Abstract
Cytolethal distending toxin (CDT) is a bacterial toxin that induces G2/M cell cycle arrest, cell distension, and/or apoptosis in mammalian cells. It is produced by several Gram-negative species and may contribute to their pathogenicity. The catalytic subunit CdtB has homology with DNase I and may act as a genotoxin. However, the mechanism by which CdtB leads to cell death is not yet clearly understood. Here, we used Saccharomyces cerevisiae as a model to study the molecular pathways involved in the function of CdtB from Aggregatibacter actinomycetemcomitans, a cause of aggressive periodontitis. We show that A. actinomycetemcomitans CdtB (AaCdtB) expression induces S/G2 arrest and death in a DNase-catalytic residue and nuclear localization-dependent manner in haploid yeasts. Yeast strains defective in homologous recombination (HR) repair, but not other DNA repair pathways, are hypersensitive to AaCdtB, suggesting that HR is required for survival upon CdtB expression. In addition, yeast does not harbor the substrate for the other activity proposed for CdtB function, which is phosphatidylinositol-3,4,5-triphosphate phosphatase. Thus, these results suggest that direct DNA-damaging activity alone is sufficient for CdtB toxicity. To investigate how CdtB induces cell death, we examined the effect of CdtB in yeast strains with mutations in apoptotic regulators. Our results suggest that yeast death occurs independently of the yeast metacaspase gene YCA1 and the apoptosis-inducing factor AIF1 but is partially dependent on histone H2B serine 10 phosphorylation. Therefore, we report here the evidence that AaCdtB causes DNA damage that leads to nonapoptotic death in yeast and the first mutation that confers resistance to CdtB.
Aggregatibacter actinomycetemcomitans is a Gram-negative bacterial species that has been implicated in the pathogenesis of periodontal diseases, especially the aggressive forms (12). This bacterium possesses many virulence factors and produces several toxins, one of which is cytolethal distending toxin (CDT). CDT induces cell distension, cell cycle arrest, and death in mammalian host cells. It is produced by several pathogenic bacterial species, including Escherichia coli, Campylobacter jejuni, Haemophilus ducreyi, Shigella dysenteriae, Helicobacter hepaticus, Salmonella enterica serovar Typhi, and others (16, 28). A. actinomycetemcomitans CDT (AaCDT) has been shown to induce G2 cell cycle arrest and/or apoptosis in many cell types, including lymphocytes (16, 26, 28, 34). Therefore, this toxin may play a role in bacterial pathogenicity via immune system evasion. Moreover, the ability to induce host cell death could lead to tissue destruction and delayed healing. In the case of periodontitis, this could lead to eventual tooth loss. Clinical isolates of A. actinomycetemcomitans from patients with periodontitis show high frequency of CDT production, further suggesting its role in the pathogenesis of the disease (43).
The genes that encode the 3 subunits of CDT, cdtA, -B, and -C, are located in the cdt locus (40). Structural and functional studies suggested that the 3 subunits form a heterotrimer of CDT holotoxin and that all subunits are required for full activity (15, 31, 33). CdtB is the enzymatically active subunit, while CdtA and CdtC are necessary for the delivery of CdtB into host cells (16). CdtB has only limited homology with DNase I, but the residues important in the catalytic activity are well conserved. DNase activity is detected in crude CDT preparation in vitro, and mutations of the residues corresponding to the DNase-active site abolish the cytotoxic effects on cultured cells (8, 14). This suggests that the DNase activity of CdtB is essential for its cytotoxicity. Nevertheless, CdtB has also been proposed to induce cell cycle arrest through its function as a phosphatidylinositol-3,4,5-triphosphate phosphatase (35). Since the residues important for both DNase and phosphatase activities largely overlap, this raises a question as to which activity is the major contributor to the effects of CDT.
Budding yeast (Saccharomyces cerevisiae) has served as an excellent model organism for the study of many biological processes, including cell cycle regulation, DNA repair, and even cell death (21). In recent years, yeast has also proven to be a useful surrogate host for the characterization of several bacterial effectors that target conserved biological processes in eukaryotic host cells (6, 39). Therefore, yeast appears to be a particularly interesting model for the elucidation of the effects of CDT on cellular processes that regulate cell cycle and death. Campylobacter jejuni CdtB (CjCdtB) has been shown to induce G2 cell cycle arrest, chromosome degradation, and loss of viability upon expression in a yeast model (10). Many genomic tools available in yeast also provide an opportunity to analyze the effects of CDT in a genome-wide fashion (13). Previous studies of yeast suggest that CdtB likely acts as a genotoxin. However, the mechanism by which CdtB induces cell cycle arrest and death has not yet been fully elucidated. Here, we used yeast as a model to characterize the mechanisms by which AaCdtB expression induces cell death.
MATERIALS AND METHODS
Plasmids and yeast strains.
Wild-type cdtB and the H160A, H274A, and Δ11aa cdtB mutants of Aggregatibacter actinomycetemcomitans with a 6-histidine tag were obtained from pET28a-CdtB (40) and pET28a-CdtB(H160A), pET28a-CdtB(H274A) (27), and pET28a-CdtB(Δ11aa) (25), respectively. The open reading frames were amplified with forward primer Kpn-Kozak-CdtB (5′-GGT ACC ACC ATG G GC AGC-3′) and the T7 reverse primer, cloned into pGEM-Teasy (Promega), and subsequently cloned into the KpnI-XhoI sites of the pYES2 vector (Invitrogen). The resulting plasmids were transformed into wild-type S. cerevisiae strains (BY4741 and W303-1A) and yeast strains with mutations in genes of interest (Table 1). Plasmids for the overexpression of human Bcl-2 and Bcl-xL, including a vector control (pFL39), were kind gifts from Stephen Manon. All recombinant DNA and yeast manipulations were performed according to standard protocols (3).
TABLE 1.
Yeast strains used in this study
| Strain(s) | Genotype | Source or reference |
|---|---|---|
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | S. Buratowski |
| YF1367 (rad51Δ) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0rad51Δ::KanMX | S. Buratowski |
| YFM 81 (ntg1Δ) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0ntg1Δ::KanMX | G. R. Fink |
| YFM 82 (ntg2Δ) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0ntg2Δ::KanMX | G. R. Fink |
| YFM 83 (rad14Δ) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0rad14Δ::KanMX | G. R. Fink |
| YFM 84 (rad23Δ) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0rad23Δ::KanMX | G. R. Fink |
| YFM 85 (yku70Δ) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0yku70Δ::KanMX | G. R. Fink |
| YFM 86 (dnl4Δ) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0dnl4Δ::KanMX | G. R. Fink |
| YFM 87 (mre11Δ) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0mre11Δ::KanMX | M. C. Keogh |
| YFM 96 (rad50Δ) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0rad50Δ::KanMX | G. R. Fink |
| YFM 97 (xrs2Δ) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0xrs2Δ::KanMX | G. R. Fink |
| YFM 98 (nej1Δ) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0nej1Δ::KanMX | G. R. Fink |
| YFM 99 (chk1Δ) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0chk1Δ::KanMX | G. R. Fink |
| YFC155, W303-1A | MATahis3-11,15 leu2-3,112 ura3-1 trp1-1 (W303 background) | E. J. Cho |
| YFC156, U960-5C (rad53Δ) | MATahis3-11,15 leu2-3,112 ura3-1 trp1-1 rad53::HIS3 sml1-1, RAD5+ (W303 background) | E. J. Cho |
| SAY2 [H2B(S10A)] | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hht1-hhf1::KAN hhf-2hht2::NAT hta1-htb1::HPH hta2-htb2::NAT pSA17[CEN LEU2 HTA1-htb1-S10A HHT2-HHF2] | 1 |
| SAY152 (yca1Δ) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 yca1Δ::kanMX6 hht1-hhf1::KAN hhf-2hht2::NAT hta1-htb1::HPH hta2-htb2::NAT pQQ18[CEN LEU2 HTA1-HTB1 HHT2-HHF2] | 1 |
| YFM88 (aif1Δ) | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0aif1Δ::KanMX | G. R. Fink |
Growth conditions and plate sensitivity assays.
Yeast cells were grown in synthetic complete medium without uracil (SC-Ura) supplemented with 2% raffinose (Fluka) to log phase (optical density at 600 nm [OD600] = 0.4 to 0.6). For the time course experiments, cells were transferred to SC-Ura supplemented with 2% galactose (Fluka) to induce the expression of CdtB. At various time points, 10-fold serial dilutions of cells were spotted on SC-Ura plates supplemented with 2% glucose to repress CdtB expression or were harvested for other assays. For plate sensitivity assays, 10-fold serial dilutions of cells were spotted on SC-Ura plates supplemented with 2% galactose for CdtB induction and on SC-Ura plates supplemented with 2% glucose as controls. In survival plating assays, approximately 500 cells were spread onto SC-Ura plates supplemented with 2% galactose and with 2% glucose. To examine the effects of different levels of CdtB expression, yeast cells were spotted or spread onto SC-Ura plates supplemented with different sugar concentrations to vary the degrees of induction, i.e., 2% glucose, 1.9% galactose plus 0.1% glucose, 0.1% galactose plus 1.9% raffinose, or 2% galactose. All experiments were performed at least in triplicate. To statistically evaluate the differences in survival rate between CdtB-expressing cells and controls or between mutant and wild-type strains, the t test was used for normal data and the Mann-Whitney U test for skewed data.
Immunoblot analysis.
Yeast cell extracts were prepared using glass bead disruption according to standard protocols and resolved on SDS-PAGE gel (3). The expression levels of CdtB were examined by immunoblotting with a mouse monoclonal antibody against the 6-His tag conjugated with horseradish peroxidase (Abcam, United Kingdom) or a rabbit anti-AaCdtB antiserum (40).
Propidium iodide staining and cell cycle analysis by flow cytometry.
Yeast cells were taken at the indicated time points after the induction of CdtB expression and were fixed with 70% ethanol at 4°C overnight or at least 1 h at room temperature. Propidium iodide staining was carried out as previously described (24). Flow cytometry was performed with a Becton Dickinson FACSCalibur and analyzed with Cell Quest Pro and ModFitLT software.
ROS and annexin V staining.
Dihydroethidium (DHE; Sigma), dihydrorhodamine (DHR; Sigma), and annexin V-Fluos (Roche) with propidium iodide (Sigma) were used to stain for accumulation of reactive oxygen species (ROS) and externalization of phosphatidylserine, respectively. The staining procedures were as previously described (18, 19). Cells treated with 3 mM hydrogen peroxide for 200 min were used as positive controls in both experiments (19).
RESULTS
Expression of AaCdtB leads to yeast cell death.
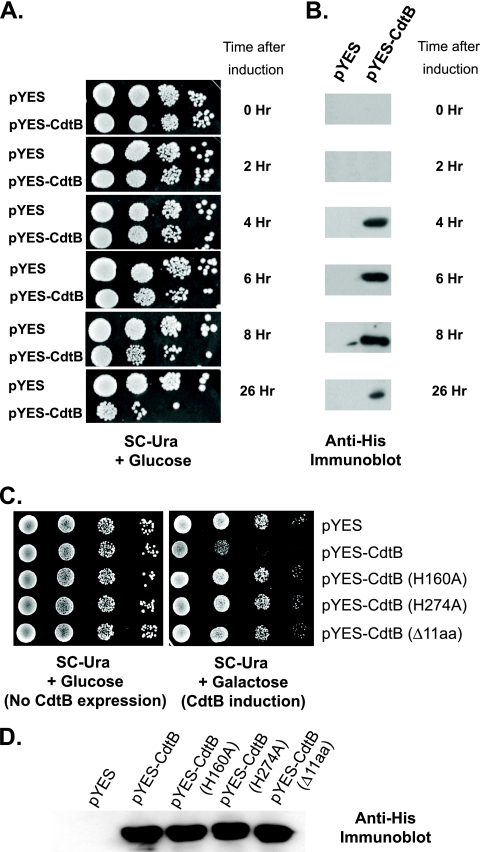
To determine if we can use yeast as a model to study the cytotoxic mechanism of AaCdtB, we first tested if AaCdtB induces yeast cell death. We transformed a haploid wild-type yeast strain (BY4741) with a yeast expression plasmid carrying the cdtB gene with a 6-histidine tag under the control of a galactose-inducible promoter (pYES-CdtB). Transformants were then grown in selective medium containing raffinose, which neither induces nor represses the galactose-inducible promoter that governs CdtB expression. When the cultures reached log phase, cells were transferred to liquid medium containing galactose to induce CdtB expression. At various time points, cells were spotted in 10-fold serial dilutions on solid medium supplemented with glucose to repress further expression of CdtB. At the same time, a portion of the cultures was harvested for protein extraction for the detection of CdtB levels by immunoblotting. As shown in Fig. 1A, cells expressing CdtB started to show lower levels of growth at 6 h after induction and the effect became more apparent at 8 h after induction. This result correlates with the expression of CdtB, which was detectable by immunoblotting at 2 to 4 h after induction (Fig. 1B).
FIG. 1.
Expression of AaCdtB is toxic to yeast cells. (A) Log-phase cultures of yeast with pYES and pYES-CdtB were transferred to medium containing galactose to induce CdtB expression. At various time points after CdtB induction, cells were spotted on medium containing glucose to repress further CdtB expression. (B) A portion of cells from the time course experiment represented in panel A was used for protein extraction and analyzed by SDS-PAGE. CdtB expression was detected by anti-His immunoblot analysis. (C) Wild-type yeast cells (BY4741) harboring pYES vector, pYES-CdtB, or pYES-CdtB mutants (H160A, H274A, or Δ11aa) were grown to log phase in liquid medium supplemented with raffinose and spotted on medium containing galactose (CdtB-inducing condition), with medium containing glucose used as a control. Significantly less growth was observed in cells expressing CdtB but not CdtB mutants. (D) Immunoblot analysis of yeast extracts from cultures expressing different forms of CdtB at 4 h after induction showed similar levels of expression. Representative photographs of results for at least three experiments are shown.
In a plate sensitivity assay, log-phase cultures of yeast cells were spotted directly on solid medium containing galactose to continuously induce CdtB expression. After 2 to 3 days of incubation, we observed markedly decreased growth in cells expressing CdtB in comparison to that in cells carrying the empty-vector control (pYES), as shown in Fig. 1C. Furthermore, the toxicity of CdtB is abolished when the conserved DNase I catalytic residues of CdtB are mutated since cells expressing the CdtB(H160A) or CdtB(H274A) mutants grew as well as those carrying the empty vector. Importantly, a deletion of the 11 amino acids (Δ11aa; aa 114 to 124) required for nuclear transport of CdtB also abolished the cytotoxicity in yeasts (25). These results are consistent with the effects observed with mammalian cell cultures (25, 27) (M. Ohara, unpublished) and suggest that the cytotoxicity of CdtB is dependent on its DNase activity and requires nuclear localization. Figure 1D shows an immunoblot analysis of yeast extracts expressing various forms of CdtB at 4 h after induction. The result indicates that all mutants were expressed at levels similar to that of wild-type CdtB, confirming that the defect in cytotoxicity was due to the mutations and not to a lower expression level.
Since the level of AaCdtB expression under the induction with 2% galactose may be very high, we examined the effects of AaCdtB expression under lower levels of induction by varying the concentrations of galactose and glucose in the medium. We found that AaCdtB inhibits yeast growth in a dose-dependent manner (data not shown). Under the conditions tested, the addition of 0.1% glucose to the medium supplemented with 1.9% galactose provided the lowest degree of induction. The level of AaCdtB expression under this condition is approximately 0.02 to 0.03 μg/10 μg total protein in yeast whole-cell extract obtained from ∼107 cells at 4 h after induction (assuming 50% extraction efficiency). This estimation was obtained by comparing the intensities of immunoblot signals of AaCdtB in the whole-cell extracts with those of purified recombinant AaCdtB. The presence of 0.1% galactose in the medium supplemented with 1.9% raffinose led to a strong induction (∼0.1 μg/10 μg total protein), but to a lesser extent than 2% galactose (∼0.15 μg/10 μg total protein). Using plate sensitivity and survival plating assays, we observed that the levels of toxicity to yeast under different inducing conditions correlate well with the levels of AaCdtB expression. At the lowest expression level examined (1.9% galactose-0.1% glucose), although the number of surviving colonies in survival plating assays was not significantly lower than that of the vector controls, the sizes of the colonies were noticeably smaller. At the higher expression levels, survival plating assays showed statistically significant reductions in the numbers of surviving colonies on inducing plates when CdtB-expressing cells and the vector controls under the same conditions were compared.
CdtB induces S/G2 cell cycle arrest.
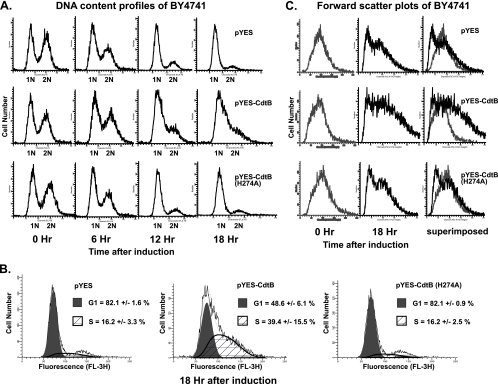
AaCDT has been shown to induce G1 or G2/M cell cycle arrest and/or apoptosis in many mammalian cell types. To examine the effects of AaCdtB expression on cell cycle progression in yeast, we performed propidium iodide staining and flow cytometric analyses of asynchronous cultures after CdtB induction. As shown in Fig. 2A, CdtB expression led to an S/G2 cell cycle arrest in our haploid yeast system. In log-phase asynchronous cultures, actively dividing cells were distributed in G1 (1-N DNA content), S (DNA content between 1 N and 2 N), and G2/M (2-N DNA content) at the time of induction (zero hour). As cultures grew toward stationary phase at 10 to 12 h after induction, the DNA content profiles of control cells showed higher proportions of cells accumulated in G0/G1 phase (1 N). On the other hand, CdtB-expressing cells started to accumulate in S and G2 phases at 10 to 12 h after induction of CdtB expression. An analysis of the DNA content profiles suggests that the proportion of cells in S phase in CdtB-expressing cells was higher than that of the control cells (means ± standard deviations [SD] = 39.4% ± 15.5% in CdtB versus 16.2% ± 3.3% in the vector control) (Fig. 2B). Furthermore, microscopic analysis showed that CdtB-expressing cells were enlarged, and the majority accumulated as large budded cells, consistent with an S/G2 arrest phenotype (data not shown). An accumulation of enlarged cells in the CdtB-expressing population was also demonstrated by flow cytometric analysis. Forward-scatter histograms, which reflect cell sizes, showed an increase in cell numbers with high forward-scatter values in cells with CdtB expression in comparison to the levels for the controls (Fig. 2C).
FIG. 2.
AaCdtB induces S/G2 arrest in haploid yeast cells. (A) DNA content profiles from flow cytometric analyses of BY4741 carrying pYES control (upper panel), pYES-CdtB (middle panel), and pYES-CdtB(H274A) (lower panel) at different time points after CdtB induction. Profiles shown are representative of at least 3 independent experiments. (B) Quantitative analysis of DNA content profiles with ModFitLT software showed a higher proportion of cells accumulated in S phase in CdtB-expressing cells. Percentages of cells in G1 (shaded in gray) and S (striped area) phases are shown (mean ± SD). (C) Forward-scatter histograms showed an accumulation of enlarged cells in the CdtB-expressing population. Light gray lines represent cells before induction (zero hour), and black lines represent cells at 18 h after induction. The rightmost panel shows 18-h histograms superimposed over 0-h histograms.
Yeast strains with defects in homologous recombination (HR) repair, but not other repair pathways, are hypersensitive to CdtB.
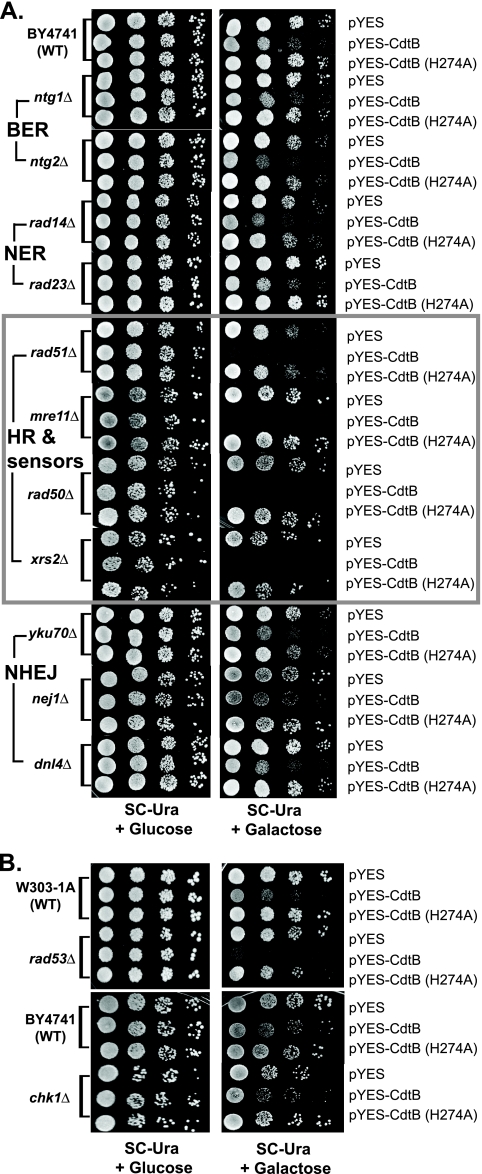
Our initial experiments showed that the DNase I catalytic residues (H160 and H274) and nuclear localization are essential for CdtB cytotoxicity (Fig. 1C). This is in agreement with the hypothesis that CdtB induces cell cycle arrest/apoptosis through its ability to generate DNA damage. To further test this hypothesis, we employed yeast strains with deletions of genes involved in various DNA repair pathways. We hypothesized that defects in DNA repair should increase the sensitivity to CdtB if the cytotoxic effect of CdtB is due to its DNA-damaging activity. Moreover, since each of the DNA repair pathways is specific for distinct types of DNA lesions, the requirement for a particular pathway for survival upon CdtB expression would also imply the type of DNA lesions CdtB generates.
Using plate sensitivity assays, we examined the cytotoxicity of CdtB expression in representative yeast strains with defects in base excision repair (BER; ntg1Δ and ntg2Δ), nucleotide excision repair (NER; rad14Δ and rad23Δ), homologous recombination (HR; rad51Δ) and components of the MRX DNA break sensor complex (mre11Δ, rad50Δ, and xrs2Δ), or nonhomologous end joining (NHEJ; yku70Δ, nej1Δ, and dnl4Δ). Remarkably, only strains with defects in the HR repair pathway and the MRX complex, not those with defects in other repair pathways, were significantly hypersensitive to CdtB (Fig. 3A). In strains with the deletion of RAD51, MRE11, RAD50, or XRS2, genes essential for homologous recombination repair of a DNA double-strand break (DSB), there was essentially no survival upon CdtB induction in galactose-containing medium. These results indicate that CdtB expression causes DNA damage, likely in the form of DNA breaks that requires HR for repair, in yeast cells and leads to eventual cell death.
FIG. 3.
Yeast strains defective in homologous recombination, but not other repair pathways, and the DNA damage checkpoint are hypersensitive to CdtB. (A) Plate sensitivity assays showed the sensitivities of different yeast strains with deletions in DNA repair pathways to CdtB in comparison to that of wild-type (WT) yeast (BY4741). Among the tested strains, only strains with the deletions of RAD51 and the MRX DNA break sensor complex components are hypersensitive to CdtB expression induced on a galactose plate. Glucose plates served as controls. BER, base excision repair; NER, nucleotide excision repair; HR, homologous recombination; NHEJ, nonhomologous end joining. (B) A plate sensitivity assay of two DNA damage checkpoint mutants showed that the rad53Δ strain is hypersensitive to CdtB in comparison to its wild-type parent strain (W303-1A) but that the chk1Δ strain is not. Representative photographs from at least three experiments are shown.
In response to DNA damage, cells also need to activate cell cycle checkpoint controls to delay cell cycle progression and allow time for the repair processes to take place. In S. cerevisiae, two DNA damage checkpoint effector kinases, Rad53 (homologue of mammalian Chk2) and Chk1, play a major role in the regulation of cell cycle checkpoints (4). If CdtB creates DNA damage, cell cycle checkpoint regulators should also be critical in the cellular responses for coping with the insults. To test this hypothesis, we examined the effects of CdtB in the rad53Δ and chk1Δ strains. As expected, the rad53Δ strain showed hypersensitivity to CdtB expression in comparison to its isogenic wild-type control, although to a lesser extent than the strains with defects in HR repair (Fig. 3B). In contrast, the chk1Δ strain showed the same level of sensitivity as its wild-type parent strain. These results suggest that Rad53 plays a more important role than Chk1 in the survival of yeast cells upon CdtB expression.
The effect of AaCdtB is enhanced in a DNA repair mutant.
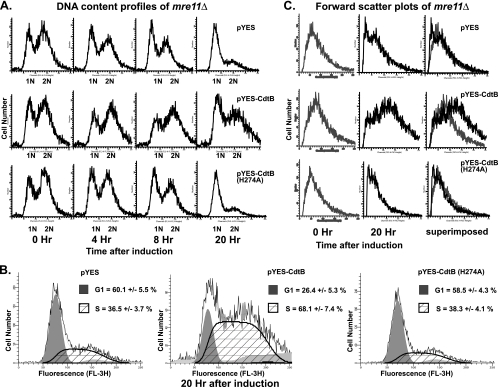
Since yeast strains with defects in HR repair are hypersensitive to CdtB, we examined the effects of CdtB expression on cell cycle progression in a repair-defective mutant strain, the mre11Δ strain. DNA content analyses showed that CdtB expression led to an accumulation of cells in the S/G2 phase of the cell cycle (Fig. 4A). Importantly, the effect was more pronounced and was noticeable at an earlier time than that observed in wild-type yeast. At 4 h after induction, DNA content profiles of CdtB-expressing cultures started to shift toward S/G2 phase (2 N), while this effect was not apparent in wild-type yeast cultures at 6 h after induction (Fig. 2A). An accumulation of cells in the S/G2 phase was clearly observed at 8 h after induction in the mre11Δ strain. An analysis of the DNA content profiles also confirmed the stronger effects in the mre11Δ strain with a high proportion of cells in accumulated in S phase (mean ± SD = 68.1% ± 7.4%) at 20 h after induction (Fig. 4B). Forward-scatter analyses also showed an accumulation of enlarged cells among the CdtB-expressing population (Fig. 4C).
FIG. 4.
S/G2 arrest is more pronounced in a DSB repair-defective strain. (A) DNA content profiles from flow cytometric analyses of the mre11Δ strain carrying the pYES control (upper panel), pYES-CdtB (middle panel), and pYES-CdtB(H274A) (lower panel) at different time points after CdtB induction. Profiles shown are representative of at least 3 independent experiments. (B) Quantitative analysis of the DNA content profile with ModFitLT software showed a higher proportion of cells accumulated in S phase in CdtB-expressing cells. Percentages of cells in G1 (shaded in gray) and S (striped area) phases are shown (mean ± SD). (C) Forward-scatter histograms showed an accumulation of enlarged cells in the CdtB-expressing population. Light gray lines represent cells before induction (zero hour), and black lines represent cells at 20 h after induction. The rightmost panel shows 20-h histograms superimposed over 0-h histograms.
AaCdtB-induced yeast cell death is independent of many apoptotic regulators but is partially dependent on histone H2B phosphorylation.
CDT can induce apoptosis in certain mammalian cell types, particularly in lymphocytes (36). Although yeast is a unicellular organism, it has been shown to undergo apoptosis-like death upon exposure to certain stress conditions (21). To determine the mode of cell death induced by CdtB, we examined if yeast death due to CdtB expression is apoptotic. We first examined if there was an accumulation of reactive oxygen species (ROS), a feature of apoptotic death, upon CdtB expression. Staining with dihydroethidium (DHE) or dihydrorhodamine (DHR) failed to demonstrate ROS accumulation after CdtB induction as analyzed by flow cytometry (data not shown). We also did not observe positive staining of annexin V, which detects the externalization of phosphatidylserine on the cell membrane (data not shown). These experiments were performed using cells treated with hydrogen peroxide, a known apoptotic inducer in yeast, as positive controls to ensure that the staining procedures were effective.
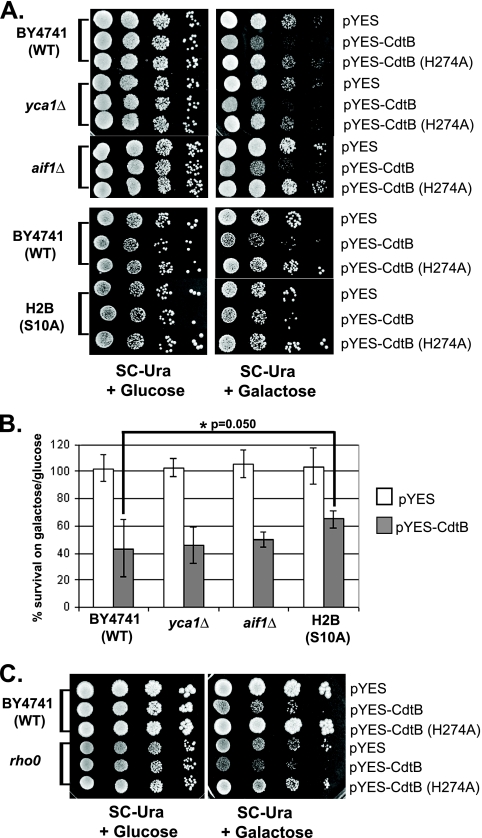
To further characterize the mode of cell death induced by CdtB, the levels of cytotoxicity in yeast strains with mutations in apoptotic regulators were determined. Yeast harbors a single metacaspase gene, YCA1, which has been shown to play an important role in apoptotic death under several conditions (20). It also has a homologue of the apoptosis-inducing factor AIF1, a key regulator of apoptosis (41). Disruption of these genes has been shown to increase survival of yeast under the conditions that induce apoptotic death. In the case of CdtB, however, plate sensitivity and survival plating assays (Fig. 5A and B, respectively) showed that neither YCA1 nor AIF1 is required for CdtB-induced cell death, since the deletion of either gene hardly had any effect on the sensitivity to CdtB.
FIG. 5.
CdtB-induced yeast cell death is independent of the yeast metacaspase, AIF, and functional mitochondria but is partially dependent on histone H2B phosphorylation. (A) Plate sensitivity assays showed the yca1Δ and aif1Δ strains exhibiting the same level of sensitivity to CdtB as wild-type yeast (BY4741), while the histone H2B(S10A) mutant showed partial resistance, with a larger colony size than the wild type. Representative photographs of at least three experiments are shown. (B) Survival plating analysis of these strains also showed similar-to-wild-type levels of survival in the yca1Δ and aif1Δ strains but a slightly higher survival rate in the histone H2B(S10A) mutant. White bars represent strains carrying the pYES vector control, while gray bars represent strains carrying pYES-CdtB. The y axis shows percent survival calculated from the number of CFU on galactose plates relative to the number of CFU on glucose control plates. Values are averages of results from triplicate experiments, with error bars representing SD. The asterisk (*) represents a statistically significant difference between the wild-type and the H2B(S10A) strains (P = 0.050; Mann-Whitney U test). (C) A plate sensitivity assay showed rho0 derivatives exhibiting a level of sensitivity to CdtB similar to that observed for wild-type yeast (BY4741).
Another conserved characteristic of apoptotic death from yeast to mammals is the phosphorylation of histone H2B during apoptosis (2). To examine the requirement for this histone modification in CdtB-induced death, a yeast strain with a point mutation in histone H2B on the serine residue that is phosphorylated upon apoptotic induction (S10A) was employed. Interestingly, the H2B(S10A) strain showed a slight but significant increase in resistance to CdtB in both plate sensitivity and survival plating assays (Fig. 5A and B). In particular, the colonies that survived CdtB intoxication were noticeably larger in the H2B(S10A) strain than in the wild type (Fig. 5A). It appears that, although the yeast metacaspase or Aif1 is not required, the phosphorylation of histone H2B at least partially plays a role in CdtB-induced death.
Our earlier study of MOLT-4 cells suggested that CDT induces both caspase-dependent and caspase-independent cell death that can be overcome by overexpression of Bcl-2 (27). Furthermore, Bcl-2 expression has also been shown to rescue yeast from death induced by many apoptotic inducers (17). Therefore, we tested if overexpression of human Bcl-2 and Bcl-xL, the negative regulators of apoptosis, can rescue yeast cells from the cytotoxic effect of CdtB. Plasmids with human Bcl2 or Bcl-xL under a galactose-inducible promoter or an empty vector were transformed into W303-1A yeast cells harboring pYES-CdtB or the pYES control. Both plate sensitivity and survival plating assays showed that neither Bcl2 nor Bcl-xL could rescue CdtB-induced death (data not shown). Furthermore, rho0 derivative strains without functional mitochondria exhibited a level of sensitivity to CdtB expression similar to that observed for the wild-type parent (Fig. 5C). These suggest that mitochondria are not required for CdtB-induced cell death in our yeast model.
DISCUSSION
Cytolethal distending toxin is one of many virulence factors of A. actinomycetemcomitans that may contribute to its pathogenicity (28). AaCDT can induce cell cycle arrest and/or apoptosis in several kinds of mammalian cells. Here, we used S. cerevisiae as a model to study the mechanisms underlying such cytotoxic effect. The results suggest that expression of CdtB, the catalytic subunit of CDT, causes DNA damage and induces cell death in yeast that is independent of the yeast metacaspase Yca1, Aif1, and mitochondria.
In our system, AaCdtB expression is regulated by a galactose-inducible promoter. Temporary expression of CdtB was achieved by transferring cells from galactose-containing medium to glucose-containing medium to repress further expression after the designated time (Fig. 1A). We observed an apparent cytotoxic effect of CdtB at 8 h after induction even when the expression was turned off afterwards. This result suggests that the cytotoxic effect of CdtB is irreversible.
Continuous expression of CdtB in a haploid wild-type yeast strain results in a significant level of cell death, although a portion of cells still survive (Fig. 1C). This result suggests that the level of cytotoxicity of AaCdtB is different from that of Campylobacter jejuni CdtB (CjCdtB) (10, 13). The recent genome-wide study reported that CjCdtB is highly toxic in haploid yeast cells but is only marginally toxic to diploid yeast cells (13). AaCdtB is highly similar to Haemophilus ducreyi CdtB, with over 95% identity at the amino acid level, but shares less than 60% homology with Escherichia coli CdtB or CjCdtB (37, 40). Therefore, it is possible that the differences in toxicity level between AaCdtB and CjCdtB are due to the differences in the primary amino acid sequences. Furthermore, because of the high toxicity level in haploid cells, the genome-wide screen for CjCdtB-sensitive yeast mutants was performed with diploid strains (13). In contrast, since AaCdtB is moderately toxic to haploid yeast cells, we can use this system to detect both hypersensitivity and resistance to AaCdtB among haploid yeast strains with mutations or deletions in various pathways.
In this study, we employed a high-copy-number plasmid for AaCdtB expression, while previous studies of yeast with CjCdtB used a low-copy-number plasmid (10, 13). This may raise a concern with regard to the high level of intracellular CdtB. We had initially expressed AaCdtB in a low-copy-number plasmid, but we did not observe any effect on yeast growth in contrast to what was observed for CjCdtB, which demonstrated a high level of toxicity, both in our hands (data not shown) and as previously reported (10, 13). Therefore, we used the high-copy-number plasmid for AaCdtB expression and examined the effects of different levels of AaCdtB by varying the concentrations of the inducer (galactose) and the repressor (glucose) in the medium. We observed a clearly dose-dependent cytotoxic response to AaCdtB expression (data not shown). These results suggest that the cytotoxic effect was observed not only at an extremely high concentration of intracellular CdtB and further provide supporting evidence that the level of toxicity of AaCdtB is lower than that of CjCdtB. There is no study to date that directly compares the levels of toxicity of the CDTs of the two bacterial species in the same cell culture system.
It is rather difficult to compare the levels of CdtB expression in our system to the levels of CDT holotoxin that are toxic to mammalian cells. Only a few studies reported the actual AaCDT protein amounts used for intoxication. The toxic doses of AaCDT previously reported ranged from 50% effective doses (ED50s) of 0.4 ng CDT/ml (equivalent to ∼0.15 ng of CdtB) and 0.04 ng CdtB/ml (for 5 × 105 cells; Jurkat cells) (32, 33) to a toxic dose of 10 to 100 ng CDT/ml (equivalent to 4 to 40 ng CdtB for 106 cells; MOLT-4 and Jurkat cells) (27), if most of the CdtB were internalized. A direct delivery of AaCdtB by microinjection was done at 1 μg/μl at 50 to 120 hPa for 0.2 s (25), but the actual amount of injected CdtB is unknown. In comparison with these data, the levels of AaCdtB expressed in our system (2 to 15 ng/106 cells) were within a similar range of magnitude. Furthermore, previous reports indicated that microinjection or electroporation of the CdtB subunit alone into mammalian cells could recapitulate all the effects of CDT treatment (7, 14, 25). Additionally, in the case of Salmonella enterica serovar Typhi, CdtB is the only CDT subunit in the genome and the polypeptide is delivered into host cells by bacterial internalization without the requirement for CdtA and CdtC (9). Therefore, we believe that our system using internally expressed AaCdtB in yeast is a reasonable model for the study of the effects of CdtB on conserved cellular processes.
It has been unclear which catalytic activities of CdtB lead to its cytotoxicity. CdtB shows homology in the catalytic residues conserved among families of enzymes with phosphoesterase activities. Two such activities have been proposed to be important for CdtB function, those of a DNase and a phosphatidylinositol-3,4,5-triphosphate [PI(3,4,5)P3] phosphatase (14, 35). Mutations of the conserved catalytic residues abolish the cytotoxicity of CdtB, as shown in Fig. 1C and previous work (8, 14, 27). Although these residues are critical to both DNase and PI(3,4,5)P3 phosphatase activities, our results indicate that DNase activity alone is sufficient for CdtB cytotoxicity. This is suggested by the requirement for a nuclear localization signal (Fig. 1C) and the fact that yeast does not contain PI(3,4,5)P3 (23). Therefore, although this does not preclude a possible role for other enzymatic activities in mammalian cells, yeast cell death likely results from the DNA-damaging activity of CdtB.
Moreover, we found that CdtB is specifically highly toxic to yeast strains lacking Rad51, a single-strand DNA binding protein crucial in homologous recombination, and the members of the MRX complex that acts as a sensor of DNA breaks (Fig. 3A). Thus, cells that cannot repair DNA breaks by homologous recombination cannot survive the damage generated by CdtB. This evidence strongly suggests that homologous recombination is critical for the repair of CdtB-induced lesions. This is in agreement with the recent genome-wide screen for CjCdtB-sensitive yeast diploid mutants, which also identified the rad51Δ, mre11Δ, rad50Δ, and xrs2Δ strains (13). However, our results differ from the screen with regard to the sensitivity of yku70Δ, which showed a weak growth defect upon CjCdtB expression but showed the same level of sensitivity to wild-type yeast in our assay. Nevertheless, our results are consistent with the screen with regard to the other genes involved in NHEJ (DNL4 and NEJ1), the deletion of which had no effect on the sensitivity to CdtB. Thus, it appears that NHEJ plays a minor role in the repair of CdtB-induced damage in yeast.
Previous studies reported a rapid induction of Rad50 and Mre11 focus formation upon CDT intoxication in mammalian cells (11, 17). We also observed a strong hypersensitivity to AaCdtB in yeast strains lacking MRE11, RAD50, or XRS2, confirming the importance of all the members of the MRX complex in the response to CdtB. It is interesting to note that the MRX complex is particularly crucial in the repair of DNA breaks that require processing, such as irradiation-induced breaks and meiotic breaks created by the Spo11 nuclease, which remains bound to the DNA ends after cleavage, but only mildly affects the repair of “clean” breaks created by the HO nuclease (38). The strongly hypersensitive phenotype observed in the MRX mutant strains implies that CdtB may also create DNA breaks that similarly require processing. Although the MRX complex is involved in both HR and NHEJ, only the defects in HR, not those in NHEJ, render the cells hypersensitive to CdtB. This suggests that HR is the major DNA break repair pathway for CdtB in yeast. It is possible that the situation could be different in mammalian cells since different species do have different preferences over the choices of repair pathways. HR is the predominant DNA break repair pathway in yeast, while higher eukaryotes display a greater use of NHEJ (38).
To ensure the integrity of genomic DNA during cell division, in response to DNA damage, cells also need to coordinate DNA repair with cell cycle progression through the function of DNA damage checkpoints. Rad53 and Chk1 are the DNA damage checkpoint effector kinases required in such response. In S. cerevisiae, Rad53, but not Chk1, is required for the S-phase DNA damage checkpoint, while both Rad53 and Chk1 play a role in the G2/M checkpoint (22). Our result showed that CdtB induces S/G2 arrest in yeast and that the rad53Δ strain, but not the chk1Δ strain, is hypersensitive to CdtB. Together, these results suggest that cell cycle arrest through Rad53 checkpoint activation is required for the survival of yeast cells upon CdtB expression. Nevertheless, since the rad53Δ strain still retains DNA break repair capability, the level of hypersensitivity is expected to be less than that of strains with defects in HR, which cannot repair the damage at all. This was actually what we observed, as shown in Fig. 3. This mutant was not identified in the genome-wide screen for strains sensitive to CjCdtB, because RAD53 is an essential gene and therefore was absent from the deletion library. The strain used in our study has a secondary mutation in SML1, which suppresses rad53Δ lethality (44).
Although CDT was shown to induce G1 or G2/M cell cycle arrest in mammalian cells, depending on the cell types affected, we found that AaCdtB induces S/G2 arrest in haploid yeast cells. The significant delay in S phase and the requirement for the major S-phase checkpoint kinase, Rad53, may imply that CdtB generates single-stranded DNA breaks (SSBs), which requires DNA replication to be converted to double-strand breaks (DSBs) in S phase. This is consistent with the findings of Kitagawa and colleagues, who identified several genes that function during S phase in the genome-wide screen with CjCdtB and proposed that CdtB likely creates SSB (13). Since haploid yeasts do not contain homologous chromosomes for homologous recombination repair except in G2 phase, S-phase delay may be more pronounced in haploid cells, as was observed in our study. The exact nature of CdtB-induced DNA damage is still not well understood. Further investigations of the yeast system with the relatively well-characterized DNA damage responses could shed new light on this unanswered question.
To examine the pathways through which CdtB induces cell death, we tested the effect of CdtB in yeast strains lacking key apoptotic genes. Both the caspases and AIF (represented by YCA1 and AIF1 in yeast [20, 41]) are important regulators of apoptosis in mammalian cells and yeasts. However, the deletion of either gene had essentially no effect on the sensitivity to CdtB. Furthermore, we could not detect ROS accumulation or phosphatidylserine staining upon CdtB expression. Overexpression of Bcl-2 or Bcl-xL or the loss of functional mitochondria also failed to rescue CdtB-induced death. These data argue against the role of apoptosis in CdtB-induced yeast cell death. Nevertheless, a yeast strain carrying the mutant form of histone H2B(S10A) shows partial resistance to CdtB expression. Since phosphorylation of histone H2B is involved in chromatin condensation (2), it is conceivable that H2B phosphorylation may play additional role in other DNA metabolic processes that affect survival. Otherwise, there may be another death-related pathway that can activate H2B phosphorylation independently of caspase and AIF.
Although DNA damage is a well-known inducer of apoptosis in mammalian cells (30), clear evidence is still lacking in yeast. It has been proposed that a yeast strain with mutation of CDC13, a telomere binding protein that prevents telomere degradation, undergoes apoptotic death at nonpermissive temperature (29). However, Wysocki and Kron (42) showed that dead yeast cells bind nonspecifically to fluorochromes used to detect caspase activity and that cell death is independent of YCA1. Therefore, to answer the questions of whether and how DNA damage induces apoptotic death, or other kinds of cell death, in yeast still requires further investigation (5). Our study suggests that, in the case of DNA damage induced by CdtB, cell death occurs via a caspase-independent process that does not show typical apoptotic characteristics.
In conclusion, we have established a yeast system in which the mechanisms of AaCdtB-induced cell death can be characterized in detail. Our data suggest that AaCdtB induces DNA breaks, S/G2 cell cycle arrest, and cell death that is likely nonapoptotic in yeast. Further characterization of the effects of CdtB in yeast will bring additional insights into both the mechanism of CdtB and the biology of yeast in response to DNA damage. Moreover, we reported here the first mutation in yeast that increases resistance to CdtB. A further search for genes required for CdtB-induced death could lead to the identification of potential targets for the development of a future therapeutic approach for CDT-related diseases.
Acknowledgments
We thank C. David Allis, Steve Buratowski, Gerald R. Fink, Michael C. Keogh, Eun-Jung Cho, Stephen Manon, and Cheunchit Boonchird for yeast strains and plasmids; Frank Madeo and David Pellman for protocols and suggestions; and Waranuch Pitiphat for statistical advices. We are grateful to Ratiboot Sallabhan for his technical assistance. We thank the Research Facilities of the Hiroshima University School of Dentistry for the use of their facility. We also appreciate helpful suggestions from the members of the Laboratory of Biotechnology, CRI, and the Laboratory of Bacteriology, Hiroshima University.
This work is supported by grant no. MRG5080154 from the Thailand Research Fund, by the Support Program for Improving Graduate School Education (BioDental Education in Hiroshima University GSBS), and by a grant-in-aid for Scientific Research (C) from the Ministry of Education, Science, Sport, Culture and Technology of Japan.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 7 December 2009.
REFERENCES
- 1.Ahn, S. H., W. L. Cheung, J. Y. Hsu, R. L. Diaz, M. M. Smith, and C. D. Allis. 2005. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell 120:25-36. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, S. H., K. A. Henderson, S. Keeney, and C. D. Allis. 2005. H2B (Ser10) phosphorylation is induced during apoptosis and meiosis in S. cerevisiae. Cell Cycle 4:780-783. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2002. Short protocols in molecular biology, 5th ed. Wiley, Hoboken, NJ.
- 4.Aylon, Y., and M. Kupiec. 2004. DSB repair: the yeast paradigm. DNA Repair (Amst.) 3:797-815. [DOI] [PubMed] [Google Scholar]
- 5.Burhans, W. C., M. Weinberger, M. A. Marchetti, L. Ramachandran, G. D'Urso, and J. A. Huberman. 2003. Apoptosis-like yeast cell death in response to DNA damage and replication defects. Mutat. Res. 532:227-243. [DOI] [PubMed] [Google Scholar]
- 6.Curak, J., J. Rohde, and I. Stagljar. 2009. Yeast as a tool to study bacterial effectors. Curr. Opin. Microbiol. 12:18-23. [DOI] [PubMed] [Google Scholar]
- 7.Elwell, C., K. Chao, K. Patel, and L. Dreyfus. 2001. Escherichia coli CdtB mediates cytolethal distending toxin cell cycle arrest. Infect. Immun. 69:3418-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elwell, C. A., and L. A. Dreyfus. 2000. DNase I homologous residues in CdtB are critical for cytolethal distending toxin-mediated cell cycle arrest. Mol. Microbiol. 37:952-963. [DOI] [PubMed] [Google Scholar]
- 9.Haghjoo, E., and J. E. Galan. 2004. Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc. Natl. Acad. Sci. U. S. A. 101:4614-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassane, D. C., R. B. Lee, M. D. Mendenhall, and C. L. Pickett. 2001. Cytolethal distending toxin demonstrates genotoxic activity in a yeast model. Infect. Immun. 69:5752-5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassane, D. C., R. B. Lee, and C. L. Pickett. 2003. Campylobacter jejuni cytolethal distending toxin promotes DNA repair responses in normal human cells. Infect. Immun. 71:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henderson, B., S. P. Nair, J. M. Ward, and M. Wilson. 2003. Molecular pathogenicity of the oral opportunistic pathogen Actinobacillus actinomycetemcomitans. Annu. Rev. Microbiol. 57:29-55. [DOI] [PubMed] [Google Scholar]
- 13.Kitagawa, T., H. Hoshida, and R. Akada. 2007. Genome-wide analysis of cellular response to bacterial genotoxin CdtB in yeast. Infect. Immun. 75:1393-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lara-Tejero, M., and J. E. Galan. 2000. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science 290:354-357. [DOI] [PubMed] [Google Scholar]
- 15.Lara-Tejero, M., and J. E. Galan. 2001. CdtA, CdtB, and CdtC form a tripartite complex that is required for cytolethal distending toxin activity. Infect. Immun. 69:4358-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lara-Tejero, M., and J. E. Galan. 2002. Cytolethal distending toxin: limited damage as a strategy to modulate cellular functions. Trends Microbiol. 10:147-152. [DOI] [PubMed] [Google Scholar]
- 17.Li, L., A. Sharipo, E. Chaves-Olarte, M. G. Masucci, V. Levitsky, M. Thelestam, and T. Frisan. 2002. The Haemophilus ducreyi cytolethal distending toxin activates sensors of DNA damage and repair complexes in proliferating and non-proliferating cells. Cell. Microbiol. 4:87-99. [DOI] [PubMed] [Google Scholar]
- 18.Madeo, F., E. Frohlich, and K. U. Frohlich. 1997. A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 139:729-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madeo, F., E. Frohlich, M. Ligr, M. Grey, S. J. Sigrist, D. H. Wolf, and K. U. Frohlich. 1999. Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 145:757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madeo, F., E. Herker, C. Maldener, S. Wissing, S. Lachelt, M. Herlan, M. Fehr, K. Lauber, S. J. Sigrist, S. Wesselborg, and K. U. Frohlich. 2002. A caspase-related protease regulates apoptosis in yeast. Mol. Cell 9:911-917. [DOI] [PubMed] [Google Scholar]
- 21.Madeo, F., E. Herker, S. Wissing, H. Jungwirth, T. Eisenberg, and K. U. Frohlich. 2004. Apoptosis in yeast. Curr. Opin. Microbiol. 7:655-660. [DOI] [PubMed] [Google Scholar]
- 22.Melo, J., and D. Toczyski. 2002. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14:237-245. [DOI] [PubMed] [Google Scholar]
- 23.Mitra, P., Y. Zhang, L. E. Rameh, M. P. Ivshina, D. McCollum, J. J. Nunnari, G. M. Hendricks, M. L. Kerr, S. J. Field, L. C. Cantley, and A. H. Ross. 2004. A novel phosphatidylinositol(3,4,5)P3 pathway in fission yeast. J. Cell Biol. 166:205-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nash, R., G. Tokiwa, S. Anand, K. Erickson, and A. B. Futcher. 1988. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 7:4335-4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishikubo, S., M. Ohara, Y. Ueno, M. Ikura, H. Kurihara, H. Komatsuzawa, E. Oswald, and M. Sugai. 2003. An N-terminal segment of the active component of the bacterial genotoxin cytolethal distending toxin B (CDTB) directs CDTB into the nucleus. J. Biol. Chem. 278:50671-50681. [DOI] [PubMed] [Google Scholar]
- 26.Ohara, M., T. Hayashi, Y. Kusunoki, M. Miyauchi, T. Takata, and M. Sugai. 2004. Caspase-2 and caspase-7 are involved in cytolethal distending toxin-induced apoptosis in Jurkat and MOLT-4 T-cell lines. Infect. Immun. 72:871-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohara, M., T. Hayashi, Y. Kusunoki, K. Nakachi, T. Fujiwara, H. Komatsuzawa, and M. Sugai. 2008. Cytolethal distending toxin induces caspase-dependent and -independent cell death in MOLT-4 cells. Infect. Immun. 76:4783-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohara, M., E. Oswald, and M. Sugai. 2004. Cytolethal distending toxin: a bacterial bullet targeted to nucleus. J. Biochem. 136:409-413. [DOI] [PubMed] [Google Scholar]
- 29.Qi, H., T. K. Li, D. Kuo, E. K. A. Nur, and L. F. Liu. 2003. Inactivation of Cdc13p triggers MEC1-dependent apoptotic signals in yeast. J. Biol. Chem. 278:15136-15141. [DOI] [PubMed] [Google Scholar]
- 30.Roos, W. P., and B. Kaina. 2006. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 12:440-450. [DOI] [PubMed] [Google Scholar]
- 31.Saiki, K., T. Gomi, and K. Konishi. 2004. Deletion and purification studies to elucidate the structure of the Actinobacillus actinomycetemcomitans cytolethal distending toxin. J. Biochem. 136:335-342. [DOI] [PubMed] [Google Scholar]
- 32.Shenker, B. J., D. Besack, T. McKay, L. Pankoski, A. Zekavat, and D. R. Demuth. 2004. Actinobacillus actinomycetemcomitans cytolethal distending toxin (Cdt): evidence that the holotoxin is composed of three subunits: CdtA, CdtB, and CdtC. J. Immunol. 172:410-417. [DOI] [PubMed] [Google Scholar]
- 33.Shenker, B. J., D. Besack, T. McKay, L. Pankoski, A. Zekavat, and D. R. Demuth. 2005. Induction of cell cycle arrest in lymphocytes by Actinobacillus actinomycetemcomitans cytolethal distending toxin requires three subunits for maximum activity. J. Immunol. 174:2228-2234. [DOI] [PubMed] [Google Scholar]
- 34.Shenker, B. J., D. R. Demuth, and A. Zekavat. 2006. Exposure of lymphocytes to high doses of Actinobacillus actinomycetemcomitans cytolethal distending toxin induces rapid onset of apoptosis-mediated DNA fragmentation. Infect. Immun. 74:2080-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shenker, B. J., M. Dlakic, L. P. Walker, D. Besack, E. Jaffe, E. LaBelle, and K. Boesze-Battaglia. 2007. A novel mode of action for a microbial-derived immunotoxin: the cytolethal distending toxin subunit B exhibits phosphatidylinositol 3,4,5-triphosphate phosphatase activity. J. Immunol. 178:5099-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shenker, B. J., R. H. Hoffmaster, A. Zekavat, N. Yamaguchi, E. T. Lally, and D. R. Demuth. 2001. Induction of apoptosis in human T cells by Actinobacillus actinomycetemcomitans cytolethal distending toxin is a consequence of G2 arrest of the cell cycle. J. Immunol. 167:435-441. [DOI] [PubMed] [Google Scholar]
- 37.Shenker, B. J., T. McKay, S. Datar, M. Miller, R. Chowhan, and D. Demuth. 1999. Actinobacillus actinomycetemcomitans immunosuppressive protein is a member of the family of cytolethal distending toxins capable of causing a G2 arrest in human T cells. J. Immunol. 162:4773-4780. [PubMed] [Google Scholar]
- 38.Shrivastav, M., L. P. De Haro, and J. A. Nickoloff. 2008. Regulation of DNA double-strand break repair pathway choice. Cell Res. 18:134-147. [DOI] [PubMed] [Google Scholar]
- 39.Siggers, K. A., and C. F. Lesser. 2008. The yeast Saccharomyces cerevisiae: a versatile model system for the identification and characterization of bacterial virulence proteins. Cell Host Microbe 4:8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugai, M., T. Kawamoto, S. Y. Peres, Y. Ueno, H. Komatsuzawa, T. Fujiwara, H. Kurihara, H. Suginaka, and E. Oswald. 1998. The cell cycle-specific growth-inhibitory factor produced by Actinobacillus actinomycetemcomitans is a cytolethal distending toxin. Infect. Immun. 66:5008-5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wissing, S., P. Ludovico, E. Herker, S. Buttner, S. M. Engelhardt, T. Decker, A. Link, A. Proksch, F. Rodrigues, M. Corte-Real, K. U. Frohlich, J. Manns, C. Cande, S. J. Sigrist, G. Kroemer, and F. Madeo. 2004. An AIF orthologue regulates apoptosis in yeast. J. Cell Biol. 166:969-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wysocki, R., and S. J. Kron. 2004. Yeast cell death during DNA damage arrest is independent of caspase or reactive oxygen species. J. Cell Biol. 166:311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamano, R., M. Ohara, S. Nishikubo, T. Fujiwara, T. Kawamoto, Y. Ueno, H. Komatsuzawa, K. Okuda, H. Kurihara, H. Suginaka, E. Oswald, K. Tanne, and M. Sugai. 2003. Prevalence of cytolethal distending toxin production in periodontopathogenic bacteria. J. Clin. Microbiol. 41:1391-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao, X., E. G. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]