Abstract
We reported previously that Treponema denticola, one of the periodontal pathogens, suppresses the expression of human β-defensins (HBDs) in human gingival epithelial cells. To identify the mechanisms involved in this suppression, immortalized and normal human gingival epithelial cells were infected with live or heat-killed T. denticola for 24 h, and then the expression of HBDs was examined by real-time RT-PCR. Live T. denticola suppressed the expression of HBD-3 substantially and also suppressed the expression of HBD-1 and HBD-2. However, heat-killed bacteria did not produce a suppressive effect but instead slightly upregulated the levels of HBD-2 and HBD-3. In contrast to live T. denticola, which reduced the activation of mitogen-activated protein kinase (MAPK) and NF-κB within an hour of infection, heat-killed bacteria did not show any inhibitory effect on the MAPK and NF-κB signaling pathways. Knockdown of Toll-like receptor 2 (TLR2) via RNA interference abolished the suppressive effect of T. denticola on the expression of HBD-3. Heat-killed T. denticola but not live bacteria could activate TLR2 in CHO/CD14/TLR2 reporter cells, suggesting that T. denticola contains a heat-labile inhibitor(s) of TLR2 in addition to ligands recognized by TLR2. Indeed, live T. denticola was able to inhibit TLR2 activation by Pam3CSK. In conclusion, T. denticola suppressed the expression of HBD-3 by inhibiting the TLR2 axis in gingival epithelial cells. These results may provide new insight into the pathogenesis of periodontitis caused by T. denticola.
Periodontitis is the inflammation of periodontal tissue that results from the interaction of plaque-associated bacteria with the host immune system. During the transition from health to periodontitis, the amount of total bacteria and the proportion of periodontopathic bacteria in the plaque microflora increase (22). Among the periodontitis-associated bacteria, the members of the so-called red complex triad (Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola) are the most well-studied periodontal pathogens, and these bacteria have been characterized as the primary causal agents of periodontal disease (10, 22). The gingival epithelium provides the first line of defense against these plaque bacteria by forming a physical and chemical barrier. The barrier function of epithelia is attributed to the unique architectural integrity of the cells and to the production of antimicrobial peptides, such as human β-defensins (HBDs) and the cathelicidin LL-37 (9). Patients with Kostmann syndrome lack LL-37 in their saliva and develop severe periodontitis in young adulthood (21), suggesting the importance of antimicrobial peptides in the maintenance of periodontal health.
The regulation of HBD expression has been studied extensively. HBD-1 is expressed constitutively, whereas the expression of HBD-2 and HBD-3 is under the control of the NF-κB and/or mitogen-activated protein kinase (MAPK) signaling pathway (11). At the receptor level, induction of HBD-2 expression is mediated via pattern recognition receptors (PRRs) and proinflammatory cytokine receptors that respond to microbe-associated molecular patterns and cytokines such as tumor necrosis factor alpha, interleukin-1 (IL-1), and IL-17 (11). PRRs that mediate HBD upregulation in various epithelial tissues include Toll-like receptors (TLRs) and NOD2 (nucleotide-binding oligomerization domain 2) (11).
In gingival epithelial tissues, components of several species of oral bacteria, including Streptococcus sanguinis, S. gordonii, Veillonella atypica, Fusobacterium nucleatum, Prevotella intermedia, and P. gingivalis, have been shown previously to induce HBD-2 and/or HBD-3 expression (6, 13, 17, 24, 29). Protease-activated receptor 2 (PAR2) has been shown to be partially involved in the upregulation of HBD-2 by P. gingivalis (7). We have recently demonstrated that NACHT-LRR- and pyrin domain-containing protein 2 (NALP2) and TLR2 mediate the induction of HBD-2 and HBD-3 by F. nucleatum in gingival epithelial cells (14).
Pathogens often subvert host immune responses, and downregulation of HBD expression by bacteria has also been demonstrated. Virulent Shigella flexneri has been shown to suppress transcription of HBD-3 in human intestinal cells (23). In our previous study, we demonstrated that T. denticola suppressed the expression of HBD-1 and HBD-3 in gingival epithelial cells (13). Compared with the mechanisms involved in bacterium-induced upregulation of antimicrobial peptides, the mechanisms for the suppression of antimicrobial peptides by pathogens are poorly understood. The aim of this study was to delineate the mechanism(s) involved in the suppression of HBD expression by T. denticola in gingival epithelial cells.
MATERIALS AND METHODS
Human materials.
The use of human materials was approved by the Institutional Review Board at the Seoul National University Dental Hospital. Normal human gingival tissue specimens were obtained with informed consent from healthy volunteers (aged 20 to 30 years) undergoing oral surgery.
Bacterial culture.
S. sanguinis NCTC 10904, S. gordonii ATCC 10558, V. atypica ATCC 17744, F. nucleatum ATCC 25586, P. intermedia ATCC 25611, P. gingivalis ATCC 49417, T. forsythia ATCC 43407, and T. denticola ATCC 33521 were obtained from the ATCC (Bethesda, MD) and cultured in the appropriate medium as described previously (13). The bacteria were enumerated by flow cytometry. Heat-killed bacteria were prepared by being heated at 95°C for 1 h. Bacterial lysates were prepared by sonication with a Sonic Dismembrator 300 (Fisher Scientific, Fair Lawn, NJ) for 15 min on ice water, and then complete destruction of bacterial cells was confirmed using a microscope. Some heat-killed bacteria were fragmented, but most maintained bacterial morphology.
Cell culture.
Normal human gingival epithelial cells (NGECs) were isolated from excised retromolar gingival tissue, and primary cultures were established in bullet kit medium comprising keratinocyte growth medium (KGM) containing 0.15 mM calcium and a supplementary growth factor bullet kit (Clonetics, San Diego, CA) as reported previously (19). Cells were subcultured when they reached 70% confluence, and second-passage cells were used for all experiments. Immortalized human gingival keratinocyte HOK-16B cells (20), a kind gift from N.-H. Park (University of California—Los Angeles, Los Angeles), were maintained in supplemented KGM. The reporter CHO cell line expressing human CD14 and human TLR2 that expresses surface CD25 as a result of TLR2-mediated NF-κB translocation (designated CHO/CD14/TLR2) (30) was kindly provided by D. Golenbock (University of Massachusetts Medical School, Worcester). The CHO/CD14/TLR2 cells were maintained in Ham's F-12 medium (Gibco, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (Gibco), 1 mg/ml G418 (Invitrogen, Carlsbad, CA), 400 U/ml hygromycin (Amresco, San Francisco, CA), and 1× penicillin-streptomycin (Gibco).
Infection of epithelial cells with bacteria.
NGECs or HOK-16B cells were plated at 6 × 104 cells/500 μl/well in triplicate in 24-well plates 1 day before infection. At 80% confluence, the cells were infected with live or heat-killed bacteria at a multiplicity of infection (MOI) of 1,000 in KGM containing 2% heat-inactivated human serum (Sigma, St. Louis, MO). The cells were cultured at 37°C in a water-saturated atmosphere of 95% air and 5% CO2 for 24 h or the time indicated below for a specific experiment. The antibiotics in the KGM were removed to prolong the survival of bacteria. Under these culture conditions, T. denticola, an absolute anaerobe, did not grow well but survived long enough to invade the cells. In addition, the viability of the HOK-16B cells was not affected. Total RNA was extracted from the cells of the combined triplicate wells using TRIzol (Invitrogen). Experiments were repeated three times.
Real-time RT-PCR.
Total RNA (2 μg) from gingival epithelial cells or CHO/CD14/TLR2 cells was subjected to reverse transcription (RT) using (dT)18 and the SuperScript II enzyme (Invitrogen) in a 25-μl reaction mixture at 42°C for 1 h. Real-time PCR was performed in a 20-μl reaction mixture containing 1 μl of template cDNA, SYBR Premix Ex Taq, ROX reference dye (Takara Bio, Otsu, Japan), and each primer (0.2 μM), as reported previously (14). Primer sequences used to amplify the CD25 gene and the hamster glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene are as follows: 5′-GAATGCAAGAGAGGTTTCCG-3′ and 5′-TGTGCATTGACATTGGTTGTC-3′ for the CD25 gene and 5′-CATGTTCCAGTATGACTCCACTC-3′ and 5′-GGCCTCACCCCATTTGATGT-3′ for the hamster GAPDH gene. Amplification was performed in a model 7500 real-time PCR fluorescence thermocycler (Applied Biosystems, Foster City, CA) under the following conditions: initial denaturation at 94°C for 1 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 15 s, and elongation at 72°C for 33 s. The specificity of the PCR product was verified by melting-curve analysis and by examination on a 3% agarose gel. The housekeeping GAPDH gene was amplified in parallel with the gene of interest. Copy numbers relative to those of the GAPDH gene were calculated using 2−CΔT. Real-time PCR was performed in triplicate for each RNA sample.
RNAi in HOK-16B cells and bacterial infection.
Stealth small interfering RNA (siRNA) duplex oligoribonucleotides against human TLR2 (corresponding to GenBank accession no. NM_003264) were designed using the BLOCK-iT RNA interference (RNAi) designer program (https://rnaidesigner.invitrogen.com/rnaiexpress/) and were synthesized by Invitrogen. The oligonucleotide sequences used for TLR2 were as follows: sense, 5′-UGAAGCAUCAAUCUCAAGUUCCUCA-3′, and antisense, 5′-UGAGGAACUUGAGAUUGAUGCUUCA-3′. The BLOCK-iT (Invitrogen) fluorescent oligonucleotide, which is not homologous to any known gene, was used as a transfection efficiency detector and as a negative control. HOK-16B cells were plated at 4 × 104 cells/well in a 24-well plate. After overnight incubation, the cells were transfected with 20 nM siRNAs using a BLOCK-iT transfection kit (Invitrogen). After culture for 24 h, the cells were retransfected with siRNA and simultaneously infected with live T. denticola at an MOI of 1,000 for 24 h. Experiments were repeated three times. The T. denticola-induced suppression of HBD-3 expression was recorded as the percentage of expression relative to the expression in the control culture without infection.
Flow cytometry.
HOK-16B cells were stained with fluorescein isothiocyanate (FITC)-conjugated mouse anti-human TLR2 monoclonal antibody (MAb) clone TL2.1 (BioLegend, San Diego, CA) and were analyzed with a FACSCalibur flow cytometer (BD Biosciences).
CHO/CD14/TLR2 cells were plated at 2 × 105 cells/well in a 24-well plate. After overnight incubation, the cells were stimulated with various live oral bacteria (MOI, 10 or 100) for 16 h. The CHO cells were also stimulated with live or heat-killed T. denticola (MOI, 100) or with T. denticola lysate in the absence or presence of 0.3 μg/ml Pam3CSK4 for 16 h. After being stained with FITC-conjugated mouse anti-human CD25 MAb clone 2A3 (BD Biosciences, San Diego, CA) or FITC-conjugated mouse anti-human TLR2 MAb clone TL2.1 (BioLegend), the cells were analyzed with a FACSCalibur flow cytometer (BD Biosciences). To examine the levels of CD25 transcripts in CHO/CD14/TLR2 cells, total RNA was extracted using an RNeasy kit (Qiagen, Germantown, MD). Experiments were repeated three times.
Examination of signaling pathways.
HOK-16B cells (1.2 × 106) plated into a 100-mm dish were stimulated with live T. denticola at an MOI of 1,000 for the times indicated below. HOK-16B cells were also stimulated with live or heat-killed T. denticola at an MOI of 1,000 for 30 min. After the cells were washed with phosphate-buffered saline, protein lysates were prepared using the lysis buffer included in the PathScan inflammation multitarget enzyme-linked immunosorbent assay (ELISA) kit (Cell Signaling, Danvers, MA). The lysates were subjected to ELISA analysis for NF-κB p65 and the phosphorylated forms of NF-κB p65, IκB, p38, Jun N-terminal protein kinase (JNK), and STAT3 in duplicate by using the kit. The nonphosphorylated forms of JNK and NF-κB p65 in the lysates were detected by Western blotting using antibodies from Santa Cruz Biotechnology (Santa Cruz, CA) and Cell Signaling, respectively. Experiments were repeated twice.
Statistics.
The differences between two groups were analyzed with a two-tailed unpaired Student t test. Data were considered statistically significant at a P value of <0.05.
RESULTS
Live but not heat-killed T. denticola suppressed HBD expression in gingival epithelial cells.
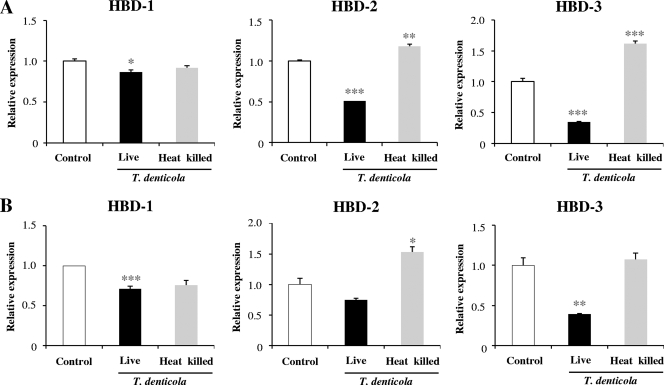
We previously demonstrated that live T. denticola but not the bacterial lysate could suppress the expression of HBD-1 and HBD-3 in immortalized human gingival epithelial (HOK-16B) cells (13). To confirm the suppressive effect of T. denticola on HBD expression in normal cells, NGECs were infected with live or heat-killed T. denticola for 24 h and the expression of HBD-1, HBD-2, and HBD-3 was examined by real-time RT-PCR. Live T. denticola suppressed the expression of HBD-3 substantially and also suppressed the expression of HBD-1 and HBD-2. In contrast, heat-killed bacteria slightly upregulated the expression of HBD-2 and HBD-3 (Fig. 1A). HOK-16B cells also experienced downregulation of HBDs only in response to live T. denticola infection (Fig. 1B). These results indicate that NGECs and HOK-16B cells respond to T. denticola similarly and that live bacteria are required to suppress HBD expression in those cells. Thus, in the subsequent experiments, regulation of HBD-3 by T. denticola was studied using HOK-16B cells and live bacteria.
FIG. 1.
Live but not heat-killed T. denticola suppressed the expression of HBDs in gingival epithelial cells. NGECs (A) or HOK-16B cells (B) were infected with live or heat-killed T. denticola at an MOI of 1,000 for 24 h. Expression of HBDs was examined by real-time RT-PCR. The means ± the standard errors of the means (SEM) of the relative copy numbers are expressed as the level of induction (n-fold) compared to expression in the control culture without bacteria. These data are representative of findings from three independent experiments with similar results. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 versus data for no infection.
Live T. denticola suppressed the NF-κB, MAPK, and STAT3 signaling pathways.
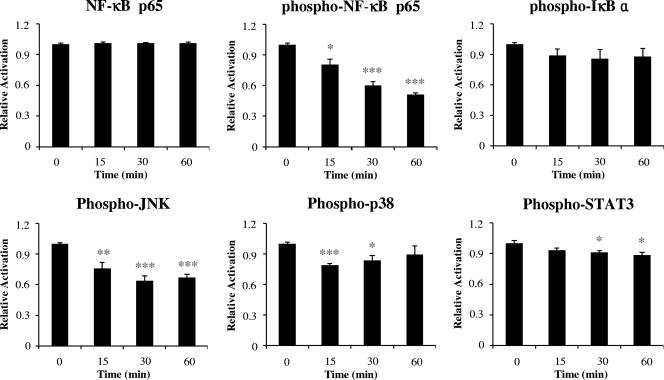
To understand the molecular basis for the suppression of HBD expression by T. denticola, the activation of several prominent signaling pathways involved in inflammation was examined. Infection with T. denticola significantly reduced the levels of the phosphorylated forms of NF-κB p65, JNK, p38, and STAT3 within 30 min (Fig. 2). These results suggest that T. denticola impedes various signaling pathways, including the NF-κB and MAPK pathways, that control HBD expression.
FIG. 2.
T. denticola suppressed various signaling pathways. HOK-16B cells plated into a 100-mm dish were stimulated with live T. denticola at an MOI of 1,000 for 0, 15, 30, or 60 min. Protein lysates (50 μg each) were prepared and subjected to ELISA analysis of NF-κB p65 and the phosphorylated forms of NF-κB p65, IkB, p38, JNK, and STAT3. The means ± the SEM of the optical densities from four assays are expressed as the levels of activation relative to that in a control culture without bacteria. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 versus data for no infection.
Suppression of HBD-3 expression occurred at early time points.
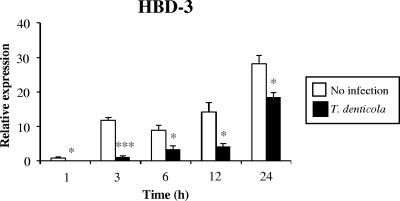
Since T. denticola affected signaling through the NF-κB and MAPK pathways within 30 min of infection, time courses for the suppression of HBD-3 were examined. The level of HBD-3 transcripts in the control culture without infection increased with time, and this increase in transcripts was suppressed by infection with T. denticola starting at a very early time point (1 h) and continuing throughout the culture period (Fig. 3).
FIG. 3.
T. denticola suppressed the expression of HBD-3 at early time points. HOK-16B cells were infected with live T. denticola at an MOI of 1,000 for 1, 3, 6, 12, and 24 h. The levels of HBD-3 transcripts were evaluated by real-time RT-PCR. The means ± the SEM of the relative copy numbers are expressed as the levels of induction (n-fold) at 1 h compared to expression in the control culture without bacteria. These data are representative of findings from two independent experiments with similar results. *, P < 0.05, and ***, P < 0.001 versus data for no infection.
Knockdown of TLR2 reversed the suppressive effect of T. denticola on HBD-3 expression.
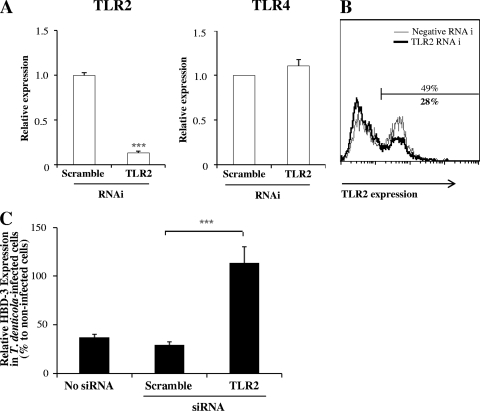
Since TLR2 mediates the induction of HBD-2 and HBD-3 by F. nucleatum in gingival epithelial cells (14), we examined whether or not TLR2 is also involved in the modulation of HBD-3 by T. denticola. TLR2-specific RNAi decreased the level of TLR2 transcripts by 80% but had no effect on TLR4 expression (Fig. 4A). We previously reported very low levels of TLR2 protein expression on HOK-16B cells (14). With the use of an antibody from a source different from that used in the previous study, 49% of HOK-16B cells stained positive for expression of TLR2 on the cell surface. This level of expression was reduced to 28% by TLR2-specific RNAi (Fig. 4B). In nontransfected cells and cells transfected with scramble siRNA, T. denticola infection reduced the levels of HBD-3 expression to 37 and 29% of that in uninfected cells, respectively. Knockdown of TLR2 reversed the reduction of HBD-3 expression by T. denticola infection, changing the level of expression from 29 to 114% of that in uninfected cells (Fig. 4C).
FIG. 4.
Knockdown of TLR2 reversed the suppressive effect of T. denticola on HBD-3 expression. HOK-16B cells were transfected with control or TLR2-specific siRNA. After a 24-h incubation, the cells were infected with live T. denticola at an MOI of 1,000 for 24 h. (A and B) Gene silencing was assessed by real-time RT-PCR (A) and flow cytometry (B) 24 h after transfection. (C) The levels of HBD-3 transcripts were evaluated by real-time RT-PCR. The means ± the SEM of results from nine real-time RT-PCR assays are given as the percentages of expression in T. denticola-infected cells compared to expression in the control culture without bacteria. ***, P < 0.001 versus results for scramble RNAi.
T. denticola inhibited TLR2 activation.
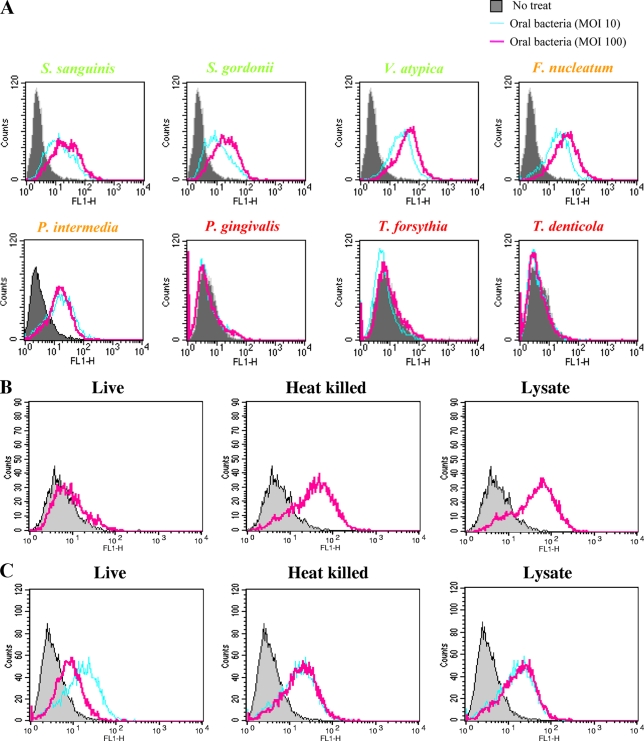
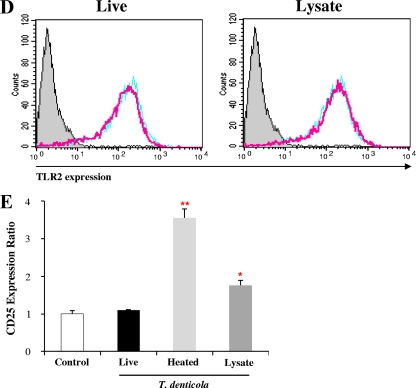
To better understand the modulation of HBD expression via TLR2, the ability of T. denticola to activate TLR2 was compared with the TLR2-activating abilities of other oral bacteria by using the CHO/CD14/TLR2 reporter cell line. When the reporter cells were stimulated with various oral bacteria at an MOI of 10 or 100, three early-colonizing species and two species belonging to the orange complex substantially activated TLR2 in a dose-dependent manner. However, the red complex bacteria, including T. denticola, did not activate TLR2 (Fig. 5A). In contrast to live T. denticola, heat-killed bacteria and bacterial lysate were able to activate TLR2 (Fig. 5B). In addition, live T. denticola but not heat-killed bacteria or lysate suppressed TLR2 activation by a synthetic ligand, Pam3CSK4 (Fig. 5C). Both live T. denticola and T. denticola lysate, which are known to be rich in proteases (28), failed to degrade the TLR2 protein expressed on the cell surface (Fig. 5D). The possibility that proteases degraded the CD25 protein was further investigated by examining the levels of transcripts. Heat-killed bacteria and lysate, but not live bacteria, upregulated CD25 transcripts (Fig. 5E).
FIG. 5.
T. denticola inhibited activation of TLR2 in CHO/CD14/TLR2 cells. (A) CHO/CD14/TLR2 cells were infected with various live oral bacterial species at an MOI of 10 or 100 for 16 h and were stained for surface CD25 expression. The plots representing the activation of CD25 expression by various oral bacteria are overlaid with that representing activation by medium alone (no treatment [no treat]). Color coding for the names of bacterial species is as follow: green, early colonizers; orange, orange complex members; and red, red complex members. (B) CHO/CD14/TLR2 cells were infected with live T. denticola, heat-killed T. denticola, or T. denticola lysate (all at an MOI of 100) for 16 h. The plots representing the activation of CD25 expression by T. denticola (pink lines) are overlaid with that representing activation by medium alone (gray shading). (C) CHO/CD14/TLR2 cells were infected with live T. denticola, heat-killed T. denticola, or T. denticola lysate (all at an MOI of 100) in the presence of Pam3CSK for 16 h. The plots representing the activation of CD25 expression by T. denticola (pink lines) are overlaid with those representing activation by medium alone (gray shading) and Pam3CSK alone (sky blue lines). (D) The expression of TLR2 on the surfaces of CHO/CD14/TLR2 cells after a 16-h incubation with medium alone (sky blue lines) or live T. denticola or T. denticola lysate (pink lines) was examined by flow cytometry. Staining with an isotype control antibody served as a negative control (gray shading). (E) The levels of CD25 transcripts in control CHO/CD14/TLR2 cells or cells stimulated with live T. denticola, heat-killed T. denticola, or T. denticola lysate for 16 h were examined by real-time RT-PCR. *, P < 0.05, and **, P < 0.01 versus data for the control. All data (in panels A to E) are representative of results from three independent experiments.
Heat-killed T. denticola did not suppress signaling pathways.
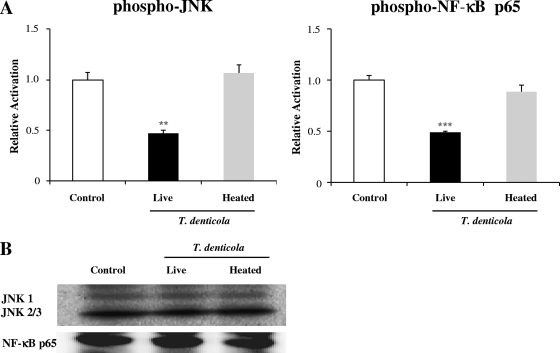
The inability of heat-killed T. denticola to suppress HBD expression was further explored by examining the activation of NF-κB and JNK, which were the most substantially inhibited proteins of those listed in Fig. 2. In contrast to live bacteria, which inhibited both signaling pathways by more than 50%, heat-killed T. denticola did not show any inhibitory effect on the NF-κB and JNK signaling pathways (Fig. 6A). Uniform levels of the nonphosphorylated forms of NF-κB and JNK proteins validated the assay (Fig. 6B). These results indicate that heat-killed T. denticola has lost the ability to inhibit activation of TLR2 and downstream signaling pathways.
FIG. 6.
Heat-killed T. denticola did not suppress signaling pathways. HOK-16B cells plated into a 100-mm dish were stimulated with live or heat-killed T. denticola at an MOI of 1,000 for 30 min, and protein lysates were prepared. (A) Protein lysates (50 μg each) were subjected to ELISA analysis of the phosphorylated forms of JNK and NF-κB p65. The means ± the SEM of the optical densities from four assays are expressed as the levels of activation compared to that in the control culture without bacteria. **, P < 0.01, and ***, P < 0.001 versus data for no infection. (B) Protein lysates (10 μg each) were subjected to Western blotting to detect nonphosphorylated forms of JNK and NF-κB p65.
DISCUSSION
In the present study, we have provided evidence that T. denticola suppresses the expression of HBD-3 in gingival epithelial cells through the inhibition of the TLR2 axis. First, T. denticola inhibited Pam3CSK-mediated activation of TLR2 in TLR2 reporter cells. Second, T. denticola significantly reduced the basal levels of activated NF-κB, JNK, and p38, which are involved in the regulation of HBD expression. Third, knockdown of TLR2 reversed the suppressive effect of T. denticola on HBD-3 expression. Brissette et al. have also shown that several strains of live T. denticola failed to induce various innate immune mediators in NGECs, including HBD-2 (3). However, Brissette et al. concluded that TLR2 does not have a role in the lack of immune modulator production by T. denticola, because they used bacterial cell wall, not live bacteria, in the testing of TLR2-stimulating ability.
The suppressive effects of T. denticola on HBD expression, TLR2 activation, and the activation of signaling pathways required live bacteria. Heat-killed T. denticola bacteria activated rather than suppressed TLR2 in the reporter cells, which explains the slight upregulation of HBD-2 and HBD-3 by these killed bacteria. Although activation of JNK and NF-κB by heat-killed T. denticola was not observed 30 min after infection, activation may occur at other time points. Activation of TLR2 by heat-killed but not by live T. denticola suggests that T. denticola contains a heat-labile inhibitor(s) of the TLR2 signaling axis in addition to ligands that activate TLR2. Since bacterial lysate suppressed neither HBD expression in gingival epithelial cells in a previous study (13) nor TLR2 activation in reporter cells in the present study, the suppressive effect of T. denticola is not likely to be attributable to the T. denticola proteases. Neither live T. denticola nor T. denticola lysate affected the level of TLR2 protein on the cell surface. The reason that live T. denticola, but not T. denticola lysate, produced a suppressive effect may be that the heat-labile inhibitor(s) targets intracellular events. T. denticola can invade gingival epithelial cells, and some of the internalized bacteria were examined in the cytosol 24 h after infection (our unpublished data). The dependence of immune evasiveness on viable bacteria has also been shown previously for Borrelia burgdorferi. Whereas B. burgdorferi bacteria in viable culture medium preferentially enhanced production of an anti-inflammatory cytokine, IL-10, by macrophages, bacteria in nonviable culture medium or heat-killed bacteria preferentially induced a proinflammatory cytokine, tumor necrosis factor alpha (18).
Although the nature of the heat-labile inhibitor(s) in T. denticola is not clear, many bacterial virulence factors that subvert the host innate immune response are known. Toll/IL-1 receptor domain-containing proteins in Brucella melitensis and in virulent uropathogenic Escherichia coli (8) that impede signaling through multiple TLRs by binding to the MyD88 adaptor protein have been identified previously. The suppression of HBD-3 by S. flexneri depends on the MixE bacterial regulator (23). The MixE-dependent proteins include the type III secretion system effectors OspF and OspG, which modify the MAPK and NF-κB pathways and remodel chromatin structure to reduce the host inflammatory response (1, 16). T. denticola may employ a similar mechanism to suppress TLR2 activation, activation of the MAPK and NF-κB pathways, and the expression of HBDs.
The reversal of T. denticola's suppressive effect by TLR2 RNAi suggests that there was an endogenous TLR2 ligand in the control culture without T. denticola. The function of HBD-3 as a ligand for TLR1/TLR2 has been reported previously (12). Thus, the interaction of HBD-3 and TLR2 in gingival epithelial cells is expected to form a positive feedback loop, unless there is something present to interfere with that loop. Indeed, the level of HBD-3 expression in the control cells rapidly increased with time, and this increase was nearly halted by T. denticola for 12 h. In addition, the suppressive effect of T. denticola on HBD expression was more prominent in the presence of 2% human serum. It is noteworthy that the basal level of HBD-3 expression increased in the presence of 2% human serum (data not shown).
It is interesting that the members of the red complex triad commonly demonstrate poor abilities to activate TLR2. We observed previously that T. forsythia also suppresses HBD-3 expression (13). By suppressing HBD expression, T. denticola is expected to perturb the chemical barrier of the gingival epithelium. A breach in the epithelial barrier would facilitate the invasion of plaque bacteria, including T. denticola, thus contributing to the pathogenesis of periodontitis. Studies using sonicates or purified components of T. denticola, such as lipooligosaccharide, peptidoglycan, or a surface protein, have reported the induction of an inflammatory response in murine macrophages, human macrophage-like cells, and fibroblasts (4, 5, 15, 26, 27). However, our results suggest that live bacteria may suppress or paralyze the inflammatory responses of these cells. Infection of murine macrophage-like J774.1 cells with live T. denticola did not induce inflammatory cytokines (25). In contrast, a coculture model composed of epithelial cell-like transformed cells and macrophage-like cells induced some inflammatory mediators in response to T. denticola infection (2). If live T. denticola can reach the lamina propria after invading the epithelium, the evaluation of the immune responses of macrophages and fibroblasts to live bacteria will be important.
In conclusion, T. denticola suppresses the expression of HBD-3 in gingival epithelial cells by inhibiting TLR2 signaling, which may provide new insight into the pathogenesis of periodontitis caused by T. denticola.
Acknowledgments
This study was supported by grants R01-2007-000-10488-0 and R13-2008-008-01003-0 from the Korea Science and Engineering Foundation.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 7 December 2009.
REFERENCES
- 1.Arbibe, L., D. W. Kim, E. Batsche, T. Pedron, B. Mateescu, C. Muchardt, C. Parsot, and P. J. Sansonetti. 2007. An injected bacterial effector targets chromatin access for transcription factor NF-κB to alter transcription of host genes involved in immune responses. Nat. Immunol. 8:47-56. [DOI] [PubMed] [Google Scholar]
- 2.Bodet, C., F. Chandad, and D. Grenier. 2006. Inflammatory responses of a macrophage/epithelial cell co-culture model to mono and mixed infections with Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia. Microbes Infect. 8:27-35. [DOI] [PubMed] [Google Scholar]
- 3.Brissette, C. A., T. T. Pham, S. R. Coats, R. P. Darveau, and S. A. Lukehart. 2008. Treponema denticola does not induce production of common innate immune mediators from primary gingival epithelial cells. Oral Microbiol. Immunol. 23:474-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi, B. K., H. J. Lee, J. H. Kang, G. J. Jeong, C. K. Min, and Y. J. Yoo. 2003. Induction of osteoclastogenesis and matrix metalloproteinase expression by the lipooligosaccharide of Treponema denticola. Infect. Immun. 71:226-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, B. K., S. Y. Moon, J. H. Cha, K. W. Kim, and Y. J. Yoo. 2005. Prostaglandin E(2) is a main mediator in receptor activator of nuclear factor-κB ligand-dependent osteoclastogenesis induced by Porphyromonas gingivalis, Treponema denticola, and Treponema socranskii. J. Periodontol. 76:813-820. [DOI] [PubMed] [Google Scholar]
- 6.Chung, W. O., and B. A. Dale. 2004. Innate immune response of oral and foreskin keratinocytes: utilization of different signaling pathways by various bacterial species. Infect. Immun. 72:352-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung, W. O., S. R. Hansen, D. Rao, and B. A. Dale. 2004. Protease-activated receptor signaling increases epithelial antimicrobial peptide expression. J. Immunol. 173:5165-5170. [DOI] [PubMed] [Google Scholar]
- 8.Cirl, C., A. Wieser, M. Yadav, S. Duerr, S. Schubert, H. Fischer, D. Stappert, N. Wantia, N. Rodriguez, H. Wagner, C. Svanborg, and T. Miethke. 2008. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat. Med. 14:399-406. [DOI] [PubMed] [Google Scholar]
- 9.Dale, B. A. 2002. Periodontal epithelium: a newly recognized role in health and disease. Periodontol. 2000 30:70-78. [DOI] [PubMed] [Google Scholar]
- 10.Feng, Z., and A. Weinberg. 2006. Role of bacteria in health and disease of periodontal tissues. Periodontol. 2000 40:50-76. [DOI] [PubMed] [Google Scholar]
- 11.Froy, O. 2005. Regulation of mammalian defensin expression by Toll-like receptor-dependent and independent signalling pathways. Cell. Microbiol. 7:1387-1397. [DOI] [PubMed] [Google Scholar]
- 12.Funderburg, N., M. M. Lederman, Z. Feng, M. G. Drage, J. Jadlowsky, C. V. Harding, A. Weinberg, and S. F. Sieg. 2007. Human β-defensin-3 activates professional antigen-presenting cells via Toll-like receptors 1 and 2. Proc. Natl. Acad. Sci. U. S. A. 104:18631-18635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji, S., Y. Kim, B.-M. Min, S. H. Han, and Y. Choi. 2007. Innate immune responses of gingival epithelial cells to nonperiodontopathic and periodontopathic bacteria. J. Periodontal Res. 42:503-510. [DOI] [PubMed] [Google Scholar]
- 14.Ji, S., J. E. Shin, Y. S. Kim, J.-E. Oh, B.-M. Min, and Y. Choi. 2009. Toll-like receptor 2 and NALP2 mediate induction of human beta-defensins by Fusobacterium nucleatum in gingival epithelial cells. Infect. Immun. 77:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jun, H. K., Y. M. Kang, H. R. Lee, S. H. Lee, and B. K. Choi. 2008. Highly conserved surface proteins of oral spirochetes as adhesins and potent inducers of proinflammatory and osteoclastogenic factors. Infect. Immun. 76:2428-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, D. W., G. Lenzen, A. L. Page, P. Legrain, P. J. Sansonetti, and C. Parsot. 2005. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. U. S. A. 102:14046-14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krisanaprakornkit, S., J. R. Kimball, A. Weinberg, R. P. Darveau, B. W. Bainbridge, and B. A. Dale. 2000. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatumin oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect. Immun. 68:2907-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazarus, J. J., M. A. Kay, A. L. McCarter, and R. M. Wooten. 2008. Viable Borrelia burgdorferi enhances interleukin-10 production and suppresses activation of murine macrophages. Infect. Immun. 76:1153-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Min, B.-M., K. M. Woo, G. Lee, and N.-H. Park. 1999. Terminal differentiation of normal human oral keratinocytes is associated with enhanced cellular TGF-β and phospholipas c-γ1 levels and apoptotic cell death. Exp. Cell Res. 249:377-385. [DOI] [PubMed] [Google Scholar]
- 20.Park, N.-H., B.-M. Min, S. L. Li, M. Z. Huang, H. M. Cherick, and J. Doniger. 1991. Immortalization of normal human oral keratinocytes with type 16 human papillomavirus. Carcinogenesis 12:1627-1631. [DOI] [PubMed] [Google Scholar]
- 21.Putsep, K., G. Carlsson, H. G. Boman, and M. Andersson. 2002. Deficiency of antibacterial peptides in patients with morbus Kostmann: an observation study. Lancet 360:1144-1149. [DOI] [PubMed] [Google Scholar]
- 22.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 23.Sperandio, B., B. Regnault, J. Guo, Z. Zhang, S. L. Stanley, Jr., P. J. Sansonetti, and T. Pédron. 2008. Virulent Shigella flexneri subverts the host innate immune response through manipulation of antimicrobial peptide gene expression. J. Exp. Med. 205:1121-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taguchi, Y., and H. Imai. 2006. Expression of beta-defensin-2 in human gingival epithelial cells in response to challenge with Porphyromonas gingivalis in vitro. J. Periodontal Res. 41:334-339. [DOI] [PubMed] [Google Scholar]
- 25.Tamai, R., X. Deng, and Y. Kiyoura. 2009. Porphyromonas gingivalis with either Tannerella forsythia or Treponema denticola induces synergistic IL-6 production by murine macrophage-like J774.1 cells. Anaerobe 15:87-90. [DOI] [PubMed] [Google Scholar]
- 26.Tanabe, S., C. Bodet, and D. Grenier. 2008. Treponema denticola lipooligosaccharide activates gingival fibroblasts and upregulates inflammatory mediator production. J. Cell. Physiol. 216:727-731. [DOI] [PubMed] [Google Scholar]
- 27.Tanabe, S. I., C. Bodet, and D. Grenier. 2009. Treponema denticola peptidoglycan induces the production of inflammatory mediators and matrix metalloproteinase 9 in macrophage-like cells. J. Periodontal Res. 44:503-510. doi: 10.1111/j.1600-0765.2008.01141.x. [DOI] [PubMed] [Google Scholar]
- 28.Uitto, V. J., D. Grenier, E. C. Chan, and B. C. McBride. 1988. Isolation of a chymotrypsinlike enzyme from Treponema denticola. Infect. Immun. 56:2717-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vankeerberghen, A., H. Nuytten, K. Dierickx, M. Quirynen, J. J. Cassiman, and H. Cuppens. 2005. Differential induction of human beta-defensin expression by periodontal commensals and pathogens in periodontal pocket epithelial cells. J. Periodontol. 76:1293-1303. [DOI] [PubMed] [Google Scholar]
- 30.Yoshimura, A., E. Lien, R. R. Ingalls, E. Tuomanen, R. Dziarski, and D. Golenbock. 1999. Cutting edge: recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J. Immunol. 163:1-5. [PubMed] [Google Scholar]