariate logistic regression analysis adjusted for age, gender, HBV DNA levels, and LVD resistance (LVDr) (L180M and M204V, but not M204I) indicated that T184 substitutions and S202G (not S202C) were a significant factor for BT (adjusted odds ratio [OR], 141.12, and 95% confidence interval [CI], 6.94 to 2,870.20; OR, 201.25, and 95% CI, 11.22 to 3608.65, respectively). Modeling of HBV reverse transcriptase (RT) by docking simulation indicated that a combination of LVDr and ETVr (T184L or S202G) was characterized by a change in the direction of the D205 residue and steric conflict in the binding pocket of ETV triphosphate (ETV-TP), by significantly longer minimal distances (2.2 Å and 2.1 Å), and by higher potential energy (−117 and −99.8 Kcal/mol) for ETV-TP compared with the wild type (1.3 Å; −178 Kcal/mol) and LVDr substitutions (1.5 Å; −141 Kcal/mol). Our data suggest that the low binding affinity of ETV-TP for the HBV RT, involving conformational change of the binding pocket of HBV RT by L180M, M204V plus T184L, and S202G, could induce BT.
Infection with hepatitis B virus (HBV) is extremely widespread and affects more than 350 million people worldwide. Chronic HBV infection leads to the development of complications, such as liver cirrhosis (LC) and hepatocellular carcinoma (HCC) (12). HBV has been classified into 8 geographically, genetically, and clinically diverse genotypes, designated alphabetically from A to H according to their order of discovery (14). Genotypes B and C are prevalent in Asia, and genotype C is associated with more serious liver disease, including LC and HCC, and a poorer response to interferon therapy than genotype B (5). The ultimate therapeutic goal when treating chronic HBV infection is to prevent the development of LC and HCC by eliminating or producing sustained suppression of HBV replication. However, lamivudine resistance (LVDr) was reported to occur in 24% of patients treated for 1 year and in 74% of those treated for 5 years (16, 26). The rate of adefovir resistance (ADVr) in nucleoside-naive hepatitis B e antigen (HBeAg)-negative patients has been reported to be 0% after 1 year, but after 5 years of treatment, the rate increases to 28% to 42% (13). Entecavir (ETV) has been shown to be more potent in vitro than either LVD or ADV. Results from clinical studies showed that the efficacy of ETV was superior to that of the direct comparator, LVD, in both nucleoside-naive and LVD-refractory patients (6, 11, 15, 18).
The persistence of LVDr substitutions in patients switched to ETV is worrisome, because LVDr was shown to enhance the risk of developing ETVr and treatment failure, defined as viral breakthrough (BT) (an increase in serum HBV DNA of at least 1 log10 copy/ml compared with the nadir value as observed during ETV therapy) (20). A recent in vitro study showed that LVDr (L180M and M204V) substitutions confer an ∼8-fold reduction in susceptibility to ETV and that additional substitutions at residues T184, S202, and M250 are needed to confer high levels of ETVr and BT (2, 3).
These analyses, however, used a limited number of patient isolates and/or laboratory HBV clones, and there has been a paucity of community-based data derived from long-term trials regarding the clinical outcomes of ETVr variants in naïve or LVD-refractory patients. Therefore, the aim of this study was to evaluate the incidence of ETVr and BT by comparing outcomes following 3-year ETV treatment in treatment-naïve patients and LVD-refractory patients. ETVr was assessed by using a recently reported line probe assay (HBV DR v.3) (7). Importantly, as the mechanism by which ETVr substitutions can induce BT during ETV therapy is largely unknown, changes in the conformation of HBV reverse transcriptase (RT) arising from LVDr and ETVr substitutions were modeled by using 3-dimensional (3D) docking simulation.
MATERIALS AND METHODS
Study design.
We conducted a cross-sectional study of 100 patients (Tables 1 and 2), 45 of whom were from Japan, 25 from the United States, and 30 from Hong Kong. The patients were subdivided into two groups; treatment-naïve (n = 59) and LVD-refractory (n = 41) patients, whose gender, age, HBeAg status, and mean HBV DNA levels are summarized in Table 1. The patients received 0.5 mg or 1.0 mg ETV. The 1.0-mg ETV once-daily (QD) dosage has been approved for use in LVD-refractory patients, and only patients treated with 1.0 mg per day were included in resistance assessments. The study protocol conformed to the 1975 Declaration of Helsinki and was approved by the Ethics Committees of the institutions, and written informed consent was obtained from each participant.
TABLE 1.
Patient characteristics of naïve and LVD-refractory patients
| Characteristic | Value |
|
|---|---|---|
| Naïve (n = 59)a | LVD refractory (n = 41)a | |
| Male/female no. | 41/18 | 34/7 |
| Mean age (yr) | 46.5 ± 8.4 | 48.6 ± 8.3 |
| HBeAg positive (%) | 33 (55.9) | 23 (56.1) |
| Mean ALT (U/liter) | 118.9 ± 108.6 | 119.8 ± 99.0 |
| Mean HBV DNA (log10 copies/ml) | 6.7 ± 1.8 | 6.8 ± 1.0 |
| Genotypes (no. A/B/C/D/E) | 3/11/43/1/1 | 5/7/28/1/0 |
Values are means ± standard deviations.
TABLE 2.
Three-year assessment (HBeAg loss, ALT normalization, and HBV-DNA 2.6) of naïve and LVD-refractory patients
| Parameter | Value for follow-up week: |
|||||
|---|---|---|---|---|---|---|
| Naïve (n = 59)a |
LVD refractory (n = 41)a |
|||||
| 48 | 96 | 144 | 48 | 96 | 144 | |
| Follow-up [n (%)] | 59 (100) | 39 (66.1) | 38 (64.4)b | 41 (100) | 40 (97.6)c | 26 (63.4)d |
| HBeAg loss [n (%)] | 5 (15.2) | 7 (24.1) | 9 (32.1) | 4 (17.4) | 7 (31.8) | 4 (22.2) |
| ALT normalization [n (%)] | 24 (40.7) | 25 (64.1) | 27 (71.1) | 16 (39.0) | 20 (50.0) | 13 (50.0) |
| HBV DNA loss [n (%)] | 24 (40.7) | 28 (71.8) | 28 (73.7) | 15 (36.6) | 19 (47.5) | 12 (46.2) |
Values are means ± standard deviations.
One naïve patient (J44) stopped ETV therapy at week 80 (ALT, 119 U/liter, and HBV DNA, 7.6 log10 copies/ml at baseline; ALT, 17, and HBV DNA, <2.6 at week 80) due to severe headache during therapy. Twenty patients in Hong Kong stopped ETV therapy between weeks 48 and 72.
One LVD-refractory patient (J37) switched from ETV therapy to LVD plus adefovir due to BTH with ETVr before week 96.
Two patients (J33 and J40) switched from ETV therapy to LVD plus adefovir due to BTH with ETVr before week 144. Twelve patients in the United States were treated with ETV for <120 weeks.
Screening for drug-resistant substitutions.
Simultaneous detection of wild-type HBV and drug-induced substitutions was performed using HBV DR v.3 and v.2 (Innogenetics, Ghent, Belgium) according to the manufacture's protocol. HBV DR v.3 and v.2 were developed for detection of ETVr-specific substitutions (T184SCGA/ILFM, S202G/C/I, and M250V/I/L), TDFr-specific substitutions (A194T), and newly reported ADVr (I233V) substitutions, as well as LVDr (L80V/I, V/G173L, L180M, and M204V/I) and ADVr (A181T/V and N236T) substitutions. The HBV DR assay consistently detected ETVr-specific substitutions present in ≥5% of the virus population when the HBV DNA concentration was ≥4 log10 copies/ml (7). The AUTOLiPA (Innogenetics, Ghent, Belgium) was used for the automated test procedure. An 867-bp-long fragment of the polymerase gene (domains A to F) was amplified using biotinylated PCR primers (HBV DR v.3 and v.2). PCR products were directly sequenced.
Statistical analyses.
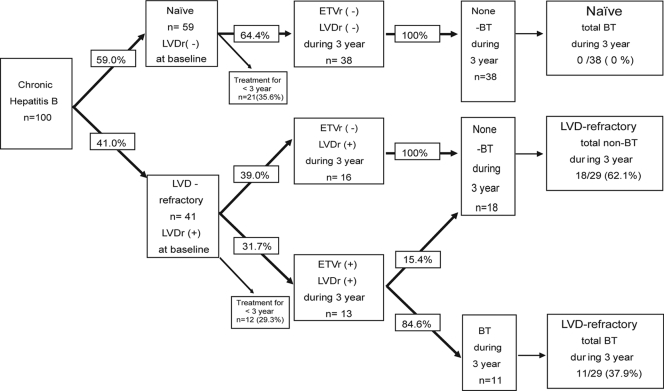
The statistical significance of observed differences was assessed using the chi-square test and the Mann-Whitney U test, where appropriate. In the 67 patients (38 naïve and 29 LVD refractory) with 3 years of ETV treatment (Fig. 1), the logistic regression model was used to assess the factors associated with BT. STATA 10 (Statacorp LP, TX) and the Statistical Program for Social Sciences (SPSS 12.0 for Windows; SPSS Inc., Chicago, IL) were used for all analyses.
FIG. 1.
Flowchart of 100 naïve/LVD-refractory patients during ETV therapy.
HBV polymerase sequencing.
HBV DNA was extracted from serum samples using a Qiagen QIAamp DNA blood minikit (Qiagen GmbH, Germany), and an 867-bp-long fragment of the polymerase gene (domains A to F) was amplified using biotinylated PCR primers (INNO-LiPA). PCR products were directly sequenced. Nucleotide mixtures were reliably detected when they were mixed at a ratio of approximately 25% or greater.
Three-dimensional-structure-based docking simulation methods.
The amino acid sequence of HBV RT was retrieved from GenBank (gene Pol product of accession no. X75665), and the 323rd to 697th residues, which correspond to the finger, palm, and thumb domains, were extracted. The sequence and that of HIV RT, retrieved from the Protein Data Bank (accession no. 1RTD), were aligned using BLASTP (1), and then the resulting alignment was modified manually to obtain a match of the RT-specific motifs in both sequences. The main-chain structure of HBV RT was built from the alignment and the 3D structure of HIV RT (accession no. 1RTD) (8) by the use of the “nest” module (17) in the JACKAL package (19), where global energy minimization was done to find the most stable backbone structure. The loop and secondary-structure regions were then refined (24), after which the side chain structure was refined by the use of the “scap” module in the package (23). The 3D structures of HBV RT containing three sets of substitutions, L180M plus M204V, L180M plus S202G plus M204V, and L180M plus T184L plus M204V, were also designed in the same manner.
The binding site of ETV was searched on the wild-type HBV RT molecule by docking simulation. First, the structure of ETV triphosphate (ETV-TP) was designed by a small-molecule-editing function in the SYBYL 8.0 package (Tripos Inc., St. Louis, MO). Then, the possible binding sites of the ligand were searched from the surface of the protein by the use of the “Surflex-Dock” (9) module in the package. Here, the docking candidate area was restricted to the surfaces of the residues that were within 3 Å from L180, T184, S202, Y203, M204, D205, or D206. The binding potential was estimated from the GOLD score calculated by the “CScore” module (4) in the package. The score was evaluated based on hydrogen bond energy, the internal energy of molecules, and complex energy between ligand and protein. The minimal distance between their molecular surfaces was also calculated.
RESULTS
Clinical efficacy.
The clinical backgrounds and the percentages of LVD-naïve and LVD-refractory patients who achieved HBeAg loss, alanine aminotransferase (ALT) normalization, and non-PCR-detectable HBV DNA levels (<2.6 log10 copies/ml) during the ETV treatment course are summarized in Table 2. There were no significant differences in clinical data at entry between the 2 groups. The rates of HBeAg loss, ALT normalization, and HBV DNA loss were significantly higher in naïve patients than in LVD-refractory patients.
Detection of substitutions responsible for ETV resistance in naive and LVD-refractory patients during treatment for 144 weeks.
The characteristics of patients who had ETVr substitutions detected by HBV DR v.3 are summarized in Table 3. The percentage of the typical LVDr (L180M, M204V, and M204I) or ETVr observed in naïve patients was 0% (0/38) during the 144-week treatment period.
TABLE 3.
Characteristics of ETVr detected by HBV DR v3 among 13 patients at week 0, 24, 48, 72, 96, and 144
| Case | Age (yr) | Sex | HBV genotype | ALT (IU/liter) |
HBV DNA (copies/ml) |
Wk |
No. of wks ETVr to BT | LVDr at baseline | ETVr at baseline | ETVr emerged during therapy | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline | Nadir | Peak | Baseline | Nadir | Peak | ETVr | BT | BTH | ||||||||
| J20 | 34 | M | C | 82 | 22 | 131 | 7.6 | 4.5 | 7.4 | <92a | 92 | 100 | <22 | M180, V204 | NDf | ILMF184, G202 |
| J27 | 61 | M | C | 79 | 38 | 150 | 7.4 | 3.9 | 7 | <128b | 128 | 144 | <32 | M180, V204 | ND | SCGA184 |
| J30 | 43 | M | C | 95 | 21 | 108 | 7.6 | 4.6 | 6.8 | 48 | 76 | 92 | 28 | I80, M180, V/I204 | ND | G202 |
| J33 | 43 | M | C | 69 | 21 | 199 | 7.6 | 5.1 | 7.1 | 96 | 116 | 128 | 18 | I80, L173, M180, VI204 | ND | (ILFM184),c G202 |
| J37d | 48 | M | C | 465 | 43 | 76 | 7.6 | 4.7 | 6 | 48 | 60 | 60 | 12 | I80, M180, V204, I204 | ND | G202 |
| J39 | 59 | M | C | 35 | 22 | 82 | 7 | 4.6 | 8 | 72 | 108 | 128 | 36 | M180, V204 | ND | SCGA/ILFM184, G202 |
| J40 | 28 | M | C | 149 | 15 | 398 | 5.3 | 3.6 | 6.6 | −8 | 64 | 72 | 72 | M180, V204 | (G+C) 202 | G202 |
| J22 | 44 | M | C | 240 | 17 | 24 | 7.6 | 3.5 | 5.6 | <140e | 144 | NO | <44 | M180, I204 | ND | G202 |
| J28 | 45 | M | B | 43 | 15 | 23 | 6.9 | 5.5 | 6.5 | 0 | 144 | NO | 144 | M180, V204 | SCGA184, V233 | SCGA184, V233 |
| U72 | 47 | F | C | 29 | 25 | 44 | 7.5 | 3.8 | 5 | <144 | 144 | NO | <48 | L + V80, M180, V204 | ND | SCGA184, (S+G)202 |
| H55 | 47 | F | C | 102 | 19 | 20 | 6.1 | 3 | 4.2 | 48 | 88 | NO | 40 | V + I80, M180, V204 | ND | ILFM184 |
| J19 | 57 | M | B | 233 | 29 | 43 | 7.4 | 2.8 | 3.3 | 24 | NO | NO | NO | V + 180, M180, I204 | ND | G202 |
| U42 | 52 | M | A | 135 | 36 | 50 | >7.6 | 5.3 | 5.8 | <80 | NO | NO | NO | M180, V204 | ND | SCGA184 |
Not tested during weeks 72 and 92.
Not tested during weeks 96 and 128.
ILFM184 was detected at week 144.
Switch to ADV/LVD at week 60.
Not tested during weeks 96 and 140.
ND, not detected.
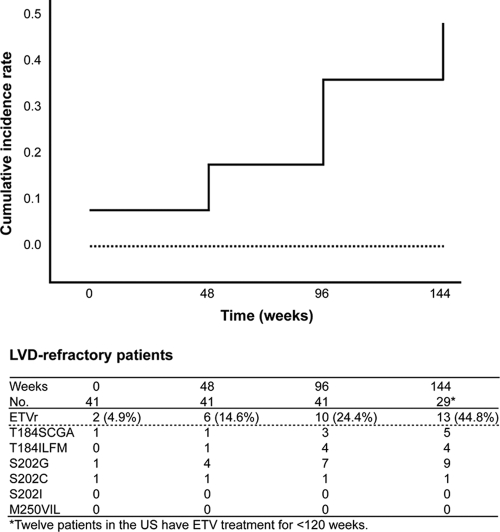
Among the patients examined at entry prior to treatment with ETV, in 41 LVD-refractory patients, M204V (30/41; 72.4%), M204I (24/41; 58.5%), L180M (38/41; 92.7%), L80V (6/41; 14.6%), L80I (18/41; 43.9%), and V173L (4/41; 9.76%) substitutions were detected. In the 41 LVD-refractory patients, the cumulative ETVr substitutions were detected in 2/41 (4.9%) at baseline and increased to 6/41 (14.6%), 10/41 (24.4%), and 13/29 (44.8%) at weeks 48, 96, and 144, respectively (Fig. 2). In the 29 patients treated with ETV for 3 years, T184SCGA, T184ILMF, S202G, and S202C were found in 5 (17.2%), 4 (13.8%), 9 (31.0%), and 1 (3.4%), respectively. Neither S202I nor M250V/I/L substitutions were detected in this population.
FIG. 2.
Kaplan-Meier plot and tabulated data for time to ETVr and cumulative ETVr patterns over 144 weeks. Pretreatment variables (solid line, LVD refractory; broken line, naïve) were analyzed in relation to the occurrence of ETVr. A previous LVD treatment was associated with a more rapid occurrence of ETVr (Breslow analysis; P < 0.001). Among the patients examined at entry prior to treatment with ETV, for the 41 LVD-refractory patients, the cumulative ETVr substitutions were detected in 2/41 (4.9%) at baseline and increased to 6/41 (14.6%), 10/41 (24.4%), and 13/29 (44.8%) at weeks 48, 96, and 144, respectively. Neither the S202I nor the M250V/I/L substitution was detected in this population.
A comparative summary of the ETVr substitutions, detected by HBV DR v.3 and direct sequencing, during week −8 (8 weeks before the start of treatment) and week 144 is presented in Table 4. HBV DR v.3 revealed ETVr substitutions earlier (up to 48 weeks) than did direct sequencing. In addition, HBV DR v.3 allowed the detection of mixed quasispecies containing different substitutions.
TABLE 4.
Detection of ETVr mutations by HBV DR v3 and direct sequencing at weeks 0, 24, 48, 72, 96, and 144
| Case | INNO-LiPA detection |
Direct-sequencing detection |
||
|---|---|---|---|---|
| Wk | ETVr | Wk | ETVr | |
| J20 | 96 | ILFM184 + G202 | 96 | L184 (L184 and G202 detected at wk 144) |
| J27 | 144 | SCGA184 + T184 | 144 | S184 + T184 |
| J30 | 48 | G202 | 96 | G202 (NDb at wk 48) |
| J39 | 72 | SCGA/ILFM184 + G202 | 96 | I184, G202 (ND at wk 72) |
| J40 | −8 | G202 | 0 | G202 (ND at wk −8) |
| J33 | 96 | G202 | 144 | G202 (ND at wk 96) |
| J19 | 24 | G202 + C202 | 48 | G202 (ND at wk 24) |
| J22 | 144 | S202 + G202 | 144 | S202 |
| J28 | 0 | SCGA184, V233 | 0 | A184, V233 |
| J37a | 48 | S202 + G202 | 48 | S202 |
| U42 | 80 | SCGA184 + T184 | 80 | T184 + A184 |
| U72 | 144 | T184 + SCGA184, S202 + G202 | 144 | T184 + A184, S202 |
| H55 | 48 | T184 + ILFM184 | 48 | T184 |
Switched from ETV to LVD plus ADV therapy at week 60.
ND, ETVr was not detected.
Viral BT during the 144 weeks on treatment.
The rates of BT among 59 naive and 41 LVD-refractory patients treated with ETV for 144 weeks are summarized in Fig. 1. There were no cases of BT in the LVD-naïve group during the 144-week treatment period, whereas in the LVD-refractory group treated with 1.0 mg ETV, 11 of 13 patients with genotypic ETVr had evidence of BT after 60 to 144 weeks of treatment, followed by 7 breakthrough hepatitis (BTH) (defined as a flare up of ALT) patients (median interval, 11.4 weeks after BT). The LVDr substitutions (L180M and M204V/I) were detected in all of the BT patients in specimens obtained at baseline (Table 3). Among the 11 patients with BT, 8 (72.7%) had an additional S202G substitution and 7 (63.6%) had a T184SCGA or T184ILMF substitution, indicating that the T184 and/or S202 substitution emerged before BT during ETV treatment (Table 3 and Fig. 2). Seven patients with BTH had LVDr (100% for both M180 and V204) at baseline and ETVr substitutions (S202G, 85.7%; T184SCGA/ILFM, 57.1%) during 3-year ETV treatment. Representative cases with BTH during ETV therapy are shown in the supplemental material.
Pretreatment status (LVD refractory or naïve) was analyzed in relation to the occurrence of ETVr, BT, and BTH during the 144-week course of ETV therapy. The log rank analysis of pretreatment variables showed that prior refractoriness to LVD was associated with more rapid occurrence of ETVr (Fig. 2), BT, and BTH (P ≤ 0.001, P < 0.001, and P = 0.0039, respectively). Additionally, among the 67 patients receiving 3-year ETV treatment, BT occurred in 10 of 52 (19.2%) patients with HBV genotype C and 1 of 8 (12.5%) with genotype B, whereas no BT was observed in patients with genotypes A, D, and E. No significant association between BT and HBV genotypes was found.
Baseline characteristics and factors associated with viral breakthrough during 3-year ETV therapy.
When non-BT and BT groups within the 67 patients treated with ETV for 144 weeks were compared, no significant baseline differences were observed in mean age, gender, serum ALT levels, HBV DNA, or HBeAg status (Table 1), while 2-log10-unit reductions in HBV DNA levels or undetectable (<2.6) HBV DNA levels at the end of year 1 were significantly higher in the non-BT group (Table 5). Interestingly, the proportion of patients refractory to LVD with both the L180M and M204V substitutions at baseline (P < 0.001) and the incidence of S202G or T184SCGA/ILMF substitutions during the 3-year ETV treatment (P < 0.001) were significantly higher in the BT group.
TABLE 5.
ORs and 95% CIs of BT according to baseline characteristics among 67 patients treated with ETV for 3 years
| Characteristic | Non-BT (n = 56)a | BT (n = 11)a | P for difference | Contrast | OR | 95% CI |
|---|---|---|---|---|---|---|
| Age (mean) | 45.4 ± 8.1 | 45.4 ± 9.4 | 0.932 | 1-yr increase | 1 | 0.92-1.08 |
| Male (%) | 71.4 | 81.8 | 0.481 | Male vs. female | 1.8 | 0.35-9.26 |
| ALT (mean) | 115.9 ± 105.6 | 126.2 ± 127.3 | 0.832 | 1-U increase | 1 | 0.99-1.00 |
| HBeAg (%) | 66.1 | 81.8 | 0.307 | Positive vs. negative | 2.31 | 0.45-11.78 |
| HBV-DNA | ||||||
| Level (mean) | 6.8 ± 1.6 | 7.1 ± 0.8 | 0.959 | 1-U increase | 1.16 | 0.72-1.89 |
| 2-log-unit reduction at 1 yr (%) | 98.2 | 63.6 | <0.001 | With vs. without | 0.03 | 0.00-0.33 |
| DNA <2.6 at 1 yr (%) | 58.9 | 0.0 | <0.001 | With vs. without | NAb | |
| LVD refractory (%) | 32.1 | 100.0 | <0.001 | With vs. without | NA | |
| Amino acid substitutions at baseline | ||||||
| V80 (%) | 3.6 | 18.2 | 0.063 | With vs. without | 6 | 0.74-48.17 |
| I80 (%) | 17.9 | 36.4 | 0.171 | With vs. without | 2.63 | 0.64-10.72 |
| L173 (%) | 3.6 | 18.2 | 0.063 | With vs. without | 6 | 0.75-48.18 |
| M180 (%) | 28.6 | 100.0 | <0.001 | With vs. without | NA | |
| V204 (%) | 19.6 | 90.9 | <0.001 | With vs. without | 10.91 | 4.72-354.28 |
| I204 (%) | 23.2 | 36.4 | 0.363 | With vs. without | 1.89 | 0.48-7.49 |
| Amino acid substitutions during ETV therapy (3 yr) | ||||||
| SCGA/ILFM184 (%) | 1.8 | 72.7 | <0.001 | With vs. without | 96.25 | 9.38-987.41 |
| G202 (%) | 1.8 | 63.6 | <0.001 | With vs. without | 146.67 | 13.55-1,587.24 |
Values are means ± standard deviations.
NA, not applicable.
None of the BT cases reached undetectable HBV DNA levels at the end of the first year of ETV treatment (BT, 0%, versus non-BT, 58.9%), but all were refractory to LVD (BT, 100%, versus non-BT, 32.1%) and had the L180M substitution at baseline (BT, 100%, versus non-BT, 28.6%) (Table 5). For other factors, additional analysis showed that ETVr substitutions (i.e., S202G, T184SCGA/ILFM, and M204V) were strongly associated with BT during the 3-year ETV treatment (OR, 146.67 [95% CI, 13.55 to 1,587.24], 96.25 [95% CI, 9.38 to 987.41], and 10.91 [95% CI, 4.72 to 354.28]) (Table 5).
After adjustment for age, gender, baseline HBV DNA, and reduction in HBV DNA, we found that ETVr substitutions (i.e., T184SCGA/ILFM and S202G) significantly increased the risk of BT among patients with LVDr (OR, 141.12 [95% CI, 6.94 to 2,870.20] and 201.25 [95% CI, 11.22 to 3,608.65], respectively).
Mechanism of ETVr assessed by 3D docking simulation.
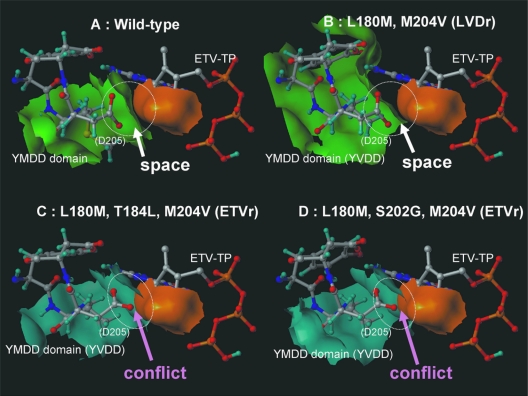
Modeling of the DNA binding cleft of HBV RT by docking simulation indicated that ETVr substitutions (T184L and S202G), which are located in the palm, were found to change the direction of the D205 residue (YMDD domain) and to narrow the binding pocket in comparison with the wild type and LVDr substitutions (M204V and L180M) (Fig. 3). The results of docking simulation showed that ETVr substitutions (T184L and S202G) plus LVDr substitutions (M204V and L180M) have significantly longer minimal distances between the molecular surfaces of the protein and the drug (2.2 Å and 2.1 Å) and higher potential energy (−118 and −99.8 Kcal/mol [smaller absolute values have a minus sign]) for ETV-TP than for the wild type (1.3 Å; −178 Kcal/mol) and LVDr substitutions (1.5 Å; −141 Kcal/mol) (Table 6). Since binding at higher potential energy creates a less stable structure, the deoxyribonucleotide triphosphate (dNTP)-binding domains of the ETVr substitutions plus the LVDr substitutions in HBV RT have lower binding affinity for ETV-TP than the wild type. Molecular docking simulation in the present study showed that the L180M, M204V, S202G, and T184L substitutions can lessen the affinity of ETV-TP for HBV RT by heightening the potential energy between them, suggesting that S202G and T184L substitutions, in addition to M204V in the YMDD motif and L180M in domain C, could affect the initial polymerase binding of dNTP analog inhibitors.
FIG. 3.
3D structures of the dNTP-binding domains of HBV RT of the wild type (A), an LVDr substitution (B), and ETVr substitutions (C and D). The molecular surfaces of the wild type and the LVDr mutant are drawn in green, those of LVDr plus ETVr mutants are drawn in blue, and that of ETV-TP is drawn in orange.
TABLE 6.
Minimal distances and binding potentials between ETV and the HBV RT domain in the wild type, one LVDr mutant, and 2 ETVr mutants
| Strain | distance (Å) | Binding potential (GOLD score [Kcal/mol]) | Reference (Fig. 3) |
|---|---|---|---|
| Wild type | 1.3 | −178.4 | A |
| L180M, 204V | 1.5 | −141.3 | B |
| L180M, T184L, 204V | 2.2 | −117.9 | C |
| L180M, S202G, 204V | 2.1 | −99.8 | D |
DISCUSSION
Based on the combination of clinical observations and 3D docking simulation, this is the first report to suggest the mechanism by which ETVr substitutions (T184SCGA/ILFM and S202G, but not S202C), in addition to LVDr (L180M and M204V, but not M204I), can induce BT during ETV therapy. First, an assessment of virological and biochemical events during a 3-year ETV treatment course showed that ETVr substitutions were absent among treatment-naïve patients but were detected in 44.8% of patients who were refractory to LVD during the preceding treatment period. Evidence of BT during ETV therapy was observed in 26.8% of LVD-refractory patients between weeks 60 and 144 of treatment. All 11 of the BT cases had both L180M and M204V/I substitutions at baseline (LVD refractory), as well as additional substitutions, such as T184 and/or S202G (not S202I/C), during the 3-year ETV treatment period.
Statistically significant risk factors for BT were the presence of LVDr (L180M and M204V) at baseline, detection of ETVr (S202G and T184SCGA/ILFM substitutions) during ETV treatment, and undetectable HBV DNA (<2.6) or more than a 2-log10-unit reduction in HBV DNA levels during the first year of ETV treatment. Detection of T184SCGA/ILFM and S202G was significantly associated with BT independent of age, gender, and LVDr (M204V and/or L180M) at baseline or nondetection or reduction in HBV DNA at the first year of treatment, indicating that these substitutions could be used as predictive markers for BT.
The mechanism by which combinations of ETVr (S202G and T184 SCGA/ILFM) and LVDr (L180M and M204V) can induce BT during ETV therapy is largely unknown. Note that T184L and S202G residues are located within domain B and domain C of the RT/polymerase, respectively, as well as L180M and M204V. The modeling of HBV RT indicated that the combination changed the direction of the D205 residue (YMDD domain) and narrowed the dNTP-binding pocket in comparison with the wild type and LVDr substitutions (M204V and L180M) (Fig. 3). The results of docking simulation of HBV RT and ETV-TP showed that the ETVr (184L and S202G) plus LVDr (L180M and M204V) substitutions had significantly longer minimal distances for ETV-TP and steric conflict with the D205 residue (Fig. 3 and Table 6). These docking simulation results suggest that nucleotide analogs that have the exocyclic alkene moiety of ETV-TP replaced by a smaller atom may retain activity against ETV-resistant mutants. Differences in the mode of binding of nucleotide inhibitors to the dNTP-binding pocket of HBV polymerase, as predicted from the current modeling studies, may account for the complementary drug resistance profiles seen for different nucleotide analogs. Interestingly, a previous in vitro study showed that ETVr substitutions (S202I and T184G), in addition to LVDr (L180M and M204V), were associated with a >1,100-fold decrease in susceptibility to ET (20). Collectively, these data indicate that nucleoside-naïve patients treated with ETV were less likely to become resistant to ETV.
In an in vitro assay, the rtA181T/V clinical-isolate genome from patients refractory to LVD/ADV induced a decrease in susceptibility to LVD, ADV, and, to a lesser extent, TDF, but sensitivity to ETV remained (22). LVDr selected by LVD exposure may lead to ETV failure. Therefore, for patients refractory to LVD/ADV, a combination of emtricitabine/TDF (10) might be an effective option. Furthermore, since sequential antiviral therapy leads to the selection of multidrug-resistant HBV and fitness or maximal viral resistance (25), combination therapy using a nucleoside together with a nucleotide analog, such as emtricitabine/TDF (10), ADV/LVD, ADV/ETV, ADV/telbivudine, or TDF, would be a more appropriate treatment strategy for patients with the LVDr substitution.
Based on HBV DR v3, T184SCGA/ILMF and S202G substitutions were present at baseline in 4.8% of patients and were detected in 14.6%, 24.4%, and 44.8% during 48, 96, and 144 weeks, respectively, of ETV therapy (Fig. 2). The prevalence of ETVr in our cohort seems to be higher than that reported in previous studies, based on assessment of ETV treatment at weeks 48, 96, 144, 192, and 240 using direct sequencing, where ETVr emerged in 6%, 15%, 36%, 47%, and 51% of LVD-refractory patients, respectively (21). The differences might be attributable to the tools used to detect HBV DNA substitutions associated with drug resistance, which differed between the studies. HBV DR v.3 and v.2 performed better than direct sequencing, and monitoring of the nucleoside mutations by HBV DR v.3 and v.2 in patients before and during ETV therapy was good for selecting effective therapeutic strategies and new combination therapies.
In conclusion, the combination of clinical observations and 3D docking simulation in the present study indicated that the low binding affinity of ETV-TP for the dNTP-binding domains of HBV RT by the ETVr plus LVDr substitutions could induce BT and provides the mechanistic foundations for a mechanism of inhibition of ETV against HBV. This modeling would be useful for designing new antiviral drugs.
Supplementary Material
Acknowledgments
This work was supported in part by a grant-in-aid from the Ministry of Health, Labor, and Welfare of Japan and a grant-in-aid from the Ministry of Education, Culture, Sports, and Science.
We thank Kenichi Fukai, Graduate School of Medicine, Chiba University, Chiba, Japan; Tatsuya Ide, Department of Internal Medicine, Kurume University School of Medicine, Kurume, Japan; Debbie Hana Yi, Department of Emergency Medicine, New York-Presbyterian Hospital Columbia/Cornell, New York, NY; and Robert G. Gish, California Pacific Medical Center, San Francisco, CA, for their help throughout this work.
We have no conflicts of interest to disclose, except for M. F. Yuen and C. L. Lai, who received research support from BMS.
Footnotes
Published ahead of print on 23 November 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Baldick, C. J., B. J. Eggers, J. Fang, S. M. Levine, K. A. Pokornowski, R. E. Rose, C. F. Yu, D. J. Tenney, and R. J. Colonno. 2008. Hepatitis B virus quasispecies susceptibility to entecavir confirms the relationship between genotypic resistance and patient virologic response. J. Hepatol. 48:895-902. [DOI] [PubMed] [Google Scholar]
- 3.Baldick, C. J., D. J. Tenney, C. E. Mazzucco, B. J. Eggers, R. E. Rose, K. A. Pokornowski, C. F. Yu, and R. J. Colonno. 2008. Comprehensive evaluation of hepatitis B virus reverse transcriptase substitutions associated with entecavir resistance. Hepatology 47:1473-1482. [DOI] [PubMed] [Google Scholar]
- 4.Brautigan, D. L., M. Brown, S. Grindrod, G. Chinigo, A. Kruszewski, S. M. Lukasik, J. H. Bushweller, M. Horal, S. Keller, S. Tamura, D. B. Heimark, J. Price, A. N. Larner, and J. Larner. 2005. Allosteric activation of protein phosphatase 2C by D-chiro-inositol-galactosamine, a putative mediator mimetic of insulin action. Biochemistry 44:11067-73. [DOI] [PubMed] [Google Scholar]
- 5.Chan, H. L., A. Y. Hui, M. L. Wong, A. M. Tse, L. C. Hung, V. W. Wong, and J. J. Sung. 2004. Genotype C hepatitis B virus infection is associated with an increased risk of hepatocellular carcinoma. Gut 53:1494-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, T. T., R. G. Gish, S. J. Hadziyannis, J. Cianciara, M. Rizzetto, E. R. Schiff, G. Pastore, B. R. Bacon, T. Poynard, S. Joshi, K. S. Klesczewski, A. Thiry, R. E. Rose, R. J. Colonno, R. G. Hindes, and the BEHoLD Study Group. 2005. A dose-ranging study of the efficacy and tolerability of entecavir in lamivudine-refractory chronic hepatitis B patients. Gastroenterology 129:1198-1209. [DOI] [PubMed] [Google Scholar]
- 7.Degertekin, B., M. Hussain, J. Tan, K. Oberhelman, and A. S. Lok. 2009. Sensitivity and accuracy of an updated line probe assay (HBV DR v. 3) in detecting mutations associated with hepatitis B antiviral resistance. J. Hepatol. 50:42-48. [DOI] [PubMed] [Google Scholar]
- 8.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 9.Jain, A. N. 2003. Surflex: fully automatic flexible molecular docking using a molecular similarity-based search engine. J. Med. Chem. 46:499-511. [DOI] [PubMed] [Google Scholar]
- 10.Keeffe, E. B., D. T. Dieterich, S. H. Han, I. M. Jacobson, P. Martin, E. R. Schiff, and H. Tobias. 2008. A treatment algorithm for the management of chronic hepatitis B virus infection in the United States: 2008 update. Clin. Gastroenterol. Hepatol. 6:1315-1341. [DOI] [PubMed] [Google Scholar]
- 11.Lai, C. L., D. Shouval, A. S. Lok, T. T. Chang, H. Cheinquer, Z. Goodman, D. DeHertogh, R. Wilber, R. C. Zink, A. Cross, R. Colonno, L. Fernandes, and the BEHoLD AI463027 Study Group. 2006. Entecavir versus lamivudine for patients with HBeAg-negative chronic hepatitis B. N. Engl. J. Med. 354:1011-1020. [DOI] [PubMed] [Google Scholar]
- 12.Lee, W. M. 1997. Hepatitis B virus infection. N. Engl. J. Med. 337:1733-1745. [DOI] [PubMed] [Google Scholar]
- 13.Marcellin, P., T. Asselah, and N. Boyer. 2005. Treatment of chronic hepatitis B. Rev. Prat. 55:624-632. [PubMed] [Google Scholar]
- 14.Miyakawa, Y., and M. Mizokami. 2003. Classifying hepatitis B virus genotypes. Intervirology 46:329-338. [DOI] [PubMed] [Google Scholar]
- 15.Ono, S. K., N. Kato, Y. Shiratori, J. Kato, T. Goto, R. F. Schinazi, F. J. Carrilho, and M. Omata. 2001. The polymerase L528M mutation cooperates with nucleotide binding-site mutations, increasing hepatitis B virus replication and drug resistance. J. Clin. Invest. 107:449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orito, E., M. Mizokami, H. Sakugawa, K. Michitaka, K. Ishikawa, T. Ichida, T. Okanoue, H. Yotsuyanagi, and S. Iino. 2001. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Japan HBV Genotype Research Group. Hepatology 33:218-223. [DOI] [PubMed] [Google Scholar]
- 17.Petrey, D., X. Xiang, C. L. Tang, L. Xie, M. Gimpelev, T. Mitors, C. S. Soto, S. Goldsmith-Fischman, A. Kernytsky, A. Schlessinger, I. Y. Y. Koh, E. Alexov, and B. Honig. 2003. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins 53:430-435. [DOI] [PubMed] [Google Scholar]
- 18.Sherman, M., C. Yurdaydin, J. Sollano, M. Silva, Y. F. Liaw, J. Cianciara, A. Boron-Kaczmarska, P. Martin, Z. Goodman, R. Colonno, A. Cross, G. Denisky, B. Kreter, R. Hindes, and the AI463026 BEHoLD Study Group. 2006. Entecavir for treatment of lamivudine-refractory, HBeAg-positive chronic hepatitis B. Gastroenterology 130:2039-2049. [DOI] [PubMed] [Google Scholar]
- 19.Szczesny, P., G. Wieczorek, and P. Zielenkiewicz. 2005. MOFOID—not only the protein modeling server. Acta Biochim. Pol. 52:267-269. [PubMed] [Google Scholar]
- 20.Tenney, D. J., R. E. Rose, C. J. Baldick, S. M. Levine, K. A. Pokornowski, A. W. Walsh, J. Fang, C. F. Yu, S. Zhang, C. E. Mazzucco, B. Eggers, M. Hsu, M. J. Plym, P. Poundstone, J. Yang, and R. J. Colonno. 2007. Two-year assessment of entecavir resistance in lamivudine-refractory hepatitis B virus patients reveals different clinical outcomes depending on the resistance substitutions present. Antimicrob. Agents Chemother. 51:902-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tenney, D. J., R. E. Rose, C. J. Baldick, K. A. Pokornowski, B. J. Eggers, J. Fang, M. J. Wichroski, D. Xu, J. Yang, R. B. Wilber, and R. J. Colonno. 2009. Long-term monitoring shows hepatitis B virus resistance to entecavir in nucleoside-naive patients is rare through 5 years of therapy. Hepatology 49:1503-1514. [DOI] [PubMed] [Google Scholar]
- 22.Villet, S., C. Pichoud, G. Billioud, L. Barraud, S. Durantel, C. Trepo, and F. Zoulim. 2008. Impact of hepatitis B virus rtA181V/T mutants on hepatitis B treatment failure. J. Hepatol. 48:747-755. [DOI] [PubMed] [Google Scholar]
- 23.Xiang, Z., and B. Honig. 2001. Extending the accuracy limits of prediction for side chain conformations. J. Mol. Biol. 311:421-430. [DOI] [PubMed] [Google Scholar]
- 24.Xiang, Z., C. Soto, and B. Honig. 2002. Evaluating conformational free energies: the colony energy and its application to the problem of loop prediction. Proc. Natl. Acad. Sci. U. S. A. 99:7432-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yim, H. J., M. Hussain, Y. Liu, S. N. Wong, S. K. Fung, and A. S. Lok. 2006. Evolution of multi-drug resistant hepatitis B virus during sequential therapy. Hepatology 44:703-712. [DOI] [PubMed] [Google Scholar]
- 26.Yuen, M. F., W. K. Seto, D. H. Chow, K. Tsui, D. K. Wong, V. W. Ngai, B. C. Wong, J. Fung, J. C. Yuen, and C. L. Lai. 2007. Long-term lamivudine therapy reduces the risk of long-term complications of chronic hepatitis B infection even in patients without advanced disease. Antivir. Ther. 12:1295-1303. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.