Abstract
We created an HIV-1 cloning vector, pNL4.3ΔIN, to generate recombinant infectious molecular clones for analysis of patient-derived HIV-1 integrase coding regions. Using this vector, we constructed a panel of clinically derived viruses with the canonical patterns of raltegravir resistance mutations and submitted the panel to the NIH AIDS Research and Reference Reagent Program. Investigational integrase inhibitors with activity against these clones are likely to retain activity against the most clinically relevant raltegravir-resistant variants.
The first licensed integrase inhibitor (INI), raltegravir, is highly effective when used with two nucleoside reverse transcriptase inhibitors (NRTIs) for initial antiretroviral (ARV) therapy and when used with at least one active drug for salvage ARV therapy (9, 13). However, when raltegravir is administered without effective accompanying ARVs, virological failure with the rapid emergence of high-level raltegravir resistance is common, raising questions about its genetic barrier to resistance (1, 6).
Several investigational INIs that retain antiviral activity against the most commonly observed INI resistance mutations have been identified, suggesting that the development of a second generation of INIs with higher genetic barriers to resistance may be feasible (5, 14, 16). This is desirable, particularly if INIs are to have an impact in low-income countries where drug resistance testing is not widely available and the selection of active inhibitors to prevent the rapid development of INI resistance may not always be possible.
A number of investigators have shown that, in vivo, raltegravir resistance is caused by a limited repertoire of primary integrase mutations, specifically, G140S together with Q148H/R/K, N155H with or without E92Q, and Y143C/R (1, 3, 6). To assist researchers in developing new INIs, we created a vector for cloning patient-derived HIV-1 integrase coding regions into a full-length HIV-1 clone lacking integrase. We then used this vector to create a panel of replication-competent prototypical raltegravir-resistant clones.
pNL4.3 with IN deleted (pNL4.3ΔIN).
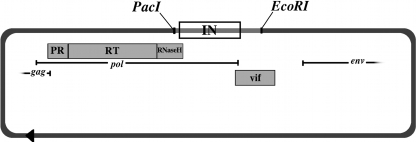
We created a unique 5′ PacI recognition sequence in pNL4.3 by introducing a silent ATA-to-ATT mutation at position 4127 (102 nucleotides upstream from the start of the integrase gene) using a QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). Successful mutagenesis was confirmed by DNA sequencing and restriction enzyme digestion. For the 3′ cloning site, we used the unique EcoRI site at pNL4.3 position 5743 (646 nucleosides downstream from the integrase gene). To create a linearized IN deletion clone, the vector is digested overnight with PacI and EcoRI and gel purified. The locations of the PacI and EcoRI sites and the integrase open reading frame within the genomic context of the pNL4.3 clone are shown in Fig. 1.
FIG. 1.
Integrase cloning vector pNL4.3ΔIN. Mutagenized pNL4.3 is digested with PacI and EcoRI to create a cloning vector with a deletion of integrase. The integrase open reading frame and flanking regions are shown between the restriction sites. The plasmid is unaltered outside this region.
Cloning and sequencing of clinical samples.
Clinical plasma samples were obtained from 15 HIV-1-positive individuals who had developed virological failure while receiving raltegravir, including four individuals in whom established raltegravir resistance mutations were detected by direct PCR sequencing. Samples from these four individuals and from six individuals with viruses lacking established raltegravir resistance mutations were chosen for cloning into pNL4.3ΔIN.
We used the high-fidelity, thermostable enzyme Pfu to amplify a 1,661-bp region from viral cDNA derived from each sample. The second-round PCR primers contained PacI and EcoRI sites to enable restriction digestion of IN-containing amplicons. PacI/EcoRI double digests of PCR products were followed by cycling T4 ligation to insert IN-containing amplicons into linearized pNL4.3ΔIN. The ligation products were transformed into competent Escherichia coli cells (Stbl3; Invitrogen, Carlsbad, CA).
The integrase sequences of 5 to 10 clones from each ligation reaction had mean numbers of uncorrected pairwise nucleotide and amino acid differences of 3.2 (range, 0 to 7.4) and 0.8 (range, 0 to 3.4), respectively. Two of the four clinical samples with raltegravir resistance mutations had clones with different patterns of mutations at INI resistance positions. Table 1 shows the 10 molecular clones selected for virus stock creation, including seven with unique patterns of established raltegravir resistance mutations and three without such mutations. One clone containing N155H alone was exempted as it was similar to other clones.
TABLE 1.
Replication characteristics and in vitro raltegravir susceptibilities of recombinant infectious molecular HIV-1 clones created using clinically derived integrase coding regions
| Clonea | Mutation(s)b |
MT2 titer/mlc | p24 concnd (ng/ml) | Fold decrease in RAL susceptibilitye | RC (%)f | ||
|---|---|---|---|---|---|---|---|
| Major | Minor | Other | |||||
| 2253 | None | None | S17N, V31I, I72V, L101I, T124A, K156N, V201I, I220M, D279G, R284G | 4.0 | 80 | 0.4 | 138 |
| 1288 | None | M154L | E11D, V31I, L101I, K160R, F181L, V201I, T206S, D256E | 3.3 | 88 | 1.06 | 212 |
| 4736_2 | N155H | T97A | D6E, S17N, R20K, D25E, S39C, L45V, L101I, V201I, S119R, T124A, K160T, K211R, D288N | 3.2 | 39 | 116 | NA |
| 4736_4 | N155H, E92Q | None | D6E, S17N, R20K, D25E, S39C, L45V, L101I, T124A, K160T, V201I, K211R, D288N | 2.8 | 28 | 96 | 58 |
| 4736_9 | N155H, E92Q | H51Y | D6E, S17N, R20K, D25E, S39C, D41H, L45V, L101I, T124A, K160T, V201I, K211R, D288N | 2.3 | 19 | 94 | 24 |
| 4736_10 | N155H | V151I, G163R | D6E, S17N, R20K, D25E, S39C, L45V, L101I, T124A, K160T, V201I, K211R, D288N | 3.3 | 12 | 48 | 116 |
| 1556 | Y143C | L74M, T97A, S230R | K7R, S17N, R20K, L101I, K111Q, T112K, S119R, T206S, D256E, V259I | 4.0 | 272 | >200 | 88 |
| 8070_1 | G140S, Y143H, Q148H | None | K14R, L101I, T112A, G193E, D229E, D279G | 3.7 | 79 | >200 | NA |
| 8070_2 | G140S, Q148H | None | K14R, L63I, L101I, T112A, G193E, V021I, D229E, D279G | 3.5 | 105 | >200 | NA |
| 4378 | None | E157Q | V31I, V32I, S39C, I72V, L101I, K160Q, F181L, V201I, Q216H, A265V | 4.6 | 141 | 1.15 | NA |
Clones shown in bold font have been submitted to the NIH AIDS Research and Reference Reagent Program.
Mutations represent amino acid differences from the consensus B wild type.
MT2 titer, log10 TCID50 after 1 week of culture in MT2 cells.
p24 antigen concentration per ml of virus stock (HIV-1 p24 ELISA; PerkinElmer, Boston, MA).
Decrease in raltegravir (RAL) susceptibility compared with that of the wild-type NL4-3 virus control.
Replication capacity (RC) was measured by virus production in a single-cycle luciferase reporter assay in the absence of inhibitor compared with that of the wild-type NL4-3 virus control (PhenoSense assay; Monogram, South San Francisco, CA) (2). “NA” indicates no data reported; it does not indicate RC below detectable limits.
Creation of recombinant infectious molecular clones.
Each of the 10 molecular clones was transfected into C8166 cells using Lipofectin (Invitrogen, Carlsbad, CA). Cells were observed daily for syncytia; once syncytia had formed in the majority of cell clusters, the C8166 cells were cocultured with SupT1 cells to increase viral titers (R.W. Shafer, unpublished data). Once the majority of SupT1 cell clusters had syncytia, 20 aliquots of cell-free virus stock were harvested. The median time from transfection to virus stock creation was 5 days (range, 5 to 8). The integrase coding region of one aliquot of cell-free virus stock was sequenced to confirm that the viral genome had not changed following transfection and virus stock creation.
Table 1 shows all amino acid differences from the consensus B sequence, as well as the p24 antigen concentrations, MT2-cell titers (50% tissue culture infective dose [TCID50] at day 7), and PhenoSense raltegravir susceptibilities of the 10 virus stocks. Replication capacity results were available for six of the recombinant infectious molecular clones. As judged by the MT2 cell titers, p24 antigen concentrations, and replication capacity results, there was no apparent difference in replication between the raltegravir-resistant and -susceptible clones. Five of the seven virus stocks with unique patterns of raltegravir resistance mutations were submitted to the NIH AIDS Research and Reference Reagent Program.
Discussion.
In contrast to mutants with site-directed mutations, a panel created from clinical isolates contains drug resistance mutations in their naturally occurring genetic context. This genetic context may include known accessory drug resistance mutations, as well as changes at positions that are not currently known to be associated with drug resistance.
The highest levels of raltegravir resistance (>200-fold decrease in susceptibility) were observed in clones containing both G140S and Q148H and in the clone containing Y143C. Lower levels of resistance (48- to 116-fold decreases in susceptibility) were found in clones containing N155H. These results are consistent with those reported by several other groups (3, 8, 10). The clone with Y143C and T97A had an extraordinarily high level of raltegravir resistance, confirming the highly synergistic interaction previously demonstrated only in site-directed mutants (4). Indeed, the level of raltegravir resistance associated with this pair of mutations is striking considering that Y143C alone reduces raltegravir susceptibility by only 3.5-fold (4) and T97A is a polymorphism with a 1 to 2% prevalence in raltegravir-naïve patients (12) that does not reduce raltegravir susceptibility (15).
Our data set contains the first reported clinical sequences with the mutations H51Y and Y143H. H51Y has been selected in vitro by elvitegravir and shown, using the PhenoSense assay, to reduce elvitegravir susceptibility 5-fold (7). A comparison of clones containing N155H/E92Q with and without H51Y suggests that H51Y has little impact on raltegravir resistance but may lower virus fitness (Table 1). Susceptibility data for Y143H have not been published previously.
E157Q, a polymorphism occurring in about 1 to 2% of INI-naïve patients (12), has been associated with virological failure during raltegravir therapy in one patient (11) and with a 3.3-fold reduction in elvitegravir susceptibility (7). However, the one sample in our panel with this mutation was fully susceptible to raltegravir.
Other than H51Y and S230R, each of the minor mutations listed in Table 1 is polymorphic, occurring at a frequency of >1.0% in the absence of therapy (12). With the exception of D41H, K160T, Q216H, I220M, and D229E, each of the mutations listed under “Other” in Table 1 is also polymorphic (12). Their potential impact, if any, on raltegravir susceptibility is not known.
In conclusion, investigational INIs which are active against the viruses in this panel are likely to retain activity against the most clinically relevant or, possibly, all raltegravir-resistant variants. Moreover, the use of a standard publicly available set of virus clones makes it possible for the relative activity of different INIs to be compared with one another. Researchers planning to create their own recombinant viruses can also do so using the pNL4.3ΔIN vector as described above. We plan to expand this panel as new INIs become licensed and new patterns of INI resistance mutations emerge.
Nucleotide sequence accession numbers.
The sequences of the clones shown in Table 1 are available in GenBank under accession nos. GU076499, GU076502, GU076504 to 8, and GU076510 to 12.
Acknowledgments
E.C.R., M.H.B., V.V., and R.W.S. were supported by NIAID grant AI46148.
This study was approved by the Institutional Review Board of Stanford University.
Footnotes
Published ahead of print on 16 November 2009.
REFERENCES
- 1.Cooper, D. A., R. T. Steigbigel, J. M. Gatell, J. K. Rockstroh, C. Katlama, P. Yeni, A. Lazzarin, B. Clotet, P. N. Kumar, J. E. Eron, M. Schechter, M. Markowitz, M. R. Loutfy, J. L. Lennox, J. Zhao, J. Chen, D. M. Ryan, R. R. Rhodes, J. A. Killar, L. R. Gilde, K. M. Strohmaier, A. R. Meibohm, M. D. Miller, D. J. Hazuda, M. L. Nessly, M. J. DiNubile, R. D. Isaacs, H. Teppler, and B. Y. Nguyen. 2008. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N. Engl. J. Med. 359:355-365. [DOI] [PubMed] [Google Scholar]
- 2.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 3.Fransen, S., S. Gupta, R. Danovich, D. Hazuda, M. Miller, M. Witmer, C. J. Petropoulos, and W. Huang. 16 September 2009, posting date. Loss of raltegravir susceptibility of HIV-1 is conferred by multiple non-overlapping genetic pathways. J. Virol. doi: 10.1128/JVI.01168-09. [DOI] [PMC free article] [PubMed]
- 4.Fransen, S., S. Gupta, A. Frantzell, C. Petropoulos, and W. Huang. 2009. HIV-1 mutations at positions 143, 148, and 155 of integrase define different genetic barriers to raltegravir resistance in vivo, abstr. 69. Abstr. 16th Conf. Retrovir. Oppor. Infect., Montreal, Canada.
- 5.Garvey, E. P., B. A. Johns, M. J. Gartland, S. A. Foster, W. H. Miller, R. G. Ferris, R. J. Hazen, M. R. Underwood, E. E. Boros, J. B. Thompson, J. G. Weatherhead, C. S. Koble, S. H. Allen, L. T. Schaller, R. G. Sherrill, T. Yoshinaga, M. Kobayashi, C. Wakasa-Morimoto, S. Miki, K. Nakahara, T. Noshi, A. Sato, and T. Fujiwara. 2008. The naphthyridinone GSK364735 is a novel, potent human immunodeficiency virus type 1 integrase inhibitor and antiretroviral. Antimicrob. Agents Chemother. 52:901-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hazuda, D., M. D. Miller, B. Y. Nguyen, and J. Zhao for the P005 Study Team Merck Research Labs. 2007. Resistance to the HIV integrase inhibitor raltegravir: analysis of protocol 005, a phase II study in patients with triple-class resistant HIV-1, abstr. 8. 16th Intl. HIV Drug Resist. Workshop, Barbados.
- 7.Jones, G., R. Ledford, F. Yu, M. Miller, M. Tsiang, and D. McColl. 2007. Resistance profile of HIV-1 mutants in vitro selected by the HIV-1 integrase inhibitor, GS-9137 (JTK-303), abstr. 627. Abstr. 14th Conf. Retrovir. Oppor. Infect., Los Angeles, CA.
- 8.Kobayashi, M., K. Nakahara, T. Seki, S. Miki, S. Kawauchi, A. Suyama, C. Wakasa-Morimoto, M. Kodama, T. Endoh, E. Oosugi, Y. Matsushita, H. Murai, T. Fujishita, T. Yoshinaga, E. Garvey, S. Foster, M. Underwood, B. Johns, A. Sato, and T. Fujiwara. 2008. Selection of diverse and clinically relevant integrase inhibitor-resistant human immunodeficiency virus type 1 mutants. Antivir. Res. 80:213-222. [DOI] [PubMed] [Google Scholar]
- 9.Lennox, J. L., E. DeJesus, A. Lazzarin, R. B. Pollard, J. V. Madruga, D. S. Berger, J. Zhao, X. Xu, A. Williams-Diaz, A. J. Rodgers, R. J. Barnard, M. D. Miller, M. J. DiNubile, B. Y. Nguyen, R. Leavitt, and P. Sklar. 2009. Safety and efficacy of raltegravir-based versus efavirenz-based combination therapy in treatment-naive patients with HIV-1 infection: a multicentre, double-blind randomised controlled trial. Lancet 374:796-806. [DOI] [PubMed] [Google Scholar]
- 10.Malet, I., O. Delelis, C. Soulie, M. Wirden, L. Tchertanov, P. Mottaz, G. Peytavin, C. Katlama, J. F. Mouscadet, V. Calvez, and A. G. Marcelin. 2009. Quasispecies variant dynamics during emergence of resistance to raltegravir in HIV-1-infected patients. J. Antimicrob. Chemother. 63:795-804. [DOI] [PubMed] [Google Scholar]
- 11.Malet, I., O. Delelis, M. A. Valantin, B. Montes, C. Soulie, M. Wirden, L. Tchertanov, G. Peytavin, J. Reynes, J. F. Mouscadet, C. Katlama, V. Calvez, and A. G. Marcelin. 2008. Mutations associated with failure of raltegravir treatment affect integrase sensitivity to the inhibitor in vitro. Antimicrob. Agents Chemother. 52:1351-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhee, S. Y., T. F. Liu, M. Kiuchi, R. Zioni, R. J. Gifford, S. P. Holmes, and R. W. Shafer. 2008. Natural variation of HIV-1 group M integrase: implications for a new class of antiretroviral inhibitors. Retrovirology 5:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steigbigel, R. T., D. A. Cooper, P. N. Kumar, J. E. Eron, M. Schechter, M. Markowitz, M. R. Loutfy, J. L. Lennox, J. M. Gatell, J. K. Rockstroh, C. Katlama, P. Yeni, A. Lazzarin, B. Clotet, J. Zhao, J. Chen, D. M. Ryan, R. R. Rhodes, J. A. Killar, L. R. Gilde, K. M. Strohmaier, A. R. Meibohm, M. D. Miller, D. J. Hazuda, M. L. Nessly, M. J. DiNubile, R. D. Isaacs, B. Y. Nguyen, and H. Teppler. 2008. Raltegravir with optimized background therapy for resistant HIV-1 infection. N. Engl. J. Med. 359:339-354. [DOI] [PubMed] [Google Scholar]
- 14.Underwood, M., B. Johns, A. Sato, T. Fujiwara, and W. Spreen. 2009. S/GSK1349572: a next generation integrase inhibitor with activity against integrase inhibitor-resistant clinical isolates from patients experiencing virologic failure while on raltegravir therapy, abstr. WEPEA098. 5th IAS Conf. HIV Pathog. Treat. Prev.
- 15.Van Baelen, K., M. Clynhens, E. Rondelez, V. Van Eygen, P. Van Den Zegel, H. Vermeiren, I. Vandenbroucke, and L. J. Stuyver. 2007. Low level of baseline resistance to integrase inhibitors L731,988 and L810,810 in randomly selected subtype B and non-B HIV-1 strains, poster 5. 16th Intl. HIV Drug Resist. Workshop, Barbados.
- 16.Wai, J., T. Fisher, M. Embrey, M. Egbertson, J. Vacca, D. Hazuda, M. Miller, M. Witmer, L. Gabryelski, and T. Lyle. 2007. Next generation of inhibitors of HIV-1 integrase strand transfer: structural diversity and resistance profiles, abstr. 87. Abstr. 14th Conf. Retrovir. Oppor. Infect., Los Angeles, CA.