Abstract
Ciprofloxacin was introduced for treatment of patients with cholera in Bangladesh because of resistance to other agents, but its utility has been compromised by the decreasing ciprofloxacin susceptibility of Vibrio cholerae over time. We correlated levels of susceptibility and temporal patterns with the occurrence of mutation in gyrA, which encodes a subunit of DNA gyrase, followed by mutation in parC, which encodes a subunit of DNA topoisomerase IV. We found that ciprofloxacin activity was more recently further compromised in strains containing qnrVC3, which encodes a pentapeptide repeat protein of the Qnr subfamily, members of which protect topoisomerases from quinolone action. We show that qnrVC3 confers transferable low-level quinolone resistance and is present within a member of the SXT integrating conjugative element family found commonly on the chromosomes of multidrug-resistant strains of V. cholerae and on the chromosomes of Escherichia coli transconjugants constructed in the laboratory. Thus, progressive increases in quinolone resistance in V. cholerae are linked to cumulative mutations in quinolone targets and most recently to a qnr gene on a mobile multidrug resistance element, resulting in further challenges for the antimicrobial therapy of cholera.
Cholera remains a major public health problem in many areas of the developing world. In addition to maintenance oral rehydration therapy, adjunctive antimicrobial therapy reduces the extent and duration of diarrhea, resulting in reduced fluid requirements and hospitalizations, reductions that are particularly important in resource-limited areas. Antimicrobial therapies have included tetracycline, azithromycin, and fluoroquinolones, such as ciprofloxacin, but the activity of fluoroquinolones has decreased in some areas, and this decreased activity has been associated with substantial reductions in the efficacy of ciprofloxacin relative to that of azithromycin (15, 20). To evaluate the evolution of ciprofloxacin resistance in Vibrio cholerae, we studied isolates from Bangladesh available over a 6-year period in which poor clinical responses of cholera patients to ciprofloxacin were recognized. We determined the presence of resistance mutations in genes encoding the subunits of the quinolone target enzymes DNA gyrase (gyrA and gyrB) and DNA topoisomerase IV (parC and parE) and the presence of qnr and other acquired genes that confer additional resistance to quinolones (25). Some qnr gene products have been shown to protect gyrase and topoisomerase IV from quinolone action in enteric bacteria (26, 27). qnr genes are usually located on mobile genetic elements, such as plasmids, that can transfer between strains but have been found on the chromosomes of some Vibrio spp. (5, 18). In V. cholerae, a qnr homolog, qnrVC1, has been described for isolates from Brazil (9) but has not been shown to confer transferable quinolone resistance or to be linked to incremental quinolone resistance and poor response to ciprofloxacin therapy of cholera. We show here that progressively higher levels of resistance in V. cholerae in Bangladesh were driven by accumulating mutations in topoisomerase target enzymes and by the acquisition of a quinolone resistance determinant, qnrVC3, which we identified as part of an SXT integrating conjugative element (4, 10) that also carried genes conferring resistance to tetracycline, trimethoprim-sulfamethoxazole, and streptomycin and accounted for transferable multidrug resistance that included ciprofloxacin in isolates positive for qnrVC3.
MATERIALS AND METHODS
Bacterial isolates.
Clinical isolates of V. cholerae O1 were recovered from patients seen at the Dhaka Hospital of the International Centre for Diarrhoeal Disease Research during the period from 2002 to 2008, as part of ongoing surveillance (24) and a prospective study of cholera patients and their household contacts (22).
Antibiotic susceptibility tests.
MICs of ampicillin, ciprofloxacin, gentamicin, levofloxacin, nalidixic acid, streptomycin, tetracycline, trimethoprim, and trimethoprim-sulfamethoxazole were determined by Etest (AB Biodisk, Solna, Sweden). Susceptibility criteria were those of the Clinical and Laboratory Standards Institute (6).
Screening for QRDR mutations and the PMQR genes.
The PCR amplifications for gyrA, gyrB, parC, and parE were carried out using the primers shown in Table 1, as previously described (1). Purified PCR products were sequenced on both strands, and quinolone resistance determining region (QRDR) DNA sequences were compared with the genome sequence of V. cholerae O1 strain N16961, available at http://www.jcvi.org/. In addition, we performed PCR screening for known plasmid-mediated quinolone resistance (PMQR) genes, including qnrA, qnrB, qnrC, qnrS, aac(6′)-Ib-cr, qepA, and oqxAB, using previously described primers (16).
TABLE 1.
Primers used in this study
| Gene | Primer | Sequence (5′→3′) | Size of PCR-amplified product (bp) |
|---|---|---|---|
| gyrA | Forward | AATGTGCTGGGCAACGACTGG | 239 |
| Reverse | GTGCGCGATTTTCGACATACG | ||
| gyrB | Forward | GGAAATGACTCGCCGTAAAGG | 309 |
| Reverse | GTTGTGATAACGCAGTTTATCTGGG | ||
| parC | Forward | GTCTGAGTTGGGTCTCTCGGC | 248 |
| Reverse | AGAATCTCGGCAAACTTTGACAG | ||
| parE | Forward | ATGCGTGCCAGCAAGAAAGTG | 268 |
| Reverse | TTATCGCTGTCAGGGTCAATCC | ||
| qnrVC | Forward | AATTTTAAGCGCTCAAACCTCCG | 521 |
| Reverse | TCCTGTTGCCACGAGCATATTTT |
Conjugation.
In order to determine whether quinolone resistance was transferable in V. cholerae, conjugation experiments were carried out in Luria-Bertani (LB) broth, with Escherichia coli J53 resistant to azide (Azr) as the recipient. Cultures of donor and recipient cells in logarithmic phase (0.5 ml of each) were added to 4 ml of fresh LB broth and incubated together overnight without shaking. Transconjugants were selected on Mueller-Hinton agar plates containing tetracycline (25 μg/ml; Sigma Chemical Co., St. Louis, MO) or sulfisoxazole (100 μg/ml) and sodium azide (100 μg/ml) for counterselection. To determine if quinolone resistance was cotransferred, MICs for the donor, recipient, and transconjugant strains were compared by Etest.
PCR amplification for the qnrVC3 gene.
We designed a pair of the primers (Table 1) based on the published sequences of the qnrVC1 gene (9) (GenBank accession number EU436855) on the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/). After detecting amplification products on the gel, we sequenced the purified PCR products on both strands. The purified PCR products were also used as the probe for Southern blotting.
Plasmid extraction and Southern hybridization for qnrVC3.
Plasmid DNAs from the donor and transconjugant strains were obtained using a plasmid midi kit (Qiagen, Valencia, CA). DNAs were subjected to electrophoresis in 1.0% agarose gel with ethidium bromide at 90 V for 3 h. After depurination, denaturation, and neutralization of the gel, DNAs were transferred to Hybond-N+ membrane by capillary blotting overnight. The membrane, which was fixed by UV exposure, was hybridized with the qnrVC3 probe labeled with horseradish peroxidase (Amersham ECL kit; Amersham Biosciences Corp., GE Healthcare, Chalfont St. Giles, United Kingdom), and then the signals were detected by exposure of the membrane to Hyperfilm ECL film (Amersham Biosciences Corp., GE Healthcare, Chalfont St. Giles, United Kingdom).
Cloning of qnrVC3.
The qnrVC3 gene from V. cholerae O1 strain 59 was cloned into expression vector pQE-60 (Qiagen, Valencia, CA) at its NcoI and BamHI sites. The gene was amplified by PCR using primers with sequences 5′-GCCATGGAAAAATCAAAGCAAT and 5′-GCGGATCCGTCAGGAACAATGATTA and ligated after digestion with NcoI and BamHI into pQE-60. Proper construction was confirmed by sequencing, and the pQE60-QnrVC plasmid was transformed into E. coli J53 Azr with selection on LB agar plates containing ampicillin.
Analysis of the qnrVC3 structures.
We sequenced the DNA adjacent to qnrVC3 in transconjugant SXT1 with a series of outward-facing primers starting from both sides of the qnrVC3 gene by the use of an inverse PCR strategy (9). Sequence analyses and comparison with known sequences were performed with the BLAST programs at the NCBI.
RESULTS AND DISCUSSION
Patterns of ciprofloxacin susceptibility.
Between 2003 and 2008, the median MIC of ciprofloxacin was 0.25 to 0.38 μg/ml, without a change over this interval (Table 2). These values were, however, 100-fold and 10-fold higher than the median MICs of ciprofloxacin for isolates of V. cholerae from Bangladesh in 1996 and 2002, respectively (15, 21). The maximum MIC, however, increased between 2003 and 2005 from 0.25 to 1.5 μg/ml and then fell to 0.25 to 0.5 μg/ml from 2006 to 2008.
TABLE 2.
Ciprofloxacin susceptibility of V. cholerae clinical isolates at the International Centre for Diarrhoeal Disease Research in Bangladesh
| Yr | MIC (μg/ml)a |
n | ||
|---|---|---|---|---|
| Mean | Median | Range | ||
| 2003 | 0.23 | 0.25 | 0.19-0.25 | 47 |
| 2004 | 0.25 | 0.25 | 0.125-0.38 | 23 |
| 2005 | 0.43 | 0.38 | 0.19-1.50 | 19 |
| 2006 | 0.25 | 0.25 | 0.25 | 10 |
| 2007 | 0.33 | 0.38 | 0.25-0.50 | 20 |
| 2008 | 0.28 | 0.25 | 0.125-0.50 | 84 |
Determined by Etest.
The progressive reduction in the quinolone susceptibility of V. cholerae has been associated with clinical failures. From 1993 to 1995, the MICs of ciprofloxacin were 0.003 μg/ml or less for all strains tested in one study that demonstrated 94% efficacy of a single 1-g dose of ciprofloxacin for treatment of cholera in adults (15). In 2001 and 2002, the MICs of ciprofloxacin were 10-fold higher (0.023 μg/ml and 0.047 μg/ml) and associated with 60% efficacy when a single dose of ciprofloxacin (20 mg/kg) was given to pediatric patients (21). By 2002 to 2004, a period that overlaps with that in which our isolates were collected, the median MIC of ciprofloxacin had risen another 10-fold increment to 0.25 μg/ml and was associated with a reduction in efficacy of a single 1-g dose of ciprofloxacin in adults to 27% (20). Discouragingly, despite the switch from ciprofloxacin to azithromycin as empirical therapy for cholera in 2005, reduced susceptibility has remained stable, and the low clinical efficacy of ciprofloxacin against strains with MICs below the clinical laboratory susceptibility breakpoint of 1.0 μg/ml raises questions about whether the breakpoint should be reevaluated. Such discrepancies between in vitro susceptibility and clinical efficacy remain mechanistically unexplained but have also been seen with doxycycline treatment of cholera patients (15) and ciprofloxacin treatment of patients with typhoid fever (7).
Evaluation of the mechanisms of reduced quinolone susceptibility.
To evaluate the mechanisms of reduced susceptibility to ciprofloxacin, 16 clinical isolates from isolates obtained in a prior study (22) were chosen for study in detail to represent the range of ciprofloxacin MICs in the period from 2002 to 2008. All isolates were V. cholerae O1, and all were El Tor Ogawa biotype, except for five (strains 10, 11, 16, MDC-4, and CLS6) that were El Tor Inaba biotype. Based on ciprofloxacin MICs (Table 2), we divided strains into three groups, group I (MICs of 0.023 to 0.032 μg/ml), group II (MICs of 0.38 to 0.5 μg/ml), and group III (MICs of >0.5 μg/ml). Isolates from group I were all from 2002; isolates from group II were from 2002 to 2008; and isolates from group III were from 2005 to 2007. Strains of V. cholerae with differing levels of susceptibility have previously been shown to have similar ribotypes, highlighting the clonal nature of V. cholerae outbreaks and the additive nature of resistance determinants in this setting (8).
No strain possessed a mutation in the QRDR (28, 29) of the gyrB or parE gene, and the reference strain N16961 also had no mutations in the gyrA or parC gene (Table 3). All other strains, including those in group I, contained a mutation in gyrA, encoding Ser83Ile, which is within the QRDR and is known to cause increased MICs of ciprofloxacin (11). Group I strains had an increase of two- to fourfold in the MIC of ciprofloxacin relative to the reference strain. Strains in group II had additional mutations in parC, encoding Ser85Leu (Table 2). qnrVC3 was not detected in group I. In contrast, one group II strain and all group III strains were positive for qnrVC3 by PCR in addition to having gyrA and parC mutations. Strains positive for qnrVC3 were found in each year of 2004 through 2007, and all were the El Tor Ogawa biotype. No other PMQR genes were found in isolates in any group. Thus, increasing MICs of ciprofloxacin correlated with sequential mutations in gyrA and parC, followed by acquisition of qnrVC3, which was present as early as 2004.
TABLE 3.
Characteristics of clinical isolates of V. cholerae and their E. coli transconjugants
| Strain | Yr of isolation | MIC (μg/ml)a |
QRDR mutation |
qnrVC PCR | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | LVX | NAL | TET | TMP | SXT | STR | AMP | GEN | gyrA | parC | |||
| E. coli ATCC 25922 | 0.008 | 0.023 | 3 | 1.5 | 0.75 | 0.094 | 4 | 6 | 0.75 | ||||
| E. coli J53 Azr | 0.012 | 0.032 | 4 | 1.5 | 0.19 | 0.032 | 3 | 6 | 0.75 | ||||
| V. cholerae N16961 | 0.015 | Ser83 | Ser85 | ||||||||||
| Clinical isolates | |||||||||||||
| Group I | |||||||||||||
| V. cholerae 10 | 2002 | 0.032 | 1.0 | >32 | 256 | 1.5 | Ile83 | − | − | ||||
| V. cholerae 11 | 2002 | 0.032 | 1.0 | >32 | 256 | 1.5 | Ile83 | − | − | ||||
| V. cholerae 12 | 2002 | 0.023 | 1.0 | >32 | 384 | 1.5 | Ile83 | − | − | ||||
| V. cholerae 13 | 2002 | 0.032 | 1.0 | >32 | 512 | 1.5 | Ile83 | − | − | ||||
| V. cholerae 16 | 2002 | 0.032 | 1.0 | >32 | 192 | 1.0 | Ile83 | − | − | ||||
| Group II | |||||||||||||
| V. cholerae 17 | 2002 | 0.38 | 1.0 | >32 | 256 | 1.5 | Ile83 | Leu85 | − | ||||
| V. cholerae 19 | 2002 | 0.38 | 1.0 | >32 | 384 | 1.5 | Ile83 | Leu85 | − | ||||
| V. cholerae 24 | 2004 | 0.5 | 12 | >32 | 256 | 1.5 | Ile83 | Leu85 | + | ||||
| V. cholerae 57 | 2005 | 0.38 | 8 | >32 | 192 | 1.5 | Ile83 | Leu85 | − | ||||
| V. cholerae CLS6 | 2006 | 0.38 | 0.38 | 0.75 | >32 | >1,024 | 0.75 | Ile83 | Leu85 | − | |||
| V. cholerae MDC126 | 2006 | 0.38 | 0.38 | 8 | >32 | 192 | 1.5 | Ile83 | Leu85 | − | |||
| V. cholerae MDC4 | 2007 | 0.5 | 0.38 | 1.5 | >32 | >32 | 384 | 1.5 | Ile83 | Leu85 | − | ||
| V. cholerae MDC125 | 2008 | 0.5 | 0.38 | 1.5 | 0.38 | 0.047 | 12 | 1.5 | Ile83 | Leu85 | − | ||
| Group III | |||||||||||||
| V. cholerae 59 | 2005 | 0.75 | 0.75 | >256 | 8 | >32 | >32 | >128 | 3 | 1.5 | Ile83 | Leu85 | + |
| V. cholerae CLS4 | 2006 | 0.75 | 0.75 | 8 | >32 | >32 | 128 | 1.5 | Ile83 | Leu85 | + | ||
| V. cholerae MDC1 | 2007 | 0.75 | 0.75 | 8 | >32 | >32 | 96 | 1.0 | Ile83 | Leu85 | + | ||
| E. coli J53 Azr transconjugants | |||||||||||||
| SXT1 (with strain 59)b | 0.047 | 0.19 | 3 | 64 | >32 | >32 | >192 | 4 | 0.75 | + | |||
| SXT2 (with strain 59)b | 0.012 | 0.032 | 3 | 64 | >32 | >32 | >192 | 4 | 0.5 | − | |||
| TET1 (with strain 59)b | 0.012 | 0.032 | 4 | 64 | >32 | >32 | >192 | 6 | 0.75 | − | |||
| TET2 (with strain 59)c | 0.064 | 0.125 | 4 | 64 | >32 | >32 | >192 | 4 | 0.5 | + | |||
| SXT13-1 (with strain 13)c | 0.006 | 1.5 | >32 | ||||||||||
| Cloned strains | |||||||||||||
| E. coli J53 (pQE60) | 0.012 | 0.032 | |||||||||||
| E. coli J53 (pQE60-QnrVC3) | 0.25 | 0.19-0.38 | |||||||||||
CIP, ciprofloxacin; AMP, ampicillin; CHL, chloramphenicol; GEN, gentamicin; KAN, kanamycin; LVX, levofloxacin; NAL, nalidixic acid; SMZ, sulfisoxazole; STR, streptomycin; SXT, trimethoprim-sulfamethoxazole; TET, tetracycline; TMP, trimethoprim.
Selected on Mueller-Hinton agar with sodium azide and sulfisoxazole.
Selected on Mueller-Hinton agar with sodium azide and tetracycline.
Mutations in gyrA and parC have previously been associated with increases in the MICs of ciprofloxacin for V. cholerae isolates from India (1), but the wide range of MICs associated with these strains made the contribution of these mutations to the MICs of ciprofloxacin difficult to determine. Other mobile quinolone resistance elements were not found in any of the isolates we studied, but the roles of other mechanisms of quinolone resistance, such as those related to expression of endogenous multidrug efflux pumps, remain to be defined (1, 12-14).
Transfer of reduced quinolone susceptibility.
To investigate if an increased MIC of ciprofloxacin could be transferred to E. coli from a group III qnrVC3-positive isolate, V. cholerae 59 was used as a donor for conjugation, and transconjugants were selected with tetracycline or sulfisoxazole and then tested for changes in susceptibility to ciprofloxacin. Reduced susceptibility to ciprofloxacin was cotransferred with other antibiotic resistance determinants in transconjugants SXT1 (selected with sulfisoxazole) and TET2 (selected with tetracycline), resulting in a four- to sixfold increase in the ciprofloxacin MIC of the E. coli J53 Azr recipient, and both transconjugants were positive for qnrVC3 by PCR (Table 3). Two other transconjugants, SXT2 and TET1, which showed no increase in the ciprofloxacin MIC, were negative for qnrVC3 by PCR. V. cholerae 13, which was negative for qnrVC3 by PCR, transferred resistance to trimethoprim-sulfamethoxazole but not to ciprofloxacin (Table 3). Thus, transfer of ciprofloxacin resistance correlated with transfer of qnrVC3.
Characterization of qnrVC3.
The nucleotide sequences of the PCR products obtained from V. cholerae 59 and its transconjugant strains showed 99% identity (654/657 nucleotides) with the nucleotide sequence of qnrVC1 reported from a clinical strain from Brazil isolated in 1998 (EU436588) (9). Among the differences, the qnrVC1 sequence had a nucleotide deletion at position 157 (relative to strain 59 reported here) and an insertion of an A at position 195, resulting in a frame-shifted gene segment. The translated sequence for QnrVC3 predicted a 218-amino acid protein with two domains of 10 and 33 pentpeptide repeats and differed from QnrVC1 in 11 amino acids. Cloning of qnrVC3 in plasmid pQE60 in E. coli J53 Azr resulted in a 16- to 32-fold increase in the MIC of ciprofloxacin (0.19 to 0.38 μg/ml versus 0.012 μg/ml) relative to E. coli J53 Azr containing the plasmid vector alone (Table 3). Thus, qnrVC3 from V. cholerae 59 is itself capable of conferring substantial increases in the MIC of ciprofloxacin when expressed from a multicopy plasmid. The increase in the ciprofloxacin MIC for E. coli transconjugants of a donor qnrVC3-containing V. cholerae strain was less (four- or fivefold) than that conferred by plasmid-cloned qnrVC3, suggesting differences in copy number or expression mechanisms in the transferable element.
Characterization of the transferable element containing qnrVC3.
Thus, to determine the nature of the transferable element that contained qnrVC3, we performed Southern blotting on whole-cell DNA from V. cholerae 59 and E. coli transconjugants SXT1, SXT2, TET1, and TET2, using the qnrVC3 PCR product as a probe. A strong hybridization signal was found with the band corresponding to chromosomal DNA from V. cholerae 59 and E. coli SXT1 and TET2, which were positive for qnrVC3 by PCR (data not shown). DNA from E. coli SXT2 and TET1, which were negative for qnrVC by PCR, and the E. coli recipient itself gave no hybridization signal. Additional experiments to identify plasmid DNA in transconjugant SXT1, using methods that can detect large plasmids were also negative (data not shown). Thus, qnrVC3 was associated with chromosomal DNA, and no plasmid DNA could be detected. The absence of an identifiable plasmid that hybridized with qnrVC3 in the E. coli recipient suggested that qnrVC3 might be transferred by an integrated conjugative element (ICE), such as the SXT element that has been found in strains of V. cholerae (4). We thus determined the DNA sequences flanking qnrV3 to evaluate its possible location within an ICE.
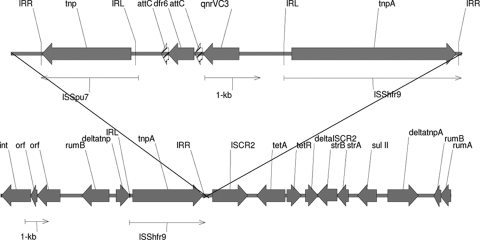
Using inverse PCR to detect flanking DNA sequences, we found (Fig. 1) that qnrVC3 in transconjugant SXT1 was situated between dfr6 and tnpA, which encodes the transposase of ISShfr9, and was linked to tetA, tetR, strA, strB, and sul11, which are identically represented in the core resistance determinants of the SXT ICE found in V. cholerae O1 El Tor strain HN1 (10). Near dfr6 and qnrVC3 is an insertion sequence (ISSpu7) first described for Shewanella putrefaciens. These three genes are bounded by directly repeated ISShfr9 units first characterized for Shewanella frigidimarina and suggest that dfr6 and qnrVC3 are located on a composite transposon, as has been found with other SXT ICEs (4, 10). A putative promoter sequence upstream of qnrVC3 was identical to that found upstream of qnrVC1 (9). Downstream from qnrVC3 and dfr6 were 126-base elements compatible with attC sites reported for mobile integrons and V. cholerae superintegrons (Fig. 1) (2, 3, 17, 19). The 126-bp element associated with qnrVC1 (9) differs by four nucleotides from that of qnrVC3 reported here. The sequence of the int gene, which encodes an integrase of the type found in mobile integrons and is distinct from the chromosomal intV gene of V. cholerae superintegrons, was identical to that found in the SXT element of V. cholerae HN1.
FIG. 1.
Genetic environment of qnrVC3. Annotated map of genes flanking qnrVC3 in E. coli transconjugant SXT1 from V. cholerae 59 in comparison to an SXT element from V. cholerae O1 strain HN1 (AB450045). IRR, inverted right repeat; IRL, inverted left repeat.
Thus, qnrVC3 may have been acquired by integron gene capture mechanisms mediated by integrases such as that encoded by the int gene found to be transferred with the SXT element. The insertion sequences flanking qnrVC3 also suggest a composite transposon-like structure, raising the additional possibility of mobilization of qnrVC3 by transposition from a yet unknown source, although transposition of such an element has not been directly demonstrated. Other Vibrio spp., including V. splendidus, V. parahaemolyticus, and V. vulnificus (5, 18), have been found to have chromosomally encoded qnr genes, but these genes differ from qnrVC3 by 35% or more and are not located on ICEs or linked to insertion sequences (ISs) (23). Other qnr genes have been found on the chromosomes of other aquatic organisms, such as Shewanella algae, and the presence in V. cholerae SXT elements of IS elements originally found in Shewanella spp. highlights the opportunities for gene exchange among bacteria living in aquatic environments.
Acknowledgments
We thank Que Chi Truong-Bolduc and Yanpeng Ding for technical advice.
This work was supported in part by grants R01 AI57576 (to D.C.H.), R01 AI43312 (to G.A.J.), U01 AI058935 (to S.B.C.), and K01 TW07144 (to R.C.L.) from United States Public Health Service, National Institutes of Health, and by a grant from the Ministry of Science and Technology, National Basic Research Program of China, 2005CB0523101 (to M.W.).
Footnotes
Published ahead of print on 30 November 2009.
REFERENCES
- 1.Baranwal, S., K. Dey, T. Ramamurthy, G. B. Nair, and M. Kundu. 2002. Role of active efflux in association with target gene mutations in fluoroquinolone resistance in clinical isolates of Vibrio cholerae. Antimicrob. Agents Chemother. 46:2676-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker, A., C. A. Clark, and P. A. Manning. 1994. Identification of VCR, a repeated sequence associated with a locus encoding a hemagglutinin in Vibrio cholerae O1. J. Bacteriol. 176:5450-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biskri, L., M. Bouvier, A. M. Guerout, S. Boisnard, and D. Mazel. 2005. Comparative study of class 1 integron and Vibrio cholerae superintegron integrase activities. J. Bacteriol. 187:1740-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burrus, V., J. Marrero, and M. K. Waldor. 2006. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid 55:173-183. [DOI] [PubMed] [Google Scholar]
- 5.Cattoir, V., L. Poirel, D. Mazel, C. J. Soussy, and P. Nordmann. 2007. Vibrio splendidus as the source of plasmid-mediated QnrS-like quinolone resistance determinants. Antimicrob. Agents Chemother. 51:2650-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CLSI. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement. CLSI M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Crump, J. A., K. Kretsinger, K. Gay, R. Hoekstra, D. J. Vugia, S. Hurd, S. D. Segler, M. Megginson, L. Luedeman, B. Shiferaw, S. S. Hanna, K. W. Joyce, E. D. Mintz, F. J. Angulo, and the Emerging Infections Program FoodNet and NARMS Working Groups. 2008. Clinical response and outcome of infection with Salmonella enterica serotype Typhi with decreased susceptibility to fluoroquinolones: a United States FoodNet multicenter retrospective cohort study. Antimicrob. Agents Chemother. 52:1278-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faruque, S. M., M. J. Islam, Q. S. Ahmad, K. Biswas, A. S. Faruque, G. B. Nair, R. B. Sack, D. A. Sack, and J. J. Mekalanos. 2006. An improved technique for isolation of environmental Vibrio cholerae with epidemic potential: monitoring the emergence of a multiple-antibiotic-resistant epidemic strain in Bangladesh. J. Infect. Dis. 193:1029-1036. [DOI] [PubMed] [Google Scholar]
- 9.Fonseca, É. L., F. dos Santos Freitas, V. V. Vieira, and A. C. P. Vicente. 2008. New qnr gene cassettes associated with superintegron repeats in Vibrio cholerae O1. Emerg. Infect. Dis. 14:1129-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hochhut, B., Y. Lotfi, D. Mazel, S. M. Faruque, R. Woodgate, and M. K. Waldor. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1SXT constins. Antimicrob. Agents Chemother. 45:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooper, D. C. 2003. Mechanisms of quinolone resistance, p. 41-67. In D. C. Hooper and E. Rubinstein (ed.), Quinolone antimicrobial agents, 3rd ed. ASM Press, Washington, DC.
- 12.Huda, M. N., J. Chen, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. Gene cloning and characterization of VcrM, a Na+-coupled multidrug efflux pump, from Vibrio cholerae non-O1. Microbiol. Immunol. 47:419-427. [DOI] [PubMed] [Google Scholar]
- 13.Huda, M. N., Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2001. Na+-driven multidrug efflux pump VcmA from Vibrio cholerae non-O1, a non-halophilic bacterium. FEMS Microbiol. Lett. 203:235-239. [DOI] [PubMed] [Google Scholar]
- 14.Huda, N., E. W. Lee, J. Chen, Y. Morita, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. Molecular cloning and characterization of an ABC multidrug efflux pump, VcaM, in non-O1 Vibrio cholerae. Antimicrob. Agents Chemother. 47:2413-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan, W. A., M. L. Bennish, C. Seas, E. H. Khan, A. Ronan, U. Dhar, W. Busch, and M. A. Salam. 1996. Randomised controlled comparison of single-dose ciprofloxacin and doxycycline for cholera caused by Vibrio cholerae O1 or O139. Lancet 348:296-300. [DOI] [PubMed] [Google Scholar]
- 16.Kim, H. B., C. H. Park, C. J. Kim, E.-C. Kim, G. A. Jacoby, and D. C. Hooper. 2009. Prevalence of plasmid-mediated quinolone resistance determinants over a nine-year period. Antimicrob. Agents Chemother. 53:639-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazel, D. 2006. Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4:608-620. [DOI] [PubMed] [Google Scholar]
- 18.Poirel, L., A. Liard, J. M. Rodriguez-Martinez, and P. Nordmann. 2005. Vibrionaceae as a possible source of Qnr-like quinolone resistance determinants. J. Antimicrob. Chemother. 56:1118-1121. [DOI] [PubMed] [Google Scholar]
- 19.Rowe-Magnus, D. A., A. M. Guerout, P. Ploncard, B. Dychinco, J. Davies, and D. Mazel. 2001. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc. Natl. Acad. Sci. U. S. A. 98:652-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saha, D., M. M. Karim, W. A. Khan, S. Ahmed, M. A. Salam, and M. L. Bennish. 2006. Single-dose azithromycin for the treatment of cholera in adults. N. Engl. J. Med. 354:2452-2462. [DOI] [PubMed] [Google Scholar]
- 21.Saha, D., W. A. Khan, M. M. Karim, H. R. Chowdhury, M. A. Salam, and M. L. Bennish. 2005. Single-dose ciprofloxacin versus 12-dose erythromycin for childhood cholera: a randomised controlled trial. Lancet 366:1085-1093. [DOI] [PubMed] [Google Scholar]
- 22.Saha, D., R. C. LaRocque, A. Khan, J. B. Harris, Y. A. Begum, S. M. Akramuzzaman, A. S. Faruque, E. T. Ryan, F. Qadri, and S. B. Calderwood. 2004. Incomplete correlation of serum vibriocidal antibody titer with protection from Vibrio cholerae infection in urban Bangladesh. J. Infect. Dis. 189:2318-2322. [DOI] [PubMed] [Google Scholar]
- 23.Sánchez, M. B., A. Hernández, J. M. Rodríguez-Martínez, L. Martínez-Martínez, and J. L. Martínez. 2008. Predictive analysis of transmissible quinolone resistance indicates Stenotrophomonas maltophilia as a potential source of a novel family of Qnr determinants. BMC Microbiol. 8:148-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoll, B., R. Glass, M. I. Huq, M. U. Khan, J. Holt, and H. Banu. 1982. Surveillance of patients attending a diarrhoeal disease hospital in Bangladesh. Br. Med. J. 285:1185-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strahilevitz, J., G. A. Jacoby, D. C. Hooper, and A. Robicsek. 2009. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22:664-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran, J. H., G. A. Jacoby, and D. C. Hooper. 2005. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 49:118-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran, J. H., G. A. Jacoby, and D. C. Hooper. 2005. Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob. Agents Chemother. 49:3050-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida, H., M. Bogaki, M. Nakamura, L. M. Yamanaka, and S. Nakamura. 1991. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents Chemother. 35:1647-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]