Abstract
Induction of mecA by ceftobiprole and oxacillin in 18 methicillin-resistant Staphylococcus aureus clinical isolates with various SCCmec cassettes was examined using reverse transcriptase PCR. The magnitude of mecA induction, 3- to 65-fold for ceftobiprole and 2- to 69-fold for oxacillin, did not correlate with ceftobiprole MICs (≤4 μg/ml), although the 11 isolates with at least 13-fold induction by oxacillin all had oxacillin MICs of ≥256 μg/ml. No correlation between magnitude of induction and SCCmec type was found.
Ceftobiprole is a cephalosporin with a broad spectrum of activity against many Gram-negative and Gram-positive pathogens including methicillin-resistant Staphylococcus aureus (MRSA) (14). It is currently under regulatory review in the United States and has been approved in several other countries for the treatment of complicated skin and skin structure infections (cSSSI).
The major mechanism of methicillin resistance in S. aureus is the acquisition and expression of the mecA gene that encodes penicillin-binding protein 2a (PBP2a) (12, 23). Unlike almost all other β-lactams, ceftobiprole exhibits tight binding to PBP2a (10), thus explaining the clinically useful activity of ceftobiprole against MRSA. However, ceftobiprole MICs are higher for MRSA isolates than for methicillin-susceptible strains (2), but all MICs are still ≤4 μg/ml (11). One possible mechanism to explain this phenomenon might be increased mecA expression, as occurs for some β-lactams such as nafcillin (6, 7), methicillin, and oxacillin (3, 4). There is ample evidence that mecA is regulated at the transcriptional level and its expression is immediately inducible in strains containing a β-lactamase plasmid (13, 19). In this study, we evaluated the ability of ceftobiprole and oxacillin to induce mecA expression in selected MRSA clinical strains with different SCCmec cassette types.
(This work was presented in part at the 108th General Meeting of the American Society for Microbiology [19a].)
From recent clinical trials and surveillance studies, we selected 18 β-lactamase-positive MRSA strains based primarily on ceftobiprole MICs (ranging from 0.5 to 4 μg/ml) and various SCCmec cassette types. MICs were determined by broth microdilution using cation-adjusted Mueller-Hinton broth according to CLSI recommendations (8, 9). Determination of the presence of the mecA gene and SCCmec typing of MRSA strains were performed primarily by a multiplex PCR method as described by Zhang et al. (24) and by other methods (15-17). Clonal relatedness was determined by pulsed-field gel electrophoresis (PFGE) (22) and spa typing (21). The induction and detection of mecA expression were performed as follows: early-log-phase cultures (optical density at 600 nm [OD600] = 0.3) were treated for 1 h with ceftobiprole and oxacillin at concentrations equal to 1× MIC or oxacillin at 10 μg/ml for strains with higher oxacillin MICs and compared to untreated cultures used as a baseline control. Total RNA was isolated using the RNeasy kit (Qiagen, Valencia, CA), and cDNA was produced using the ImProm II reverse transcriptase system (Promega, Madison, WI). The primers used to amplify the 152-bp mecA sequence were 5′-CTCAGGTACTGCTATCCACC-3′ (forward) and 5′-GGAACTTGTTGAGCAGAGG-3′ (reverse), and those for the 100-bp 16S rRNA sequence were 5′-CCAGCAGCCGCGGTAAT-3′ (forward) and 5′-CGCGCTTTACGCCCAATA-3′ (reverse). Reverse transcriptase PCRs (RT-PCRs) were performed using the SYBR green Mastermix kit (Qiagen) and the Applied Biosystems 7300 real-time PCR system (Applied Biosystems, Foster City, CA). The cycling conditions were initial denaturation at 95°C for 15 min, followed by 40 cycles of 95°C for 15 s, 50°C for 10 s, and 72°C for 30 s with fluorescence measurement at the 72°C segment. The cycle numbers at threshold were determined for each sample, and relative levels of mecA mRNA expressed as copy numbers were normalized against internal control 16S rRNA gene expression. The fold of induction was calculated relative to baseline expression, which was mecA expression in uninduced culture, and data are summarized in Table 1. The values were averages calculated from three independent induction experiments. In most instances, two- to threefold differences among the replicates were observed.
TABLE 1.
Effect of ceftobiprole and oxacillin on mecA expression in MRSA clinical isolatesa
| Isolate | MIC (μg/ml) |
PFGE SmaI pattern | Spa type | SCCmec type |
mecA expression |
|||
|---|---|---|---|---|---|---|---|---|
| Baselineb | Fold induction (mean ± SD) |
|||||||
| BPR | OXA | BPR | OXA | |||||
| 11218 | 0.5 | 32 | ND | ND | II | 97 | 3 ± 2 | 3 ± 1 |
| 11521 | 0.5 | 256 | F | 2 | II | 6 | 49 ± 22 | 13 ± 12 |
| 11617 | 0.5 | 8 | USA300 | 1 | IVa | 10 | 30 ± 5 | 5 ± 1 |
| 12639 | 0.5 | 64 | H | 35 | IVa | 21 | 5 ± 1 | 2 ± 1 |
| 12662 | 0.5 | 256 | A1 | 1001 | III | 3 | 22 ± 10 | 18 ± 11 |
| 12868 | 0.5 | 4 | D | 7 | V | 1 | 20 ± 8 | 2 ± 1 |
| 14145 | 1 | 16 | USA300 | 1 | IVa | 2 | 14 ± 9 | 5 ± 2 |
| 14154 | 1 | 16 | USA300 | 1 | IVa | 1 | 20 ± 6 | 6 ± 2 |
| 15855 | 1 | 256 | G | 2 | II | 2 | 57 ± 38 | 25 ± 16 |
| 11631 | 2 | >256 | B | 442 | I | 3 | 65 ± 14 | 66 ± 31 |
| 12192 | 2 | >256 | B1 | 442 | I | 4 | 53 ± 39 | 41 ± 27 |
| 12575 | 2 | >256 | B | 442 | I | 3 | 52 ± 17 | 69 ± 12 |
| 12597 | 2 | >256 | A | 1001 | III | 11 | 24 ± 14 | 31 ± 10 |
| 12617 | 2 | >256 | A | 1001 | III | 3 | 38 ± 12 | 50 ± 28 |
| 14567 | 2 | 256 | A2 | 3 | III | 5 | 27 ± 7 | 27 ± 7 |
| 17256 | 4 | 256 | C1 | NEW | II | 4 | 3 ± 2 | 4 ± 2 |
| 17329 | 4 | 256 | E | 1001 | NT | 2 | 4 ± 1 | 50 ± 35 |
| 17357 | 4 | >256 | C | 2 | II | 4 | 3 ± 2 | 15 ± 10 |
Abbreviations: BPR, ceftobiprole; OXA, oxacillin; ND, not determined because isolate was not available for this analysis; NT, nontypeable; NEW, not found in current published database.
mecA copy number per 10,000 copies of 16S rRNA transcripts.
MICs for ceftobiprole ranged from 0.5 to 4 μg/ml while MICs for oxacillin were ≥256 μg/ml in 12 out of 18 isolates and 4 to 64 μg/ml against the other 6 strains (Table 1). All isolates were positive for β-lactamase as measured by nitrocefin hydrolysis in whole cells. Five different SCCmec types were identified, and PFGE/spa typing confirmed nine PFGE patterns, including the predominant community-associated USA300 strain, and eight spa types among the study isolates (Table 1).
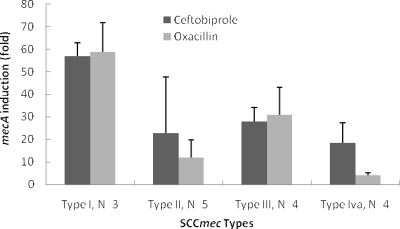
Both ceftobiprole and oxacillin induced mecA expression in all 18 MRSA strains, with similar increases in expression of 3- to 65-fold for ceftobiprole and 2- to 69-fold for oxacillin. The induction of mecA expression was strain dependent and tended to be highest for SCCmec type I and lowest for SCCmec type IVa (Fig. 1). In addition, in the same strain, ceftobiprole and oxacillin did not always induce mecA expression to the same extent. For example, ceftobiprole induced a fourfold increase in mecA expression versus a 50-fold increase for oxacillin in strain 17329, but ceftobiprole had a 20-fold induction versus twofold for oxacillin in strain 12868 (Table 1). These results suggest that ceftobiprole and oxacillin interact differently with the various components regulating mecA induction, such as mecR1/mecI or blaR1/blaI. The higher basal-level mecA expression observed in strains such as 11218 and 12639 suggests constitutive production, although low levels of induction could be measured.
FIG. 1.
Average induction of mecA expression according to SCCmec types. Ceftobiprole-treated cells are indicated by black bars and oxacillin-treated cells by gray bars.
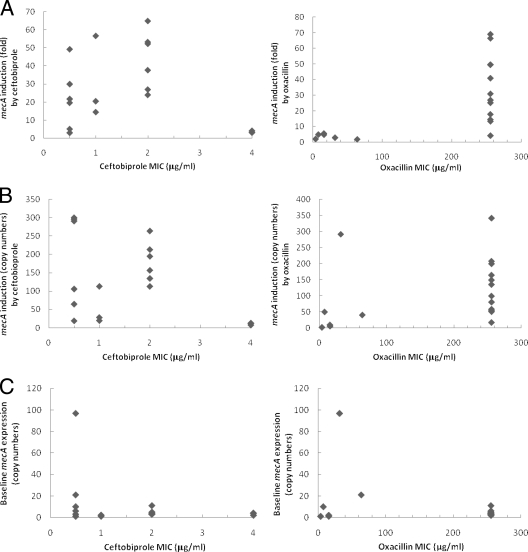
In this study, we wanted to investigate whether there was a dose-response correlation between the levels of mecA expression and ceftobiprole MICs. There was no strong correlation between increased ceftobiprole MICs and greater mecA induction measured, as either fold increases in mecA expression compared to no-drug control (Fig. 2A) or total expression of mecA (Fig. 2B). In contrast, for 11 of 12 isolates with oxacillin MICs of ≥256 μg/ml, mecA was induced by oxacillin at least 13-fold (Table 1; Fig. 2A). The exception was isolate 17256, with low baseline mecA production and only a fourfold mecA induction by oxacillin (Table 1; Fig. 2A). The MICs did not correlate well with baseline mecA expression for either agent (Fig. 2C). These results were consistent with previous studies from Chambers and Hackbarth and from Murakami and Tomasz, who had also observed noncorrelation between β-lactam MICs and cellular concentrations of PBP2a (6, 18).
FIG. 2.
Induction of mecA expression as a function of ceftobiprole and oxacillin MICs. (A) Fold of mecA induction relative to uninduced control. (B) Induction of mecA expression expressed as copy numbers per 10,000 copies of 16S rRNA transcripts in ceftobiprole- or oxacillin-treated cultures. (C) Baseline mecA expression expressed as copy numbers per 10,000 copies of 16S rRNA transcripts in uninduced cultures.
Production of PBP2a, i.e., expression of mecA, is under the control of both mec and β-lactamase regulatory genes (5) and is known to be inducible by many β-lactam antibiotics (4, 20), with oxacillin being a more potent inducer than methicillin in some strains (3, 4). However, according to Chambers and Hackbarth, cellular levels of PBP2a do not correlate with levels of methicillin resistance (6). These findings were similar to what we have seen from our experiments with ceftobiprole, although the causes may be different for the two β-lactams. Overproduction of PBP2a is irrelevant for penicillins, as there is greatly reduced binding to the protein, resulting in high penicillin MICs with no phenotypic change. Although ceftobiprole binds very tightly to PBP2a (10), these data indicate that even increases in PBP2a up to 65-fold are not directly correlated with decreased susceptibilities. This is consistent with the observation that other genetic determinants must be involved in order to attain high-level ceftobiprole resistance (1).
Our results showed that ceftobiprole and oxacillin induced mecA expression in MRSA isolates that had various SCCmec types and diverse genetic backgrounds and that the level of mecA expression did not correlate with ceftobiprole MICs. Due to the high binding affinity of ceftobiprole for PBP2a (10), ceftobiprole MICs remained ≤4 μg/ml while oxacillin had little activity with MICs of ≥256 μg/ml in most of these strains. These results indicate that induction of mecA is not expected to compromise the activity of ceftobiprole.
Acknowledgments
Many thanks to Anne Marie Queenan for her kind help in proofreading and expert comments.
This work was supported by Johnson & Johnson Pharmaceutical Research & Development, L.L.C.
Footnotes
Published ahead of print on 30 November 2009.
REFERENCES
- 1.Banerjee, R., M. Gretes, L. Basuino, N. Strynadka, and H. F. Chambers. 2008. In vitro selection and characterization of ceftobiprole-resistant methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 52:2089-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogdanovich, T., L. M. Ednie, S. Shapiro, and P. C. Appelbaum. 2005. Antistaphylococcal activity of ceftobiprole, a new broad-spectrum cephalosporin. Antimicrob. Agents Chemother. 49:4210-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyce, J. M., A. Bertasso, N. H. Georgopapadakou, and A. A. Medeiros. 1992. Differing abilities of oxacilllin (OXA) and methicillin (Met) to induce low-affinity penicillin-binding protein in strains of methicillin-resistant Staphylococcus aureus, abstr. 878. Abstr. 32nd Intersci. Conf. Antimicrob. Agents Chemother.
- 4.Boyce, J. M., A. A. Medeiros, E. F. Papa, and C. J. O'Gara. 1990. Induction of β-lactamase and methicillin resistance in unusual strains of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 25:73-81. [DOI] [PubMed] [Google Scholar]
- 5.Chambers, H. F. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers, H. F., and C. J. Hackbarth. 1987. Effect of NaCl and nafcillin on penicillin-binding protein 2a heterogeneous expression of methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 31:1982-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers, H. F., B. J. Hartman, and A. Tomasz. 1985. Increased amounts of a novel penicillin-binding protein in a strain of methicillin-resistant Staphylococcus aureus exposed to nafcillin. J. Clin. Invest. 76:325-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; M7-A7, approved standard, 7th ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- 9.Clinical and Laboratory Standards Institute. 2007. Performance standards for antimicrobial susceptibility testing; 18th informational supplement, M100-S17, vol. 27, no. 1. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.Davies, T. A., M. G. P. Page, W. Shang, T. Andrew, M. Kania, and K. Bush. 2007. Binding of ceftobiprole and comparators to the penicillin-binding proteins of Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae. Antimicrob. Agents Chemother. 51:2621-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fritsche, T. R., H. S. Sader, and R. N. Jones. 2008. Antimicrobial activity of ceftobiprole, a novel anti-methicillin-resistant Staphylococcus aureus cephalosporin, tested against contemporary pathogens: results from the SENTRY Antimicrobial Surveillance Program (2005-2006). Diagn. Microbiol. Infect. Dis. 61:86-95. [DOI] [PubMed] [Google Scholar]
- 12.Georgopapadakou, N. H. 1993. Penicillin-binding proteins and bacterial resistance to beta-lactams. Antimicrob. Agents Chemother. 37:2045-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hackbarth, C. J., and H. F. Chambers. 1993. blaI and blaR1 regulate beta-lactamase and PBP 2a production in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 37:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hebeisen, P., I. Heinze-Krauss, P. Angehrn, P. Hohl, M. G. P. Page, and R. L. Then. 2001. In vitro and in vivo properties of Ro 63-9141, a novel broad-spectrum cephalosporin with activity against methicillin-resistant staphylococci. Antimicrob. Agents Chemother. 45:825-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milheirico, C., D. C. Oliveira, and H. de Lencastre. 2007. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex.’ J. Antimicrob. Chemother. 60:42-48. [DOI] [PubMed] [Google Scholar]
- 17.Milheirico, C., D. C. Oliveira, and H. de Lencastre. 2007. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:3374-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murakami, K., and A. Tomasz. 1989. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J. Bacteriol. 171:874-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryffel, C., F. H. Kayser, and B. Berger-Bächi. 1992. Correlation between regulation of mecA transcription and expression of methicillin resistance in staphylococci. Antimicrob. Agents Chemother. 36:25-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Shang, W. F., T. A. Davies, R. K. Flamm, and K. Bush. 2008. Abstr. 108th Gen. Meet. Am. Soc. Microbiol., abstr. A-085.
- 20.Song, M. D., S. Maesaki, M. Wachi, T. Takahashi, M. Doi, F. Ishino, Y. Maeda, K. Okonogi, A. Imada, and M. Matsuhashi. 1988. Primary structure and origin of the gene encoding the beta-lactam-inducible penicillin-binding protein responsible for methicillin resistance in Staphylococcus aureus, p. 352-359. In P. Actor, L. Daneo-Moore, M. L. Higgins, M. R. J. Salton, and G. D. Shockman (ed.), Antibiotic inhibition of bacterial cell surface assembly and function. American Society for Microbiology, Washington, DC.
- 21.Strommenger, B., C. Kettlitz, T. Weniger, et al. 2006. Assignment of staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J. Clin. Microbiol. 44:2533-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tenover, F. C., R. D. Arbeit, R. V. Goering, et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Utsui, Y., and T. Yokota. 1985. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 28:397-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, K., J.-A. McClure, S. Elsayed, T. Louie, and J. M. Conly. 2005. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. J. Clin. Microbiol. 43:5026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]