Abstract
Aspergillus fumigatus is the causal agent of the life-threatening disease invasive aspergillosis. A. fumigatus laeA deletants, aberrant in toxin biosynthesis and spore development, are decreased in virulence. Among other characteristics, the decreased virulence is associated with increased spore susceptibility to macrophage phagocytosis. Three characteristics, cell wall microbe-associated molecular patterns (MAMPs), secreted metabolites, and rodlet content, thought to be important in macrophage-Aspergillus spore interactions were examined. Flow cytometry analysis of wild-type and ΔlaeA spores did not reveal any differences in surface-accessible MAMPs, including β-(1,3)-glucan, α-mannose, chitin, and other carbohydrate ligands. Blocking experiments with laminarin and mannan supported the conclusion that differences in cell wall carbohydrates were not responsible for enhanced ΔlaeA spore phagocytosis. Aspergillus spores have been reported to secrete metabolites affecting phagocytosis. Neither spent culture exchange, transwell, nor coincubation internalization experiments supported a role for secreted metabolites in the differential uptake of wild-type and ΔlaeA spores. However, sonication assays implicated a role for surface rodlet protein/hydrophobin (RodAp) in differential spore phagocytosis. A possible role of RodAp in enhanced ΔlaeA spore uptake was further assessed by RodAp extraction and quantification, where wild-type spores were found to contain 60% more RodAp than ΔlaeA spores. After removal of the surface rodlet layer, wild-type spores were phagocytosed at similar rates as ΔlaeA spores. We conclude that increased uptake of ΔlaeA resting spores is not associated with changes in secreted metabolite production of this mutant or surface carbohydrate availability but, rather, due to a decrease in the surface RodAp content of ΔlaeA spores. We theorize that RodAp acts as an antiphagocytic molecule, possibly via physicochemical means and/or by impeding MAMP recognition by macrophage receptors.
Infectious fungal diseases are an increasingly intractable cause of mortality worldwide. The most common filamentous agents of disease are members of the genus Aspergillus. These species are primarily saprophytic molds that can produce a wide spectrum of diseases in both plants and animals. The species responsible for more than 90% of human disease is A. fumigatus (7, 12, 15, 18). Disease symptoms run the spectrum from allergic bronchopulmonary aspergillosis to invasive aspergillosis (IA), the latter particularly on the rise owing to increased numbers of immunocompromised patients, including those afflicted with HIV/AIDS, malignancy, and organ dysfunction. High aspergillosis mortality rates emphasize the need to find improved means to treat these diseases. Our overall goal is to develop a better understanding of both fungal and host mechanisms contributing to disease development as a platform toward developing enhanced protective therapies.
The internalization of spores by macrophages is a key component of the early immune defense against fungal infections, including Aspergillus infections. A compilation of studies of spore phagocytosis has presented a partial understanding of this complex process. Recognition of the spore is a key requirement for both the phagocytosis of the spore and the subsequent activation of macrophages. Surface molecules, now termed microbe-associated molecular patterns (MAMPs) (8), are critical for recognition of spores. The best-characterized fungal MAMP is β-(1,3)-glucan, recognized by the mammalian receptor dectin-1 (5). Low amounts of surface-accessible β-(1,3)-glucans on resting spores of A. fumigatus are associated with decreased phagocytosis indexes compared to swollen spores, which display increased amounts of surface β-(1,3)-glucans (13). Similarly, increased internalization of resting spores of the A. fumigatus pksP mutant (lacking spore pigments) is associated with higher concentrations of surface β-(1,3)-glucans (11, 13).
In addition to β-(1,3)-glucan recognition, other molecules have been speculated to play a role in spore phagocytosis. Although not well defined, metabolites diffused from spores have been reported to affect macrophage handling of A. fumigatus spores (14), including inhibition of phagocytosis (2). Spore architecture influences cellular internalization as well; the outermost cell wall layer of Aspergillus spores is decorated with interwoven proteinaceous microfibrils called rodlets. These are hydrophobins encoded primarily by RODA that confer physiochemical properties to mediate spore dispersal and, possibly, cellular interactions (9, 20). In particular ΔrodA mutants, lacking the rodlet layer, are more sensitive to macrophage killing (16). Spore pigments, encoded by developmentally regulated clustered genes including pksP, confer resistance to macrophage phagocytosis and intracellular killing (10, 21-23), seemingly through enhanced β-(1,3)-glucan availability as discussed above (13). Null mutants of the dioxygenase PpoC, showing altered spore morphology, are also taken up more readily by macrophages, although the mechanism underlying this increased internalization has not been uncovered.
Previously, we reported on an A. fumigatus mutant, ΔlaeA, that displayed decreased virulence. Among other characteristics, decreased virulence was associated with increased susceptibility to macrophage phagocytosis. Spores harvested from ΔlaeA strains produced diminished amounts of at least one metabolite in vitro, and surface features of spores harvested from ΔlaeA strains showed an overall loss of prominent protrusions, which was speculated to be associated with abnormalities in rodlet formation (3). Here, our goal was to elucidate fungal modifications in ΔlaeA conidia related to phagocytosis. We provide evidence that increased uptake of ΔlaeA resting spores is not associated with changes in secreted metabolite production of this mutant nor with β-(1,3)-glucan (or other surface carbohydrate) availability but, rather, is associated with decreased RodAp content of ΔlaeA spores.
MATERIALS AND METHODS
Animal use.
All animal studies were performed under the approval of the University of Wisconsin animal care committee. C57BL/6 mice were used in all experiments. In preliminary experiments, alveolar macrophages and thioglycolate-elicited peritoneal macrophages behaved similarly under several functional parameters, including phagocytosis of wild-type (WT) and ΔlaeA conidia. Thus, thioglycolate-elicited peritoneal macrophages were used for all experiments. For peritoneal macrophages, mice (n = 4 to 8 mice per experiment, with 1 mouse per replicate) were injected intraperitoneally with 1 ml of 3% thioglycolate solution, and peritoneal fluids were collected after 5 days using 3 ml of phosphate-buffered saline (PBS). Cells were washed twice in RPMI 10 (RPMI 1640 [HyClone, Logan, UT] supplemented with 10% heat-inactivated fetal bovine serum [Invitrogen, Carlsbad, CA] and gentamicin [Sigma, St. Louis, MO]) prior to use. Macrophage preparations routinely exceeded 90% viability based on trypan blue exclusion.
Aspergillus preparation.
Aspergillus fumigatus Af293 and the ΔlaeA deletion strain TJW55.2 (3) were used in these studies. Strains were maintained in glycerol stocks and cultured on glucose minimal medium (GMM) (17) at 37°C to obtain spores. Spores were collected in H2O-Tween 20 (0.01%) and stored at 4°C. Swollen spores were obtained by shaking at 300 rpm at 37°C for 7 h in RPMI 10, followed by two washes in RPMI 10. For fixation, spores were incubated in a 4% solution of electron microscopy-grade formaldehyde (Polysciences, Inc., Warrington, PA)-PBS at room temperature for 30 min, and aldehydes were quenched with 100 mM glycine. Spores were washed three times in H2O-Tween 20 and twice in RPMI 10 before use. Conidial death was confirmed by plating aliquots on GMM and incubating at 37°C for 3 days.
Secondary metabolite extraction.
Wild-type and ΔlaeA spores were point inoculated onto GMM plates (30 per strain) and grown at 37°C for 5 days. The solid medium containing both conidia and mycelium was homogenized in a blender with 200 ml of methanol and agitated on a stir plate for 1 h. Fungal biomass was filtered out, and the methanol extract was evaporated. Extracts were weighed, resuspended in methanol to 1 mg/ml, and kept frozen at −80°C.
Phagocytosis assays.
Primary cells were washed twice in RPMI 10, counted, and plated at 106/well in 12-well tissue culture plates containing coverslips. After a 2-h adherence period, spores (5 × 105/well) were added to each well. Plates were spun at 1,200 rpm for 5 min to bring spores in contact with macrophages and then incubated at 37°C for 1 h. Wells were washed twice with RPMI 10 and incubated an additional 2 h to allow for complete uptake. Coverslips fixed in 2% formaldehyde were washed and mounted onto glass slides for microscopy. Extracellular spores were labeled with calcofluor white and excluded from analysis.
In blocking experiments, soluble laminarin or mannan derived from Saccharomyces cerevisiae (0.5 mg/ml final; Sigma) was added to the macrophages 30 min prior to the addition of spores. In experiments to determine the role of metabolites in phagocytosis, methanol-derived secondary metabolite extracts were added at 10 μg/ml to the macrophage culture immediately prior to spore addition. In preliminary experiments, macrophage viability was tested against 10-fold dilutions of metabolite extracts to identify the highest metabolite concentration (10 μg/ml) that did not affect cell viability. For the competition assay, wild-type conidia were labeled with Alexa 488 and ΔlaeA conidia were labeled with Alexa 594 (Invitrogen) according to the manufacturer's instructions for protein labeling. Conidia of each strain were easily distinguished by fluorescence microscopy using this labeling method.
For the transwell assay, phagocytosis was performed as above with the addition of each well containing a transwell insert (0.2-μm filter; Corning) containing medium alone or excess ΔlaeA conidia (100-fold increase; 5 × 107/transwell). For sonication experiments, conidia (5 × 107/ml) were sonicated at 120 W for four 5-min intervals with incubation on ice between sonications. This method has been shown to remove rodlet proteins from the conidial surface without affecting conidial morphology (16). Untreated and sonicated conidia were washed at least five times before use in phagocytosis experiments.
Flow cytometry.
Resting spores were washed twice in ice-cold PBS, and 100 μl of spores (107/ml) was incubated with anti-β-(1,3)-glucan primary antibody (0.5 μg/106 conidia; Biosupplies Australia Pty. Ltd., Victoria, Australia) for 20 min followed by an additional 20-min incubation with phycoerythrin-conjugated goat anti-mouse antibody. This staining regimen labels swollen conidia and germlings (positive control). For calcofluor white and lectin staining, spores were washed twice in lectin buffer (HEPES-buffered PBS with Mg2+/Ca2+) and labeled with lectins as recommended in the manufacturer's protocol (Vector Laboratories, Burlingame, CA). The lectins tested were concanavalin A (ConA), Dolichos biflorus lectin (DBA), peanut agglutinin (PNA), Ricinus communis agglutinin (RCA120), soybean agglutinin (SBA), Ulex europaeus agglutinin I (UEA I), wheat germ agglutinin (WGA), Griffonia simplicifolia lectin I (GSL I), Lens culinaris agglutinin (LCA), Phaseolus vulgaris agglutinin E (PHA-E), Phaseolus vulgaris agglutinin L (PHA-L), Pisum sativum agglutinin (PSA), Sophora japonica agglutinin (SJA), Datura Stramonium lectin (DSL), Erythrina cristagalli lectin (ECL), Griffonia simplicifolia lectin II (GSL II), Jacalin, Lycopersicon esculentum lectin (LEL), Solanum tuberosum lectin (STL), and Vicia villosa lectin (VVL).
Rodlet (RodAp) extraction method.
The rodlet layer was extracted from the spore surface by incubating dry spores with hydrofluoric acid for 72 h at 4°C (1). The contents were centrifuged (10,000 rpm, 10 min), and the supernatant obtained was dried under N2. The dried material was reconstituted in H2O, and protein was estimated by Bradford assay and also subjected to SDS-PAGE (15% gel). This method releases only RodAp, which was confirmed by matrix-assisted laser desorption ionization-mass spectrometry and tandem mass spectrometry analyses (data not shown). Spores stripped of RodAp by this method were fixed and then assessed in phagocytosis assays.
Statistical analysis.
Data were analyzed by Student's t test or, for multiple comparisons, a one-way analysis of variance with Tukey's honest significant difference test using Prism 5 software (Graphpad Software, Inc., La Jolla, CA). Data are presented as means ± standard errors of the means (SEM), and differences were considered significant at P values of <0.05.
RESULTS
Cell wall analysis of wild-type and ΔlaeA spores.
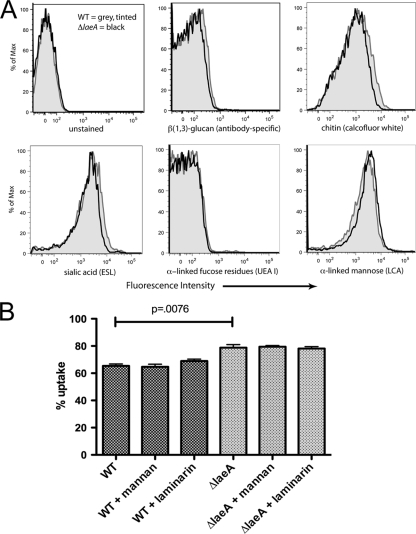
A previous study by Bok et al. (3) revealed that reduced virulence of a ΔlaeA mutant correlated with an altered spore surface and enhanced spore phagocytosis by macrophages. From these observations, we hypothesized that cell wall carbohydrate ligands, some of which are involved in spore phagocytosis [e.g., β-(1,3)-glucan], were altered on resting spores of the ΔlaeA strain. To compare the carbohydrate accessibility on wild-type and ΔlaeA spores, spores were stained with antibodies or lectins and analyzed by flow cytometry. The panel of 19 carbohydrates tested included chitin (detected with calcofluor white), β-(1,3)-glucan [detected with a β-(1,3)-glucan-specific antibody], sialic acid (detected with elderberry bark lectin [EBL]), α-linked fucose (detected with UEA I), and α-linked mannose (detected with LCA). As seen in Fig. 1A (and data not shown), wild-type and ΔlaeA spores displayed nearly identical surface labeling. To confirm that accessibility of ligands did not contribute to differential uptake, blocking experiments were undertaken using laminarin (a linear polymer of β-glucans) and mannan (derived from Saccharomyces cerevisiae). Incubation of macrophages with soluble carbohydrates during the phagocytosis assay did not alter the difference in phagocytosis observed between ΔlaeA and wild-type strains (Fig. 1B).
FIG. 1.
Surface carbohydrate analysis of A. fumigatus wild-type and ΔlaeA spores. (A) Flow cytometry was used to assess carbohydrate accessibility on the cell wall of resting spores. Shown are the results of six carbohydrate moieties analyzed (19 total). The gray lined and tinted histogram represents relative signal on wild-type spores, whereas the unfilled black histogram represents signal on ΔlaeA spores. At least 10,000 spores were counted per histogram, and the percentage of maximum on the x axis refers to the percentage of total counts for a given fluorescence intensity. Relative fluorescence intensity is shown on the y axis. (B) Soluble laminarin and mannan were used in phagocytosis assays to block peritoneal macrophage receptor recognition of carbohydrate ligands (n = 4 mice). Results are presented as the mean ± SEM of two independent experiments.
Here we note that for all experiments at least 100 macrophages were counted from triplicate wells per sample. The conidia/macrophage ratio used in these experiments resulted in equivalent numbers of wild-type or ΔlaeA conidia per macrophage but demonstrated a difference in the percentage of macrophages engulfing wild-type or ΔlaeA conidia. At larger conidia/macrophage ratios, the percent uptake was equivalent between strains, but increased numbers of ΔlaeA conidia were phagocytosed compared to wild-type conidia (data not shown). Since either ratio demonstrated increased phagocytosis of ΔlaeA conidia, we chose to examine one ratio for all experiments.
Investigating the role of secreted factors in spore uptake.
Having ruled out some common cell wall-associated carbohydrates often involved in phagocytosis, we suspected that wild-type or ΔlaeA spores may secrete a factor(s) that influences phagocytosis. To determine the potential for a secreted factor to alter uptake, we killed spores by formaldehyde fixation and subjected both live and killed spores to phagocytosis. Formaldehyde fixation had little effect on phagocytosis of spores, causing only a small decrease in both ΔlaeA and wild-type spores (Fig. 2). Although uptake of WT and WT fixed spores was significantly different (P = 0.0007), this could be the result of formaldehyde affecting the antigenic sites on WT spores more than ΔlaeA spores. Regardless, the reduced uptake of WT spores compared to ΔlaeA spores was retained whether the spores were live or killed.
FIG. 2.
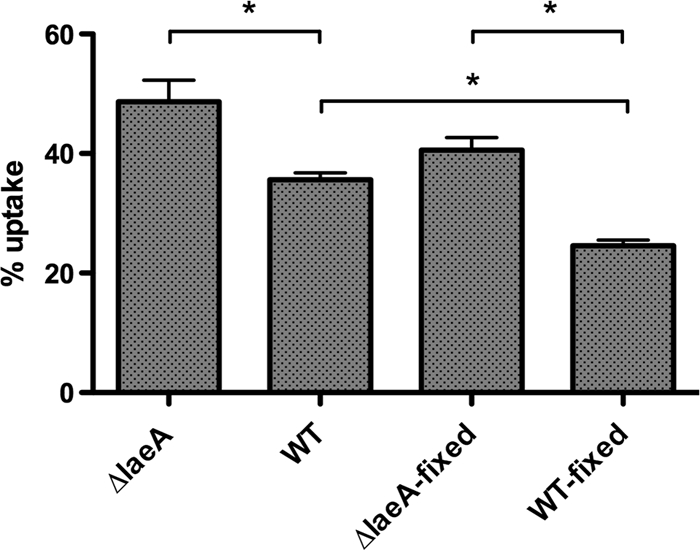
Macrophage phagocytosis comparison of live and dead spores. Spores were killed by formaldehyde fixation and washed at least five times before use. Peritoneal macrophages (n = 4 mice) were combined with spores for 1 h, washed, and incubated an additional 2 h before counterstaining with calcofluor white. Percent uptake was calculated as follows: (number of macrophages containing 1 or more spores/total number of macrophages counted) × 100. Results are presented as the mean percent uptake ± SEM of two independent experiments. *, P < 0.05.
We then examined uptake of live spores from both strains in the presence of methanol-derived metabolite extracts. Previous studies suggested a role for fungal metabolites as modulators of macrophage phagocytosis (2, 14); however, we were unable to identify a contribution of either ΔlaeA or wild-type metabolites to uptake at several concentrations tested (Fig. 3A and data not shown). To further identify whether ΔlaeA-associated products could influence uptake, we utilized a transwell system in which small, soluble compounds are able to migrate between an upper transwell and lower chamber, whereas spores are prevented from moving between chambers. In the lower chamber, macrophages adhered to coverslips were combined with live or dead wild-type spores, and live ΔlaeA spores (100-fold more than wild type) were placed in the upper transwell. Regardless of the presence of live ΔlaeA spores, wild-type spore uptake was unaffected (Fig. 3B). Phagocytosis assays were also performed to assess whether ΔlaeA or wild-type spores directly affect the rate of phagocytosis. Wild-type and ΔlaeA spores were labeled with different fluorophores and added in equivalent amounts to the same well containing macrophages. Phagocytosis of ΔlaeA spores remained significantly higher than wild type (Fig. 3C), suggesting that increased uptake of ΔlaeA spores is mediated by ΔlaeA-specific interactions.
FIG. 3.
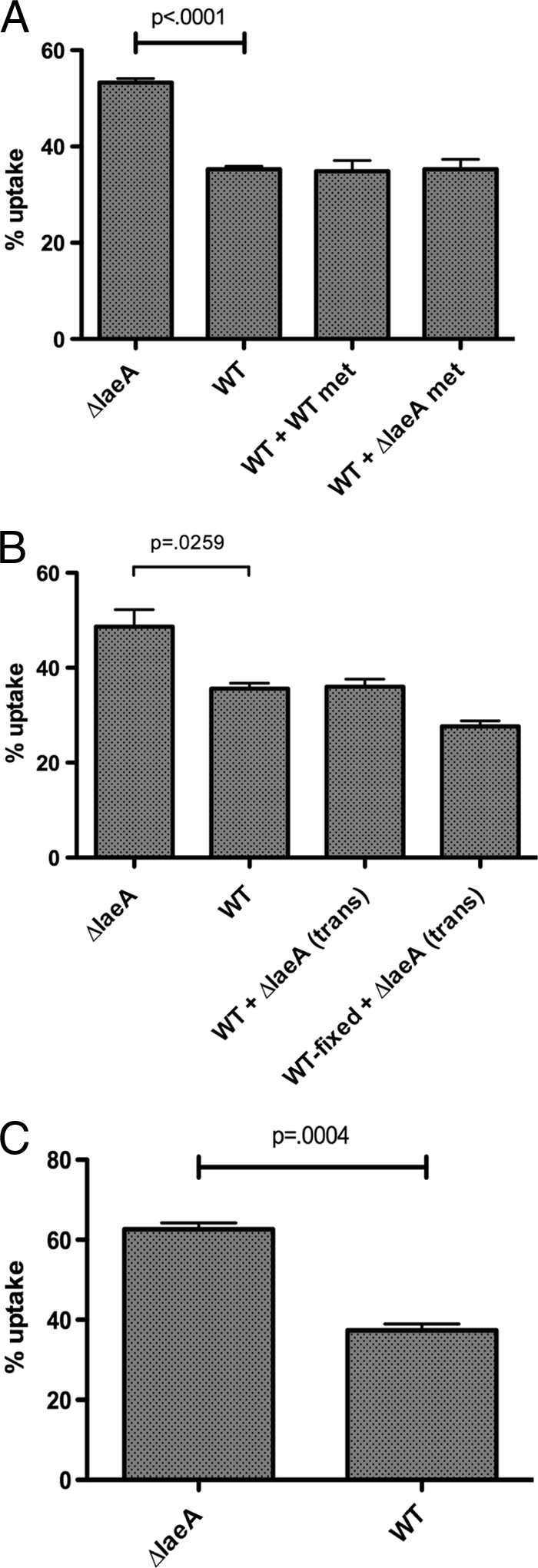
Increased uptake of ΔlaeA spores is not related to a secreted product. (A) Secondary metabolites extracted from wild-type or ΔlaeA GMM had no effect on uptake of wild-type spores. Results are shown for wild-type metabolite (WT met) and ΔlaeA metabolite (ΔlaeA met) extracts added to wild-type spores. Similarly, wild-type and ΔlaeA extracts added to ΔlaeA spores had no effect (data not shown). (B) At a 100-fold excess, ΔlaeA spores did not produce transwell-diffusible components to influence the phagocytosis of either live or dead wild-type spores. (C) When ΔlaeA and wild-type spores were combined in the same well with macrophages, phagocytosis of ΔlaeA spores remained significantly higher than wild type. At least two independent experiments were performed with peritoneal macrophages (n = 4 to 8 mice) for each panel. Results are presented as the mean percent uptake ± SEM.
The rodlet layer is disrupted in ΔlaeA spores.
A. fumigatus spores are enveloped in a hydrophobic rodlet layer comprised of RodA protein (e.g., RodAp) that enhances dispersal and protects against environmental stresses; another putative hydrophobin, RodBp, plays little or no role in the rodlet layer and hydrophobicity. Scanning electron microscopy analysis performed by Bok et al. (3) suggested an alteration in the spore cell wall, and this observation, coupled with the mischeduled RODA expression (albeit under liquid growth conditions, in contrast to use of solid growth medium in this study), warranted further investigation. During swelling, conidia release their rodlet layer, exposing other carbohydrate and protein moieties. To determine whether an altered rodlet layer in ΔlaeA spores may be contributing to differential uptake, we first compared uptake of resting and swollen spores from wild-type and ΔlaeA strains. In contrast to resting spores, phagocytosis of swollen ΔlaeA and wild-type spores was similar, suggesting that differential uptake could be specific to the rodlet layer of resting spores (Fig. 4A).
FIG. 4.
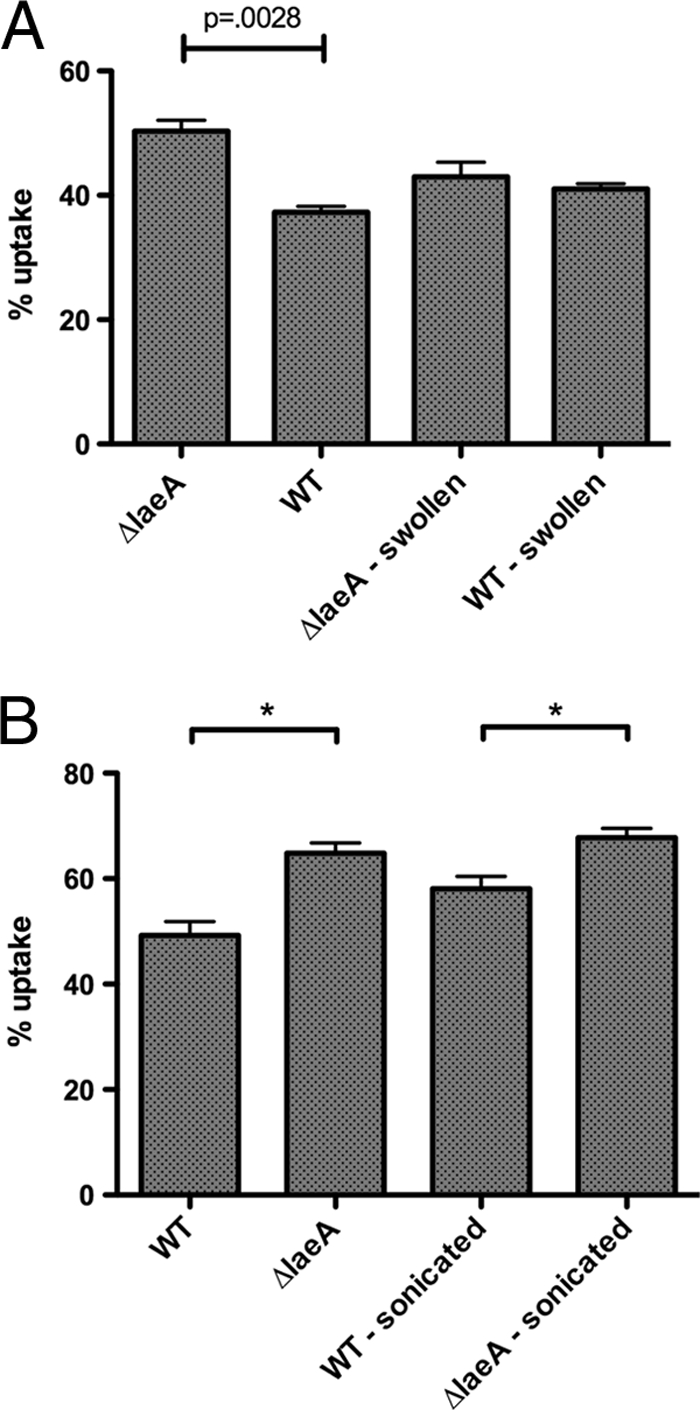
An altered rodlet layer may be responsible for increased uptake of ΔlaeA spores. (A) Swollen spores were prepared by incubating in RPMI 10 for 7 h at 300 rpm. Conidial swelling, which results in loss of the rodlet layer, eliminated the difference in peritoneal macrophage uptake between wild-type and ΔlaeA spores. (B) Resting spores were sonicated to remove the rodlet layer, and uptake of sonicated spores was compared to resting spores. The difference in uptake between wild-type and ΔlaeA spores decreased following sonication. Results of two independent experiments per figure (n = 4 mice per experiment) are shown as the mean percent uptake ± SEM. *, P < 0.05.
To further examine the possible role of an impaired rodlet layer in the enhanced ΔlaeA phagocytosis, we sonicated resting spores to release rodlet proteins from the conidial surface (modified from the method described by Paris et al. [16]). However, sonication did not significantly alter spore uptake of WT or ΔlaeA conidia (Fig. 4B). Additionally, the difference between wild-type and ΔlaeA spore uptake was not eliminated following sonication, which may have been due to the incomplete elimination of the rodlet layer of wild-type spores by this method.
Previous studies of rodlet mutants demonstrated the importance of RodAp, but not RodBp, in formation of the rodlet layer and protection against macrophage killing (16). We examined the total RodAp content in wild-type and ΔlaeA spores (Fig. 5). RodAp extracted from wild-type spores using hydrofluoric acid (HF) (1) was significantly greater (∼60%; P > 0.05) than from ΔlaeA spores, as demonstrated by Bradford assay (where 1 × 108 wild-type conidia contained 24.9 ± 0.3 μg of RodAp and ΔlaeA contained 10.3 ± 0.4 μg as determined in three individual experiments) and silver staining (Fig. 5) of the total protein extracted from each strain. To specifically address the contribution of rodlets to the differential uptake, HF-treated conidia lacking surface hydrophobins were tested for their phagocytosis. Macrophage uptake of HF-treated conidia was similar between wild-type and ΔlaeA strains (Fig. 6), in contrast to uptake of living or fixed conidia (Fig. 2). Together, these results implicate perturbations of the hydrophobic rodlet layer in the increased phagocytosis of ΔlaeA spores.
FIG. 5.
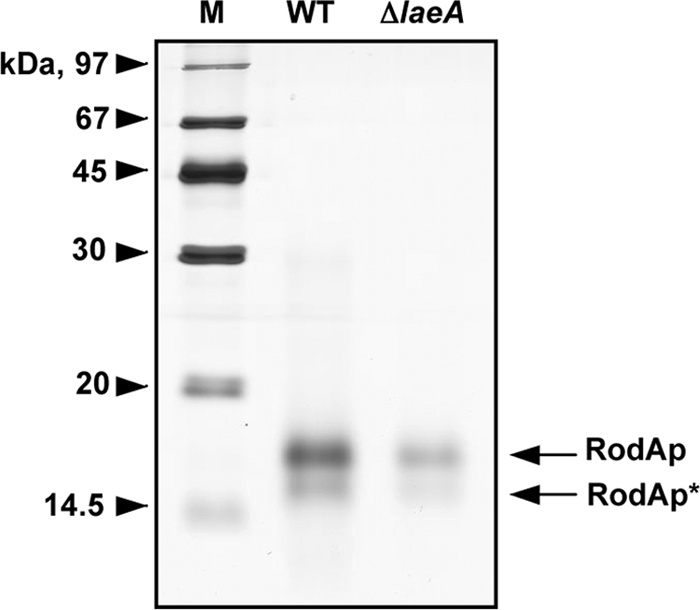
RodAp content is reduced in ΔlaeA spores compared to wild type. An SDS-PAGE (15%) profile is shown for the rodlets extracted from 1 × 107 wild-type or ΔlaeA spores using hydrofluoric acid, wherein the RodAp level was decreased in the ΔlaeA spores.
FIG. 6.
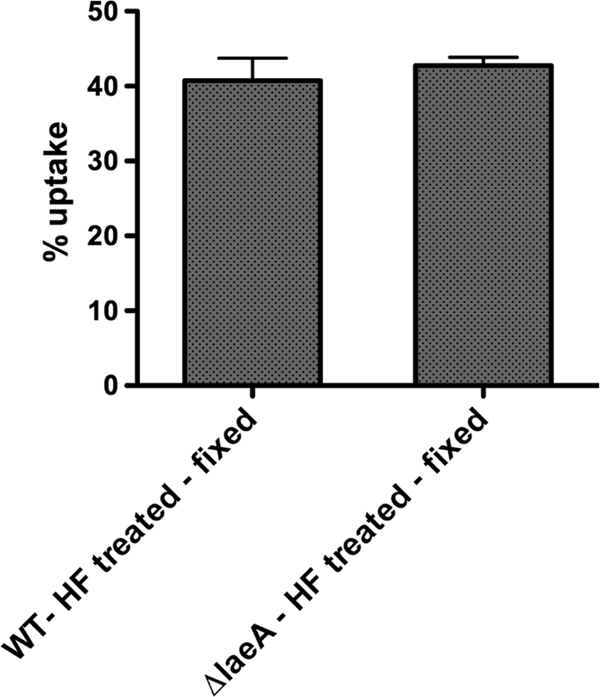
Removal of the RodAp layer from wild-type spores results in similar phagocytosis rates as with ΔlaeA spores. Resting wild-type and ΔlaeA conidia were treated with hydrofluoric acid to remove the surface rodlet layer, fixed with p-formaldehyde, and examined for phagocytic uptake by peritoneal macrophages (n = 5 mice). Results from two independent experiments are shown as the mean percent uptake ± SEM.
DISCUSSION
A primary role of the innate immune response is phagocytosis of microbes. The typical route of infection by A. fumigatus is through pulmonary exposure, where alveolar macrophages are responsible for the phagocytosis and killing of Aspergillus spores. Accordingly, a decrease or increase in spore phagocytosis may be associated with decreased or increased virulence, respectively. Strides in understanding the fungal factors involved in the engulfment process have been largely achieved through immunological and biochemical studies of Aspergillus mutant, and wild-type, host-cell interactions.
Several A. fumigatus mutations have resulted in altered in vitro and in vivo macrophage interactions. Our focus here was on the A. fumigatus ΔlaeA deletant. Removal of the laeA gene results in reduced virulence of two distinct strains of A. fumigatus (3, 19). In the Af293 background, spore morphology was affected and ΔlaeA spores were phagocytosed at strikingly higher rates (3). At the time, we speculated that an altered profile of secreted metabolites from the ΔlaeA spore could impact the ability of host phagocytic cells to ingest spores and/or, because expression of RODA was altered in the ΔlaeA mutant, this could contribute to aberrancies in engulfment. The similarity of gross appearance of ΔlaeA spores to ΔpksP (alb1) spores also suggested the possibility that increased β-(1,3)-glucan accessibility could be the root cause of increased uptake of ΔlaeA resting spores based on findings from Luther et al. (13) on increased ΔpksP phagocytosis.
The possibility that differences in common cell wall ligands could be responsible for the enhanced phagocytosis of the ΔlaeA spores was examined for a multitude of carbohydrates, including β-(1,3)-glucans, α-mannose, chitin, sialic acid, and other less commonly found ligands. However, flow cytometry results did not support this hypothesis, because both wild-type and ΔlaeA spores exhibited nearly identical cell wall properties (Fig. 1A). Rejection of the hypothesis was strengthened for two highly significant MAMPs, β(1,3)-glucans and α-mannose, by receptor blocking assays in which uptake of spores was not altered in ΔlaeA (Fig. 1B). These data led us to conclude that surface carbohydrate content does not contribute to ΔlaeA spore susceptibility to phagocytosis.
LaeA was first identified as a global regulator of secondary metabolite production (3, 4), and the possibility of loss of an antiphagocytic or gain of a prophagocytic metabolite(s) also seemed a plausible explanation for laeA mutant uptake. In support of this hypothesis, at least two studies had previously implicated diffused factors important in conidial engulfment (2, 14). However, we found no evidence for a role for secreted metabolites in the differential uptake of ΔlaeA and wild-type spores in either spent medium exchange, transwell, or mixed inoculum studies (Fig. 3A to C, respectively). While this study did not rule out a role for small molecule mediation of conidial engagement, it did not lend support for a role for LaeA in such a function.
An important clue in ΔlaeA spore-macrophage interactions came from observations of differential phagocytosis of resting and swollen conidia. In contrast to the consistent enhanced uptake of resting ΔlaeA spores, there was no statistical difference in the percent uptake of swollen ΔlaeA spores compared to comparable wild-type cells (Fig. 4A). Other studies have attributed an increase in engulfment of wild-type swollen spores to the increased exposure of β-(1,3)-glucans (13), mediated, at least in part, by the loss of spore rodlet layers (16). A. fumigatus contains two hydrophobins, RodAp and RodBp, but it is RodAp that solely contributes to the hydrophobin layer of the spore (16). Most recently, RodAp has been shown to be immunologically inert, blocking host cell activation (1). This finding, together with its hydrophobic properties and masking of β-(1,3)-glucans, implicates RodAp as a deterrent to host immune defenses. Because earlier studies had indicated mischeduled expression of RODA in the ΔlaeA mutant—although under different growth conditions than in this study—it seemed possible that RodAp could be playing a role in ΔlaeA mutant phagocytosis. We first asked whether sonication of the resting spores, presumed to remove RodAp, would affect the differential uptake of mutant and wild-type spores (Fig. 4B). Although there were no significant differences between sonicated wild-type and ΔlaeA spores, there was a trend for the wild type to approach ΔlaeA levels after 20 min of sonication. More importantly, protein analysis of ΔlaeA spores showed an approximately 60% decrease in the RodAp level compared to that of wild-type spores (Fig. 5).
As a final examination of a role for RodAp in abberant ΔlaeA spore uptake, we compared phagocytosis of untreated fixed conidia (Fig. 2) and hydrofluoric acid-treated fixed conidia (Fig. 6). The removal of the rodlet layer by this latter method eliminated the difference in uptake between wild-type and ΔlaeA conidia, supporting our hypothesis that a decreased rodlet layer contributes to the increased uptake of ΔlaeA conidia.
The RodA protein is one of two hydrophobins found in A. fumigatus spores. The resting spore is covered with a layer of hydrophobic rodlets composed of RodAp, which is disrupted and released during germination of the spore (6). The second hydrophobin, RodBp, is not required for rodlet formation (16). Deletion of RODA but not RODB results in increased spore killing by macrophages (16) and, most recently, RodAp has been demonstrated to block host cell recognition of A. fumigatus spores (1). Our data here suggest that RodAp also contributes to the ability of macrophages to internalize A. fumigatus spores. This “antiphagocytic” property of RodAp appears to be independent of β-(1,3)-glucan availability as assessed in flow cytometry and blocking experiments, although some alteration of β-(1,3)-glucan presentation to a receptor(s) by RodAp cannot be ruled out. Decreased hydrophobicity of the ΔlaeA spores due to decreased RodAp content could contribute to the enhanced uptake of the mutant spore as reported previously (3). Regardless of mechanism, and with the knowledge that we cannot completely rule out all other possible phenomena (e.g., specific ΔlaeA spore uptake enhanced by natural opsonins), the results as presented here strongly implicate RodAp in microbial evasion of phagocytic processes of the host cell. We suggest hydrophobin masking may be a common avoidance tactic of fungal pathogens.
Acknowledgments
We thank Angie Pollard and Laura Knoll for assistance with animal and cell preparations and John Bogdanske and the University of Wisconsin RARC staff for assistance in confirmatory experiments.
This research was funded by NIH 1 R01 Al065728-01 to N.P.K. and EU-STREP Fungwall LSHB-CT-2004-511952 to J.-P.L.
Editor: G. S. Deepe, Jr.
Footnotes
Published ahead of print on 16 November 2009.
REFERENCES
- 1.Aimanianda, V., J. Bayry, S. Bozza, O. Kniemeyer, K. Perruccio, S. R. Elluru, C. Clavaud, S. Paris, A. A. Brakhage, S. V. Kaveri, L. Romani, and J.-P. Latgé. 2009. Surface hydrophobin prevents immune recognition of airborne fungal spores. Nature 460:1117-1121. [DOI] [PubMed] [Google Scholar]
- 2.Bertout, S., C. Badoc, M. Mallie, J. Giaimis, and J. M. Bastide. 2002. Spore diffusate isolated from some strains of Aspergillus fumigatus inhibits phagocytosis by murine alveolar macrophages. FEMS Immunol. Med. Microbiol. 33:101-106. [DOI] [PubMed] [Google Scholar]
- 3.Bok, J. W., S. A. Balajee, K. A. Marr, D. Andes, K. F. Nielsen, J. C. Frisvad, and N. P. Keller. 2005. LaeA, a regulator of morphogenetic fungal virulence factors. Eukaryot. Cell 4:1574-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bok, J. W., and N. P. Keller. 2004. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell 3:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, G. D., and S. Gordon. 2001. Immune recognition. A new receptor for beta-glucans. Nature 413:36-37. [DOI] [PubMed] [Google Scholar]
- 6.Dague, E., D. Alsteens, J. P. Latge, and Y. F. Dufrene. 2008. High-resolution cell surface dynamics of germinating Aspergillus fumigatus conidia. Biophys. J. 94:656-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26:781-803. [DOI] [PubMed] [Google Scholar]
- 8.Didierlaurent, A., J. C. Sirard, J. P. Kraehenbuhl, and M. R. Neutra. 2002. How the gut senses its content. Cell. Microbiol. 4:61-72. [DOI] [PubMed] [Google Scholar]
- 9.Girardin, H., S. Paris, J. Rault, M. N. Bellon-Fontaine, and J. P. Latge. 1999. The role of the rodlet structure on the physicochemical properties of Aspergillus conidia. Lett. Appl. Microbiol. 29:364-369. [DOI] [PubMed] [Google Scholar]
- 10.Jahn, B., K. Langfelder, U. Schneider, C. Schindel, and A. A. Brakhage. 2002. PKSP-dependent reduction of phagolysosome fusion and intracellular kill of Aspergillus fumigatus conidia by human monocyte-derived macrophages. Cell. Microbiol. 4:793-803. [DOI] [PubMed] [Google Scholar]
- 11.Langfelder, K., B. Jahn, H. Gehringer, A. Schmidt, G. Wanner, and A. A. Brakhage. 1998. Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med. Microbiol. Immunol. 187:79-89. [DOI] [PubMed] [Google Scholar]
- 12.Lin, S. J., J. Schranz, and S. M. Teutsch. 2001. Aspergillosis case-fatality rate: systematic review of the literature. Clin. Infect. Dis. 32:358-366. [DOI] [PubMed] [Google Scholar]
- 13.Luther, K., A. Torosantucci, A. A. Brakhage, J. Heesemann, and F. Ebel. 2007. Phagocytosis of Aspergillus fumigatus conidia by murine macrophages involves recognition by the dectin-1 beta-glucan receptor and Toll-like receptor 2. Cell. Microbiol. 9:368-381. [DOI] [PubMed] [Google Scholar]
- 14.Mitchell, C. G., J. Slight, and K. Donaldson. 1997. Diffusible component from the spore surface of the fungus Aspergillus fumigatus which inhibits the macrophage oxidative burst is distinct from gliotoxin and other hyphal toxins. Thorax 52:796-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan, J., K. A. Wannemuehler, K. A. Marr, S. Hadley, D. P. Kontoyiannis, T. J. Walsh, S. K. Fridkin, P. G. Pappas, and D. W. Warnock. 2005. Incidence of invasive aspergillosis following hematopoietic stem cell and solid organ transplantation: interim results of a prospective multicenter surveillance program. Med. Mycol. 43(Suppl. 1):S49-S58. [DOI] [PubMed] [Google Scholar]
- 16.Paris, S., J. P. Debeaupuis, R. Crameri, M. Carey, F. Charles, M. C. Prevost, C. Schmitt, B. Philippe, and J. P. Latge. 2003. Conidial hydrophobins of Aspergillus fumigatus. Appl. Environ. Microbiol. 69:1581-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stevens, D. A., V. L. Kan, M. A. Judson, V. A. Morrison, S. Dummer, D. W. Denning, J. E. Bennett, T. J. Walsh, T. F. Patterson, and G. A. Pankey. 2000. Practice guidelines for diseases caused by Aspergillus. Infectious Diseases Society of America. Clin. Infect. Dis. 30:696-709. [DOI] [PubMed] [Google Scholar]
- 19.Sugui, J. A., J. Pardo, Y. C. Chang, A. Mullbacher, K. A. Zarember, E. M. Galvez, L. Brinster, P. Zerfas, J. I. Gallin, M. M. Simon, and K. J. Kwon-Chung. 2007. Role of laeA in the regulation of alb1, gliP, conidial morphology, and virulence in Aspergillus fumigatus. Eukaryot. Cell 6:1552-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thau, N., M. Monod, B. Crestani, C. Rolland, G. Tronchin, J. P. Latge, and S. Paris. 1994. Rodletless mutants of Aspergillus fumigatus. Infect. Immun. 62:4380-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai, H., M. Wheeler, Y. Chang, and K. Kwon-Chung. 1999. A developmentally regulated gene cluster involved in conidial biosynthesis in Aspergillus fumigatus. J. Bacteriol. 181:6469-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai, H.-F., R. G. Washburn, Y. C. Chang, and K. J. Kwon-Chung. 1997. Aspergillus fumigatus arp1 modulates conidial pigmentation and complement deposition. Mol. Microbiol. 26:175-183. [DOI] [PubMed] [Google Scholar]
- 23.Tsai, H. F., Y. C. Chang, R. G. Washburn, M. H. Wheeler, and K. J. Kwon-Chung. 1998. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J. Bacteriol. 180:3031-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]