Abstract
In order to contribute to the knowledge of the architecture and epidemiology of class 2 integrons, we performed a class 2 integron molecular survey in which we analyzed 726 isolates in two bacterial populations from environmental and nonepidemiologically related clinical samples, respectively, collected from 1982 to 2007. We recovered the intI2 gene from 130 of 726 isolates, most of which were clinical isolates, and only 1 (a psychrophilic Pseudomonas sp.) was from a water sample. Unlike the widespread distribution of class 1 integrons within Gram-negative bacilli, only Acinetobacter baumannii and Enterobacter cloacae harbored class 2 integrons at a high frequency in our collection. Class 2 integrons with six novel cassette arrays were documented. Characterization of the transposition module of Tn7, the genetic platform in which class 2 integrons have always been reported, showed tns modules with a mosaic genetic structure. A bioinformatic analysis performed with the tns genes present in sequence databases, the finding of intI2 not associated with tns genes, and the genetic examination of novel tns-like genes found in three isolates indicated the possibility of the independent evolution of the two components related to horizontal gene transfer, the class 2 integrons and the Tn7 transposons.
Integrons are genetic elements that contain the components of a site-specific recombination system that recognizes and captures mobile gene cassettes (12). The basic structure of an integron possesses a gene for an integrase (intI), a recombination site (attI), and a promoter (PC) that permits the expression of the gene cassettes incorporated in the variable region (11). Class 1 and 2 integrons are both usually associated with antimicrobial resistance gene cassettes in bacterial isolates from clinical samples (11, 14, 23, 27). In this regard, the detection of an intI1 gene was significantly associated with multidrug resistance in clinical isolates of the Enterobacteriaceae family independently of the origin of the species or isolate (14). Some information regarding the origin of class 1 integrons and the evolutionary pathways that lead to its association with several mobile elements has also been published (31).
In contrast, there is less knowledge of the architecture and the epidemiology of class 2 integrons. The Tn7 transposon, in which class 2 integrons have been found to be embedded, has been well studied from a functional point of view (25, 34, 36). Tn7 can be identified as a sophisticated mobile element containing a transposition module, formed by the tnsA, tnsB, tnsC, tnsD, and tnsE genes (5, 8, 34), coding for two recombinational pathways. In the main transposition pathway, the transposition is directed by tnsD, which recognizes a specific sequence called attTn7, found in the chromosomes of many bacteria (16, 20). The mobilization of Tn7 through this pathway is nondeleterious to the bacterial host due to the fact that the insertion is downstream of the region encoding the glmS gene. By using another pathway, directed by tnsE, the transposition is directed to conjugative plasmids and to the genomes of filamentous bacteriophages (7, 25, 34, 36).
The most widespread class 2 integron, linked to the five tns genes of the transposition module of Tn7, carries the dfrA1 gene cassette, which confers resistance to trimethoprim; the sat2 gene cassette, which confers resistance to streptothricin; the aadA1 gene cassette, which confers resistance to spectinomycin and streptomycin; and a putative cassette called orfX, which corresponds to the ybeA gene, which carries a truncated attI2 site (13). Between the orfX and the tnsE genes, there is a region containing three additional genes (ybfA, ybfB, and ybgA) (13, 29). The intI2 gene is usually not functional because of the presence of an internal stop codon (13), which could explain, at least in part, the low level of diversity of cassette arrays carried by class 2 integrons documented in the literature (Fig. 1). Recently, two alleles of a functional intI2 gene have also been described (3, 24) (Fig. 1).
FIG. 1.
Schematic representation of class 2 integrons. (a) Variable region of the class 2 integrons described in the GenBank database with the corresponding accession number and/or the given name. The thin vertical closed bar represents the attI2 site, and the ovals represent the attC sites of the gene cassettes. Both recently described alleles of a functional intI2 gene correspond to the sequences with GenBank accession numbers DQ533990 and EU780012. The rest of the intI2 genes exhibited a stop codon of 179 amino acids. The graphic is not to scale. (b) Description of the PCR cartography of class 2 integrons used in this study, showing as an example the organization of the class 2 integron Tn7::In2-8. The genetic localization of each primer is indicated by an arrow. The numbers in parentheses correspond to the expected sizes of the products amplified by PCR. In order to search for hybrid genetic structures between the class 1 and 2 integrons described previously (34), the sequences were also investigated for both of the 3′ ends usually found for class 1 integrons (the 3′ conserved segment and tniC). The primers are indicated by arrows. Dashed lines represent the expected amplification product. The thin vertical open bar represents the attI2 site, and the ovals represent the attC sites of the gene cassettes. ND, not positive for amplification; there is not a previous report of this positive amplification for this pair of primers.
Although the distribution of class 2 integrons in the bacterial population has not been well established, some epidemiological studies have shown a high frequency of class 2 integrons in Shigella sonnei isolates from Australia (19); Helicobacter pylori isolates from Argentina (6); Acinetobacter baumannii isolates from Chile (9); and, at a lower frequency, different serovars of Salmonella spp. from Japan, Argentina, and Spain (1, 23, 29).
The objectives of the study described here were the molecular characterization of class 2 integrons in a large collection of clinical and environmental isolates, as well as bioinformatic analysis, in order to contribute to their clinical epidemiology and genetic architecture.
MATERIALS AND METHODS
Bacterial isolates.
The study was carried out with 726 isolates. These included 564 nonepidemiologically related clinical isolates from 14 hospitals in Buenos Aires, Argentina, collected between the years 1982 and 2007 and 162 isolates from environmental samples from diverse sources. The clinical and environmental isolates were identified by standard biochemical tests, by use of a microbiological test strip (API 20NE system; bioMérieux), and by sequencing of the 16S RNA, when necessary, by using the primers described by Weisburg et al. (35).
Among the clinical isolates (n = 564), the species and genera identified were Pseudomonas spp. (n = 87), Acinetobacter spp. (n = 215), Burkholderia cepacia complex (n = 39), Ralstonia pickettii (n = 2), Achromobacter xylosoxidans (n = 1), Shewanella putrefaciens (n = 2), Dysgonomonas sp. (n = 1), Stenotrophomonas maltophilia (n = 11), Escherichia coli (n = 24), Citrobacter freundii (n = 9), Klebsiella pneumoniae (n = 25), Serratia marcescens (n = 19), Proteus mirabilis (n = 19), Proteus vulgaris (n = 1), Raoultella terrigena (n = 1), Providencia stuartii (n = 3), Enterobacter spp. (n = 32), Shigella spp. (n = 14), Vibrio cholerae (n = 8), Bordetella bronchiseptica (n = 4), Bordetella spp. (n = 1), Mycobacterium chelonae (n = 1), Mycobacterium tuberculosis (n = 1), Mycobacterium fortuitum (n = 1), Mycobacterium avium (n = 1), Mycobacterium intracellulare (n = 2), Mycobatcerium abscessus (n = 2), Staphylococcus aureus (n = 28), Streptococcus pyogenes (n = 1), and Streptococcus spp. (n = 9).
For the Gram-negative bacilli, the MICs of sulbactam, piperacillin, imipenem, meropenem, gentamicin, amikacin, ciprofloxacin, trimethoprim-sulfamethoxazole, and colistin were determined by the agar dilution method (4).
The environmental strains (n = 162) were recovered from industrial sites (n = 11), sewage (n = 10), groundwater (n = 22), and plants (n = 7) and also from areas in Argentina with low levels of anthropogenic disturbance (n = 112). For the recovery of samples with low levels of anthropogenic disturbance, we performed two surveys, one conducted in January 2005 and the second conducted in February 2006, at 10 sites from Isla Grande de Tierra del Fuego, Argentina. Shallow freshwater sediment and soil samples were plated onto nutritive agar medium (Britania, Buenos Aires, Argentina), cetrimide agar (Britania), and Levine agar (Britania). The plates were incubated at 4°C for 8 days, after which all individual colonies from each environment and from each plate were again plated in the respective agar and incubated at 4°C for 4 days in order to obtain psychrophilic strains as well. Each colony was then placed into Luria-Bertani broth and incubated at 4°C for 48 h. The isolates were identified as Pseudomonas spp. (n = 77), Serratia spp. (n = 2), Vibrio spp. (n = 6), Shewanella spp. (n = 7), Agrobacterium tumefaciens (n = 1), Colwellia spp. (n = 3), Pseudoalteromonas spp. (n = 7), Bacillus spp. (n = 5), Sinorhizobium sp. (n = 1), Azospirillum sp. (n = 1), Bradyrhizobium sp. (n = 1), Alcaligenes spp. (n = 2), Burkholderia cepacia complex (n = 3), Chryseomonas spp. (n = 2), Aeromonas sp. (n = 1), Formosa sp. (n = 1), Planococccus sp. (n = 1), Enterococcus spp. (n = 2), Rhodococcus spp. (n = 2), Nocardia sp. (n = 1), and Gram-negative bacilli not identified yet (n = 35).
DNA techniques.
Total DNA was extracted and PCR amplifications were carried out in 50-μl volumes containing 10 ng of DNA, 10 μl of 5× PCR buffer, 0.5 μl of 10× deoxynucleoside triphosphate mix (2 mM each dATP, dCTP, dGTP, and dTTP), 2 μl of each primer stock solution (2.5 pmol of each primer per μl), and sterile distilled water. Taq DNA polymerase (1 μl of a 3-U/μl diluted solution; Promega) was added. The thermocycler used was from Perkin-Elmer Cetus, Emeryville, CA, and a three-step profile was utilized. Specific primers were used to carry out the PCRs for the intI2 gene (23), and degenerate primers were also tested in order to search for other intI2-like genes (Table 1). For the gene cassettes found in the variable region of Tn7 or Tn7-like elements, the PCR cartography was performed with different combinations of primers (Table 1 and Fig. 1) (23, 27, 28). We designed and used primers for all gene cassettes described to date in the context of class 2 integrons and also for the gene cassettes widely found in the context of class 1 integrons in Argentina (23) (Table 1). By PCR with specific primers, we also investigated the isolates for the presence of the genes found in the transposition module of Tn7 (tnsA, tnsB, tnsC, tnsD, and tnsE) (27) and of the 3′ conserved segment of the class 1 integron (qacEΔ1 and sul1) and tniC gene in order to identify the hybrid structures between class 2 and class 1 integrons that are putatively present (Table 1 and Fig. 1).
TABLE 1.
Oligonucleotides designed for this study
| Primer name | Sequence (5′-3′) | Target | GenBank accession no. |
|---|---|---|---|
| 125′CS | TTTTTGTGCTGCCATATCCGTG | intI2 gene | DQ176450 |
| dfrA1 | AGCTGTTCACCTTTGGC | dfrA1 gene | DQ176450 |
| aadBF | GTAACACGCAAGCACGATGA | aadB gene | DQ176450 |
| aadBR | GCCTGTAGGACTCTATGTGC | aadB gene | DQ176450 |
| aadA1 | TCGATGACGCCAACTAC | aadA1 gene | DQ176450 |
| 23′CS | TGGGCTGAGAGAGTGGT | tnsE gene | DQ176450 |
| satR | TCATCCTGTGCTCCCGAG | sat2 gene | DQ176450 |
| CarB4F | GCAAATAGTTCAAAGTTTCAAC | blaCARB-4 gene | AY913771 |
| CarB4R | GATTGGGGATTGAGCTTCAC | blaCARB-4 gene | AY913771 |
| Inti2SonR | TRTGTCTAACRGKCYAKTTKT | intI2 and intISon genes | NC_002525/NC_004347 |
| Inti2SonF | CCGTYAKRSACRGTRAWGSA | intI2 and intISon genes | NC_002525/NC_004347 |
| ereA-R | GTCCAAGATGGGTGAATG | ereA gene | AY183453 |
| ereA-COOH | CGAATCAGGAAGAAAACTGC | ereA gene | AY183453 |
| R-tniC | CCGGTCACGGTGCGGCG | tniC gene | NC_001735 |
| qacEΔ1 | AGCCCCATACCTACAAAGCC | qacEΔ1 | AY162283 |
| Tn7RF | GTGTGGGCGGACAATAAAG | IR right of Tn7 | NC_002525 |
| Tn7RR | ACGACGGGCAATTTGCAC | IR right of Tn7 | NC_002525 |
| Tn7LF | CAGATAAGTGAAATCTAGTTC | IR left of Tn7 | NC_002525 |
| Tn7LR | GAAATGGAGTTTTTAAGG | IR left of Tn7 | NC_002525 |
DNA sequencing.
Several PCR products were sequenced after purification with a Wizard SV gel and PCR cleanup system kit, according to the manufacturer's directions (Promega). Sequencing of both DNA strands was performed with ABI Prism 3100 BioAnalyzer equipment.
Computer analysis data.
The nucleotide sequences were analyzed by using Genetics Computer Group (GCG) software and BLAST (version 2.0) software (http://www.ncbi.nlm.nih.gov/BLAST/).
Genetic analysis was performed with Artemis software (http://www.webact.org/WebACT/home).
Nucleotide sequence accession numbers.
The sequences of the class 2 integrons have been submitted to the GenBank database and can be found under accession numbers EF042190, FJ785524, FJ785525, FJ785526, FJ785527, and FJ792804.
RESULTS AND DISCUSSION
Class 2 integrons in a large collection of clinical isolates.
PCR with primers for the intI2 gene was performed with all clinical isolates and showed that 129/564 (23%) isolates harbored this gene. A high frequency of class 2 integrons was identified in two species: A. baumannii (103/203) and Enterobacter cloacae (9/27). Lower proportions of class 2 integrons was found in E. coli (2/24), Pseudomonas spp. (1/87), and P. mirabilis (3/19) (Table 2). We found that three of nine C. freundii isolates harbored the intI2 gene; in this case, we consider that more isolates are needed to arrive at a conclusion concerning the frequency of this gene in this species, as was shown for A. baumannii and E. cloacae. Some epidemiological studies have also shown that the frequency of class 2 integrons in A. baumannii isolates from Chile is high (9). This is in contrast to the few class 2 integrons detected in isolates of this species from Europe and South Korea (9, 21, 30, 33). It is likely that a clonal lineage of A. baumannii carrying class 2 integrons could be spreading within the Southern Cone of Latin America. Therefore, the distribution of class 2 integrons within Gram-negative bacilli from different continents appears to vary. This is the first main difference that we identified when we compared them with class 1 integrons, which were described to be similarly disseminated among almost all Gram-negative species from patients with nosocomial infections (15, 18).
TABLE 2.
Description of the class 2 integron variable region and the tns gene composition in the 86 isolates fully characterizeda
| Isolate(s)b | Class 2 integron variable region | Transposition gene(s) | Integron name |
|---|---|---|---|
| Ecl90, Cf701, Rt701, P408, Sp2, A1, A12, A49, A58, A300, A301, A19603, A3, A12, A16, A306, A20, A25, A26, A27, A44, A92, A94, A96, A127, A138, A139, A145, A151, A301, AABF1, A5001, ACBC, A177, A179 | intI2-dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA | tnsE, tnsD, tnsC, tnsB, tnsA | Tn7::In2-7 |
| Ecl302, A182 | intI2-dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA | tnsE, tnsD, tnsB, tnsA | ΔTn7::In2-7 |
| Ecl303 | intI2-dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA | tnsE, tnsC, tnsB, tnsA | ΔTn7::In2-7 |
| Ecl702, A125, A154, A156, A312 | intI2-dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA | tnsD, tnsC, tnsB, tnsA | ΔTn7::In2-7 |
| Ecl503, A15, A113 | intI2-dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA | tnsE, tnsD, tnsC, tnsB | ΔTn7::In2-7 |
| A5, A21 | intI2-dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA | tnsE, tnsC, tnsA | ΔTn7::In2-7 |
| Ec511, Cf703 | intI2-dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA | tnsE, tnsB, tnsA | ΔTn7::In2-7 |
| A42 | intI2-dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA | tnsE, tnsC, tnsB | ΔTn7::In2-7 |
| A310, A317 | intI2-dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA | tnsE, tnsD, tnsC | ΔTn7::In2-7 |
| ACAC | intI2-dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA | tnsD, tnsC, tnsB | ΔTn7::In2-7 |
| A152 | intI2-dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA | tnsD, tnsC, tnsA | ΔTn7::In2-7 |
| Cf702 | intI2-dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA | tnsB, tnsA | ΔTn7::In2-7 |
| Ear701 | intI2-dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA | tnsE | ΔTn7::In2-7 |
| E1-05 | intI2::IS1-dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA | tnsE, tnsD, tnsC, tnsB, tnsA | Tn7::In2-7::IS1 |
| A101 | intI2-dfrA1-sat2-aadB-catB2(ΔattC)-dfrA1-blaCARB-4-aadA1-orfX-ybfA-ybfB-ybgA | tnsE, tnsD, tnsC, tnsB, tnsA | Tn7::In2-5 |
| A112 | intI2-sat2-aadA1-orfX::IS26 | tnsC, tnsB, tnsA | ΔTn7::In2-6::IS26 |
| A32, A43 | intI2-dfrA1-sat2-aadA1 | tnsE, tnsD, tnsC, tnsB, tnsA | Tn7::In2-4 |
| A181 | intI2-sat2 | tnsE, tnsD, tnsC, tnsB, tnsA | Tn7::In2-1 |
| A106, A107 | intI2-sat2 | Negative | In2-1 |
| Ecl401 | intI2-dfrA1 | tnsC, tnsB | ΔTn7::In2-2 |
| Ecl703 | intI2 | tnsB, tnsA | ΔTn7::In2 |
| A14 | intI2-dfrA1 | tnsE, tnsD, tnsC, tnsB, tnsA | Tn7::In2-2 |
| A16 | intI2-dfrA1 | tnsD, tnsC, tnsB, tnsA | ΔTn7::In2-2 |
| A21C, A102 | intI2-dfrA1-sat2 | tnsD, tnsC, tnsB, tnsA | ΔTn7::In2-3 |
| AG1320 | intI2 | tnsD, tnsB, tnsA | ΔTn7::In2 |
| AG323 | intI2 | tnsD, tnsC, tnsB, tnsA | ΔTn7::In2 |
| A106 | intI2 | Negative | In2 |
| AG324 | intI2-dfrA1 | tnsD, tnsB, tnsA | ΔTn7::In2-2 |
| A1504 | intI2 | tnsB | ΔTn7::In2 |
| A1524 | intI2-dfrA1 | Negative | In2-2 |
| A05-6 | intI2-dfrA1 | tnsE, tnsB | ΔTn7::In2-2 |
| A61 | intI2-dfrA1-sat2 | tnsD, tnsC, tnsB | ΔTn7::In2-3 |
| P9An1 | intI2 | tnsA | ΔTn7::In2 |
| A3018, A157 | intI2 | tnsB, tnsA | ΔTn7::In2 |
| A11, A13, A16 | intI2-dfrA1-sat2 | tnsE, tnsD, tnsC, tnsB, tnsA | Tn7::In2-3 |
| A7 | intI2-dfrA1 | tnsE, tnsD, tnsC, tnsB | ΔTn7::In2-2 |
The name of the corresponding class 2 integron was determined according to the variable region and the tns gene composition.
Cf, C. freundii; Ear, E. amnigenus; Ecl, E. cloacae; Ec, E. coli; Pm, P. mirabilis; Pv, P. vulgaris; Rt, R. terrigena; Ab, A. baumannii; AG3, Acinetobacter genomic species 3; AG13, Acinetobacter genomic species 13; Al, Acinetobacter lwoffii; Anob, non-A. baumannii Acinetobacter sp.; P, Pseudomonas spp.; Sp, S. putrefaciens.
Diversity of cassette arrays in class 2 integrons.
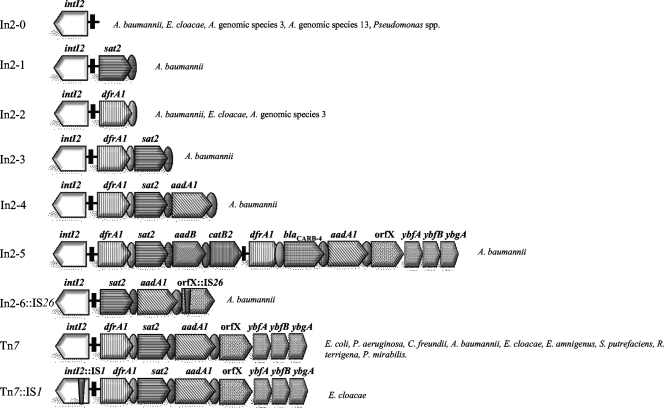
In order to study the content of cassettes within the variable region of class 2 integrons, we choose 122 isolates from our intI2-positive isolate collection and performed PCR cartography by using specific primers (Table 1) (27, 28). We found (i) PCR amplicons for class 2 integrons of the expected size that are usually described in Tn7 (dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA) (n = 80 isolates); (ii) amplicons of other sizes that were characterized by using different combinations of primers and sequencing (n = 32 isolates); and also (iii) class 2 integrons in which we could not identify any gene cassette within the variable region (n = 10) (Fig. 2 and Table 2). Tn7 was found in 80 clinical isolates of E. coli, Pseudomonas aeruginosa, C. freundii, A. baumannii, E. cloacae, Enterobacter amnigenus, S. putrefaciens, R. terrigena, and P. mirabilis. The 32 arrays that were different from the array in Tn7 could be grouped into eight types of class 2 integrons (Table 2 and Fig. 2). Some of the arrays were already described, such as those from the Tn1826 transposon (intI2-sat2-aadA1-orfX) and In2-1 (intI2-sat2), previously found in an isolate of Burkholderia cenocepacia (28, 32) (Fig. 2). A. baumannii showed most of the novel class 2 integron arrays within the variable region (n = 5), and even the β-lactamase gene blaCARB-4 that confers resistance to carbenicillin was found. This is the first time that a gene cassette encoding resistance to β-lactams has been described in a class 2 integron. For convenience, the class 2 integrons that were found are designated “In2-X” in this paper, where “X” corresponds to the gene cassette array. The A101 isolate of A. baumannii contained the Tn7::In2-5 element, which carried in its structure seven determinants of resistance, dfrA1-sat2-aadB-catB2(ΔattI2)-dfrA1-blaCARB-4-aadA1-orfX-ybfA-ybfB-ybgA, that involved four families of antibiotics and that is the longest class 2 integron described so far (Fig. 2) (GenBank accession no. FJ785525). We also found a novel class 2 integron called Tn7::IS1 (GenBank accession no. EF042190) in a clinical isolate of Enterobacter cloacae. It includes an IS1 element inserted at the beginning of the integrase gene, followed by the gene cassettes described above for Tn7 (Fig. 2). In A. baumannii isolate A112, we identified an IS26 insertion in the cassette array (sat2-aadA1-orfX::IS26) (Fig. 2).
FIG. 2.
Schematic representation of arrays of class 2 integrons found among the bacterial population (n = 126) analyzed in the present survey. Integrons In2-0, In2-2, In2-3, In2-5, Tn7::IS1, and In2-6::IS26 were described for the first time in this study. The thin vertical closed bar represents the attI2 site, and the ovals represent the attC sites of the gene cassettes. All intI2 genes from these arrays were sequenced, and they revealed the usual internal stop codon.
Taking into account the year of isolation of each class 2 integron-bearing isolate, we found that novel arrays as well as the widespread occurrence of Tn7 were present throughout the time period of our study (1983 to 2006) (Table 2).
Unlike the large amount of antimicrobial gene cassettes described so far in class 1 integrons all over the world, overall we identified few antimicrobial resistance gene cassettes in the class 2 integron context. From a functional point of view, this feature is the second difference between the two classes of integrons. While the high degree of diversity of gene cassettes within the variable region of class 1 integrons is mainly the result of the activity of the IntI1 integrase, the formation of the novel arrays of class 2 integrons found in our bacterial population could be explained by four mechanisms: (i) the insertion of insertion sequences, particularly IS1 and IS26, such as in the case of Tn7::IS1 and In2-6::IS26, respectively; (ii) an illegitimate integrase intermolecular recombination event, as has been suggested for Tn7::In2-8 [sat2-aadB-catB2(ΔattI2)-dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA] (27) and Tn7::In2-5 [dfrA1-sat2-aadB-catB2(ΔattI2)-dfrA1-blaCARB-4-aadA1-orfX-ybfA- ybfB-ybgA] from the present study; (iii) the activity of an integrase, probably provided in trans, since novel gene cassettes such as blaCARB-4 in Tn7::In2-5 and aadB in Tn7::In2-8 and Tn7::In2-5 were found; and (iv) a combination of the three mechanisms mentioned above, as was shown in Tn7::In2-5, in which mechanisms (ii) and (iii) described above probably took place.
On the other hand, the frequency and the nature of the gene cassettes within the variable region of class 2 integrons would provide less contribution to multidrug resistance phenotypes of clinical relevance than those of class 1 integrons would.
Class 2 integrons in a large collection of environmental isolates.
The last main difference between the two classes of integrons was established with environmental samples. While a intI1 gene frequency of 3% in cultivable bacteria from environmental samples has been reported (31), in our study we detected the intI2 gene in only 1 of 162 isolates (0.6%). Besides, no other gene or related intI2 integrase-like gene was found when degenerate primers were used with these isolates (Table 1). The positive strain was a Pseudomonas sp. recovered from a water sample from a river in Isla Grande de Tierra del Fuego. When a search for other class 2 integron-associated determinants in this isolate was performed, we found that the tnsA gene had 100% identity over a length of 822 bp to the gene previously found in the Tn7 transposon. However, none of the other transposition genes or gene cassettes usually described in the context of class 2 integrons were found (3, 10, 13). The intI2 gene from this environmental sample and 80 intI2 genes associated or not associated with the tns module and found in different species from clinical samples were sequenced; all of the genes exhibited the stop codon at amino acid 179, as described previously (13). It is likely that the association of the intI2 nonfunctional gene within Tn7 and the harboring of the typical array of cassettes (dfrA1-sat2-aadA1-orfX-ybfA-ybfB-ybgA) may have occurred before it spread in clinical samples in the antibiotic era.
Analysis of transposition modules associated with class 2 integrons.
In order to conduct a deeper analysis of the genetic association of class 2 integrons that were usually found to be embedded within the Tn7 transposon, we selected isolates of each species with different arrays of gene cassettes in the variable region and different years and origins of isolation. We searched by PCR analysis for the presence of each gene of the tns module in 86 representatives of 130 intI2-positive isolates. We found that 95.3% of the class 2 integrons were accompanied by at least one of the Tn7 transposition genes. However, a high degree of variability of mosaic structures was identified in the region corresponding to the transposition module, including some cases in which only one of the five transposition genes was found (Table 2). All Tn7 transposition genes (tnsA, tnsB, tnsC, tnsD, and tnsE) were detected in 51.16% (44/86) of the isolates (Table 2). Given the previous finding of a hybrid structure between class 2 and class 1 integrons in an A. baumannii isolate (26), we also investigated isolates in which all tns genes were missing (n = 4; strains A106, A107, A109, and A1524) for their presence (Fig. 1 and Table 2), but similar structures were not found. In summary, we found 18 combinations of the transposition genes in our bacterial population (Table 2).
The four A. baumannii clinical isolates with the intI2 gene with no association with the tns genes exhibited the following properties: (i) one of the isolates did not possess the gene cassettes previously found in class 2 integrons (dfrA1, sat2, aadA1, aadB, catB2, ereA, blaCARB-4), (ii) two isolates contained only the sat2 gene cassette in the variable region, and (iii) the other isolate harbored the dfrA1 gene cassette (Table 2). Neither inverted repeats (IRs) of Tn7 (Table 1) nor hybrid structures between class 1 and 2 integrons, as described before (22, 26), were detected in these samples, suggesting that class 2 integrons are not always embedded within a Tn7 transposon platform.
In addition, 270 isolates with negative results by PCR for the intI2 gene were also tested for the presence of the tns genes. We found that three isolates (0.6%) harbored at least one tns gene. One was a Pseudomonas sp. isolate that harbored the tnsA gene that had 100% identity over a length of 822 bp to the tnsA gene with GenBank accession no. NC_002525, and another was a clinical isolate, Ralstonia pickettii (strain RPB1), which had tnsB and tnsD genes with 100% identity to the tnsB and tnsD genes of Tn7 over lengths of 2,109 bp and 1,527 bp, respectively. Finally, the third one was a Shewanella algae isolate (strain Sa78) that contained the tnsB gene with 99% identity to the tnsB gene of Tn7.
We also searched for Tn7-related sequences in the GenBank database (NCBI database) using BLAST analysis, and we performed comparative sequence analysis using Artemis software to elucidate the genomic architecture of the sequences of the Tn7 and Tn7-like elements available in the GenBank database. We found a wide variety of mosaic genetic structures (Fig. 3) which were consistent with the diversity of arrays in the transposition module found in the isolates involved in our study. Sequence analysis also showed that the tns genes were widespread in the chromosomes of diverse species, including both Gram-negative and Gram-positive bacteria, some of which shared less than 30% amino acid identity (Fig. 3).
FIG. 3.
Bioinformatic analysis of the Tn7 and Tn7-like elements showing the genetic architecture found in diverse bacterial species. The Tn7 genes are represented in different colors. The different patterns of filling indicate the percent amino acid identity of the proteins codified by the Tn7-like elements found in the chromosomes of the listed species compared to the Tn7 sequence with GenBank accession number NC_002525. Vertical lines indicate less than 30% amino acid identity, diagonal lines denote less than 50% identity, and horizontal lines indicate less than 70% identity. The open reading frames are arbitrarily named orf simply to indicate the genes for hypothetical proteins. Integrases, represented by Int, not related to the integron integrase gene are indicated in reddish orange. The transposases of the IS3/IS911 family are represented as Tnp and are orange. Genes related to the integron integrase were not found in the flanking sequences (boundaries of 10 kb each), except for the class 2 integron from Shigella sonnei Ss046. A search for genes related to the glmS gene in the 10-kb flanking sequences was also performed. Percent amino acid identities and similarities were calculated by use of the same criteria used by the BLAST subroutine (NCBI database). The figure is not to scale; small arrows are used to indicate significant deletions in some of the Tns proteins.
The duplication of tns-like genes (e.g., tnsC-like genes in Ralstonia metallidurans CH34), the absence of some tns-like genes (e.g., tnsE-like genes in nine platforms, a tnsD-like gene in Ralstonia metallidurans CH34, and a tnsA-like gene in Fremyella diplosiphon), and especially noteworthy, the insertion of elements of the IS3/IS911 family were identified in two platforms in the 18 genetic contexts analyzed. We searched the flanking sequences (boundaries of 10 kb each) for genes related to the integron integrase and found negative results for all genetic structures except Tn7 from Shigella sonnei Ss046 (accession number CP000038.1) (Fig. 3).
Bioinformatic analysis revealed a high degree of heterogeneity of structures with genes related to those in the Tn7 tns region. Moreover, experimental data showed novel arrays of cassettes linked to different combinations of tns genes (Table 2), as well as intI2 genes not linked to tns genes. In addition, two novel alleles of the intI2 gene have also recently been described from different sources (2, 17). One of them is embedded in a Tn7-like structure (2), and the sequences of the tnsE and tnsD genes from the P. stuartii isolate shared 90% and 92% identities with the tnsE and tnsD genes from Tn7, respectively. Together, these results suggest that the class 2 integrons and the transposition module of Tn7 could have evolved independently. This suggestion is also in agreement with a recent proposal by Parks and Peters, who proposed that the tns module of Tn7 forms functionally diverse genomic islands at the specific site of Tn7 insertion, adjacent to glmS (24). However, on the basis of our experimental and genomic data, we argue that this event is achieved by the presence of the transposition module without a specific location, since several arrays of Tn7-like elements were found at target chromosomal sites different from glmS (Fig. 3 and data not shown).
In conclusion, the most exact comprehension of where the class 1 and 2 integrons converge lies in the fact that each system has an assembly in a specific mobile element, and at present these seem to be Tn402-like and Tn7-like elements, respectively, which allows each of them to be spread by different target affinities in clinical and environmental species. In the particular case of class 2 integrons, their association with Tn7 elements enhances their role as a genetic reservoir for the integration and dissemination of antimicrobial resistance gene cassettes in the nosocomial environment.
Acknowledgments
M.S.R. is a recipient of a CONICET fellowship. D.C. is a member of the Carrera del Investigador Científico, CONICET, Argentina. This study was supported by grant BID/OC 1723 ANPCyT 0690 from the Agencia Nacional de Promoción de Ciencia y Técnica, Buenos Aires, Argentina, and by grant UBACYT M008 to D.C.
The members of the Argentinian Integron Study Group are Sara Kaufman, Sección Microbiología, Hospital Fernández, Buenos Aires; Jaime Kovensky, Laboratorio Hospital de Quemados, Buenos Aires; Carlos Vay, Laboratorio de Bacteriología Clínica, Departamento de Bioquímica Clínica, Hospital de Clínicas José de San Martín, Facultad de Farmacia y Bioquímica, UBA, Buenos Aires; Marta Tokumoto, Laboratorio de Microbiología, ICYCC, Fundación Favaloro, and Servicio de Bacteriología e Infectología, Sanatorio Modelo Quilmes, Buenos Aires; Analía Fernández, Laboratorio de Microbiología, ICYCC, Fundación Favaloro, Buenos Aires; Angela Famiglietti, Laboratorio de Bacteriología Clínica, Departamento de Bioquímica Clínica, Hospital de Clínicas José de San Martín, Facultad de Farmacia y Bioquímica, UBA, Buenos Aires; Elsa Couto, Hospital Francés, Buenos Aires; Horacio Lopardo, Servicio de Microbiología, Hospital de Pediatría Prof. Dr. Juan P. Garrahan, Buenos Aires; Jorgelina Smayevsky, Laboratorio de Microbiología, CEMIC, Buenos Aires; Marisa Almuzara, Laboratorio de Bacteriología Clínica, Departamento de Bioquímica Clínica, Hospital de Clínicas José de San Martín, Facultad de Farmacia y Bioquímica, UBA, Buenos Aires; Adriana Procopio, Hospital de Niños Ricardo Gutiérrez, Buenos Aires; Myriam Vázquez, Hospital de Niños Ricardo Gutiérrez, Buenos Aires; Nelda Olivera, CENPAT-CONICET, Puerto Madryn, Chubut; José De Grossi, Cátedra de Higiene y Sanidad, Facultad de Farmacia y Bioquímica, UBA, Buenos Aires; Mabel Ferreiro, Hospital General de Agudos Carlos G. Durand, Buenos Aires; Marta Flaibani, Hospital General de Agudos Carlos G. Durand, Buenos Aires; Rosa Favre, Hospital General de Agudos Carlos G. Durand, Buenos Aires; Marcelo Hernán Cassini, Departamento de Ciencias Básicas, Universidad Nacional de Luján, Luján; Mercedes Iglesias, Hospital Gral. de Agudos Dr. Teodoro Álvarez, Buenos Aires; Ana Di Martino, Sanatorio Trinidad Mitre, Ciudad Autónoma de Buenos Aires, Buenos Aires; Mariana Catalano, Departamento de Microbiología, Parasitología e Inmunología, Facultad de Medicina, UBA, Buenos Aires; Liliana Jordá Vargas, Laboratorio de Análisis Clínicos, MANLAB, Buenos Aires; Adriana De Paulis, Departamento de Microbiología, Instituto de Investigaciones Médicas Alfredo Lanari, Facultad de Medicina, UBA, Buenos Aires; and Silvia Predari, Departamento de Microbiología, Instituto de Investigaciones Médicas Alfredo Lanari, Facultad de Medicina, UBA, Buenos Aires.
We are very grateful to María Paula Quiroga for revision of the manuscript.
The views presented in this article do not necessarily reflect those of the U.S. Food and Drug Administration
Footnotes
Published ahead of print on 16 November 2009.
REFERENCES
- 1.Ahmed, A. M., H. Nakano, and T. Shimamoto. 2005. Molecular characterization of integrons in non-typhoid Salmonella serovars isolated in Japan: description of an unusual class 2 integron. J. Antimicrob. Chemother. 55:371-374. [DOI] [PubMed] [Google Scholar]
- 2.Barlow, R. S., and K. S. Gobius. 2006. Diverse class 2 integrons in bacteria from beef cattle sources. J. Antimicrob. Chemother. 58:1133-1138. [DOI] [PubMed] [Google Scholar]
- 3.Biskri, L., and D. Mazel. 2003. Erythromycin esterase gene ere(A) is located in a functional gene cassette in an unusual class 2 integron. Antimicrob. Agents Chemother. 47:3326-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Craig, N. L. 1991. Tn7: a target site-specific transposon. Mol. Microbiol. 5:2569-2573. [DOI] [PubMed] [Google Scholar]
- 6.Crespo, O., M. Catalano, S. Piñeiro, M. Matteo, A. Leanza, and D. Centrón. 2005. Tn7 distribution in Helicobacter pylori: a selective paradox. Int. J. Antimicrob. Agents 25:341-344. [DOI] [PubMed] [Google Scholar]
- 7.Finn, J. A., A. R. Parks, and J. E. Peters. 2007. Transposon Tn7 directs transposition into the genome of filamentous bacteriophage M13 using the element-encoded TnsE protein. J. Bacteriol. 189:9122-9125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flores, C., M. I. Qadri, and C. Lichtenstein. 1990. DNA sequence analysis of five genes; tnsA, B, C, D and E, required for Tn7 transposition. Nucleic Acids Res. 18:901-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.González, G., K. Sossa, H. Bello, M. Domínguez, S. Mella, and R. Zemelman. 1998. Presence of integrons in isolates of different biotypes of Acinetobacter baumannii from Chilean hospitals. FEMS Microbiol. Lett. 161:125-128. [DOI] [PubMed] [Google Scholar]
- 10.Grape, M., A. Farra, G. Kronvall, and L. Sundstrom. 2005. Integrons and gene cassettes in clinical isolates of co-trimoxazole-resistant Gram-negative bacteria. Clin. Microbiol. Infect. 11:185-192. [DOI] [PubMed] [Google Scholar]
- 11.Hall, R. M. 1997. Mobile gene cassettes and integrons: moving antibiotic resistance genes in gram-negative bacteria. Ciba Found. Symp. 207:192-202. [DOI] [PubMed] [Google Scholar]
- 12.Hall, R. M., and C. M. Collis. 1995. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol. Microbiol. 15:593-600. [DOI] [PubMed] [Google Scholar]
- 13.Hansson, K., L. Sundstrom, A. Pelletier, and P. H. Roy. 2002. IntI2 integron integrase in Tn7. J. Bacteriol. 184:1712-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leverstein-van Hall, M. A., H. E. M. Blok, A. R. T. Donders, A. Paauw, A. C. Fluit, and J. Verhoef. 2003. Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. J. Infect. Dis. 187:251-259. [DOI] [PubMed] [Google Scholar]
- 15.Leverstein-Van Hall, M. A., A. Paauw, A. T. Box, H. E. Blok, J. Verhoef, and A. C. Fluit. 2002. Presence of integron-associated resistance in the community is widespread and contributes to multidrug resistance in the hospital. J. Clin. Microbiol. 40:3038-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lichtenstein, C., and S. Brenner. 1982. Unique insertion site of Tn7 in the E. coli chromosome. Nature 297:601-603. [DOI] [PubMed] [Google Scholar]
- 17.Márquez, C., M. Labbate, A. J. Ingold, P. R. Chowdhury, M. S. Ramírez, D. Centrón, G. Borthagaray, and H. W. Stokes. 2008. Recovery of a functional class 2 integron from an Escherichia coli strain mediating a urinary tract infection. Antimicrob. Agents Chemother. 52:4153-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martínez-Freijo, P., A. C. Fluit, F. J. Schmitz, V. S. Grek, J. Verhoef, and M. E. Jones. 1998. Class I integrons in Gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J. Antimicrob. Chemother. 42:689-696. [DOI] [PubMed] [Google Scholar]
- 19.McIver, C. J., P. A. White, L. A. Jones, T. Karagiannis, J. Harkness, D. Marriott, and W. D. Rawlinson. 2002. Epidemic strains of Shigella sonnei biotype g carrying integrons. J. Clin. Microbiol. 40:1538-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKown, R. L., K. A. Orle, T. Chen, and N. L. Craig. 1988. Sequence requirements of Escherichia coli attTn7, a specific site of transposon Tn7 insertion. J. Bacteriol. 170:352-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh, J. Y., K. S. Kim, Y. W. Jeong, J. W. Cho, J. C. Park, and J. C. Lee. 2002. Epidemiological typing and prevalence of integrons in multiresistant Acinetobacter strains. APMIS 110:247-252. [DOI] [PubMed] [Google Scholar]
- 22.Oppon, J. C., R. J. Sarnovsky, N. L. Craig, and D. E. Rawlings. 1998. A Tn7-like transposon is present in the glmUS region of the obligately chemoautolithotrophic bacterium Thiobacillus ferrooxidans. J. Bacteriol. 180:3007-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orman, B. E., S. A. Piñeiro, S. Arduino, M. Galas, R. Melano, M. I. Caffer, D. O. Sordelli, and D. Centrón. 2002. Evolution of multiresistance in nontyphoid Salmonella serovars from 1984 to 1998 in Argentina. Antimicrob. Agents Chemother. 46:3963-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parks, A. R., and J. E. Peters. 2007. Transposon Tn7 is widespread in diverse bacteria and forms genomic islands. J. Bacteriol. 189:2170-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters, J. E., and N. L. Craig. 2001. Tn7: smarter than we thought. Nat. Rev. Mol. Cell Biol. 2:806-814. [DOI] [PubMed] [Google Scholar]
- 26.Ploy, M. C., F. Denis, P. Courvalin, and T. Lambert. 2000. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob. Agents Chemother. 44:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramírez, M. S., C. Quiroga, and D. Centrón. 2005. Novel rearrangement of a class 2 integron in two non-epidemiologically related isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:5179-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramírez, M. S., L. J. Vargas, V. Cagnoni, M. Tokumoto, and D. Centrón. 2005. Class 2 integron with a novel cassette array in a Burkholderia cenocepacia isolate. Antimicrob. Agents Chemother. 49:4418-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodríguez, I., M. C. Martin, M. C. Mendoza, and M. R. Rodicio. 2006. Class 1 and class 2 integrons in non-prevalent serovars of Salmonella enterica: structure and association with transposons and plasmids. J. Antimicrob. Chemother. 58:1124-1132. [DOI] [PubMed] [Google Scholar]
- 30.Seward, R. J. 1999. Detection of integrons in worldwide nosocomial isolates of Acinetobacter spp. Clin. Microbiol. Infect. 5:308-318. [DOI] [PubMed] [Google Scholar]
- 31.Stokes, H. W., C. L. Nesbo, M. Holley, M. I. Bahl, M. R. Gillings, and Y. Boucher. 2006. Class 1 integrons potentially predating the association with Tn402-like transposition genes are present in a sediment microbial community. J. Bacteriol. 188:5722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tietze, E., J. Brevet, H. Tschape, and W. Voigt. 1988. Cloning and preliminary characterization of the streptothricin resistance determinants of the transposons Tn1825 and Tn1826. J. Basic Microbiol. 28:129-136. [DOI] [PubMed] [Google Scholar]
- 33.Turton, J. F., M. E. Kaufmann, J. Glover, J. M. Coelho, M. Warner, R. Pike, and T. L. Pitt. 2005. Detection and typing of integrons in epidemic strains of Acinetobacter baumannii found in the United Kingdom. J. Clin. Microbiol. 43:3074-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waddell, C. S., and N. L. Craig. 1988. Tn7 transposition: two transposition pathways directed by five Tn7-encoded genes. Genes Dev. 2:137-149. [DOI] [PubMed] [Google Scholar]
- 35.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolkow, C. A., R. T. DeBoy, and N. L. Craig. 1996. Conjugating plasmids are preferred targets for Tn7. Genes Dev. 10:2145-2157. [DOI] [PubMed] [Google Scholar]