Abstract
Vibrio cholerae is the agent of the severe diarrheal disease cholera, and it perpetuates in aquatic reservoirs when not in the host. Within the host's intestines, the bacteria execute a complex regulatory pathway culminating with the production of virulence factors that allow colonization and cause disease. The ability of V. cholerae to form biofilms is thought to aid its persistence in the aquatic environment and passage through the gastric acid barrier of the stomach. The transcriptional activators VpsR and VpsT are part of the biofilm formation-regulatory network. In this study, we screened a V. cholerae genomic library in Escherichia coli cells containing a PvpsT-luxCDBAE transcriptional fusion reporter and found that a plasmid clone containing the aphA gene activates the expression of vpsT in E. coli. AphA is a master virulence regulator in V. cholerae that is required to activate the expression of tcpP, whose gene products in turn activate all virulence genes including those responsible for the synthesis of the toxin-coregulated pilus (TCP) and cholera toxin through the activation of toxT. AphA has a direct effect on the vpsT promoter, as gel shift experiments demonstrated that AphA binds to the vpsT promoter region. Furthermore, V. cholerae aphA mutants exhibit significantly lower levels of vpsT expression as well as reduced biofilm formation. AphA thus links the expression of virulence and biofilm synthesis genes.
Vibrio cholerae is the causative agent of the devastating diarrheal disease cholera. Human infection normally begins with the oral ingestion of food or water contaminated with V. cholerae. Bacteria that survive the acid shock of the stomach proceed to penetrate the mucus layers of the intestinal epithelium, which leads to the production of an array of virulence factors that facilitate adherence and colonization. The coordinate expression of V. cholerae virulence genes results from the activity of a cascading system of regulatory factors (25). Many virulence genes are controlled directly by ToxT. The transcription of toxT is regulated by the ToxRS and TcpPH proteins in response to environmental signals (10). The additional transcriptional regulators AphA and AphB function together to activate tcpPH expression (16, 17, 32). The expression of aphA is repressed by quorum sensing through the master quorum-sensing regulator HapR (18).
Between epidemics, V. cholerae cells live in natural aquatic habitats in association with various plankton and zooplankton, often in the form of biofilms. Various studies suggested that biofilm-mediated attachment to abiotic surfaces may be important for V. cholerae survival in the environment (37, 38, 41). Biofilm formation in V. cholerae is a multistep developmental process that is controlled by several regulatory pathways (37, 42). The surface attachment of V. cholerae activates the transcription of the vps (Vibrio polysaccharide synthesis [VPS]) genes, which are responsible for the synthesis of the VPS exopolysaccharide (13, 28, 41). VPS production is critical for biofilms, as a deletion in any vps gene completely abolishes biofilm formation. The regulation of VPS synthesis in V. cholerae is very complex. Environmental signals such as monosaccharides, nucleosides, indole, and osmolarity have been identified as being activators of vps gene transcription and biofilm formation (11, 13, 29, 31). VpsT and VpsR are positive regulators of vps expression (1, 34, 39). VpsT and VpsR positively autoregulate their own expression and also form a complex regulatory network by positively regulating each other's expression (1, 40). Quorum sensing negatively regulates biofilm formation, as mutations in the master quorum-sensing regulator HapR increase vps expression and biofilm formation (8, 40, 43). HapR directly represses the expression of vpsT (36) and also regulates the expression of vpsR and one of the vps genes (35, 40). Recently, it was reported that a hybrid sensor histidine kinase, VpsS, uses components of the quorum-sensing pathway to modulate biofilm formation (30). Moreover, the transcription of genes involved in biofilm formation is enhanced by the cytoplasmic signaling molecule cyclic di-GMP (c-di-GMP) and negatively regulated by the cyclic AMP (cAMP)-cAMP receptor protein (CRP) regulatory complex (5, 12, 19-22, 34).
Bacterial biofilms have been demonstrated to be clinically significant for many human pathogens because biofilm-associated bacteria are less susceptible to host immune responses and antimicrobial agents (3). Biofilm formation is also important for V. cholerae pathogenesis. Several lines of evidence suggested that V. cholerae may enter hosts in the form of biofilms. The concentration of V. cholerae required to induce symptomatic cholera is estimated to be approximately 104 to 106 total cells, a concentration easily achieved by bacteria within biofilms accumulated on biotic or abiotic surfaces (2). Evidence to support this was provided when the number of cholera cases in a Bangladeshi village dramatically declined following crude water filtration through sari cloth (2). Furthermore, free-living V. cholerae bacteria are highly sensitive to low pH, while biofilm-associated V. cholerae bacteria are more acid resistant, resulting in the hypothesis that this increased resistance to acid may promote survival during passage through the stomach (43). However, it is unclear how V. cholerae regulates its biofilm formation during infection. In this study, we performed a genetic screen and found that the virulence activator AphA enhances biofilm formation through the activation of vpsT expression, which provides a link between the pathways of virulence factor production and biofilm formation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
All V. cholerae strains used in this study were derived from E1 Tor C6706 (33) and were propagated in Luria broth (LB) medium containing appropriate antibiotics at 37°C. For biofilm formation, test tubes containing V. cholerae cultures were incubated at 22°C without shaking. Transcriptional fusion reporters were constructed by cloning promoter sequences of the genes of study into pBBR-lux, which contains a promoterless luxCDABE reporter (9), digested with BamHI and SacI. The Lux reporter containing a deletion of the putative AphA binding site TATACA in the vpsT promoter region was constructed by overlapping PCR and cloned into plasmid pBBR-lux as described above. The plasmids containing Ptac-aphA or Ptac-aphB were constructed by cloning aphA and aphB coding sequences into pMal-c2x (New England Biolabs) digested with NdeI and HindIII (the malE gene is replaced by aphA or aphB). The plasmid containing PBAD-aphA was constructed by cloning aphA coding sequences into pBAD24 (7) digested with EcoRI and HindIII. The plasmid overexpressing recombinant AphA was constructed by cloning the aphA coding sequences into pET32a (EMD Biosciences) digested with NdeI and XhoI. The resulting plasmid that produces the AphA-His6 C-terminal fusion was then introduced into Escherichia coli strain BL21(DE3) (Promega). In-frame deletions of aphA, vpsT, vpsR, lrp, and cdgC were constructed by cloning the regions flanking the target genes into suicide vector pWM91 containing a sacB counterselectable marker (27). The resulting plasmids were introduced into V. cholerae by conjugation, and deletion mutants were selected for double homologous recombination events.
Screening for vpsT activators in E. coli.
The V. cholerae genomic library was constructed by cloning partially HincII-digested V. cholerae genomic DNA fragments into pEZ-Seq-Amp (Lucigen). E. coli DH5α transformants containing library plasmids and the plasmid harboring PvpsT-luxCDABE were spread onto LB agar plates containing the appropriate antibiotics. Bright colonies were restreaked, and plasmids were purified for sequencing.
Measurement of transcriptional expression using Lux reporters.
Bacterial cultures were grown without shaking at 22°C for 24 h, and the luminescence of either planktonic cells or biofilm-associated cells was read by using a Bio-Tek Synergy HT spectrophotometer and normalized for growth against the optical density at 600 nm (OD600). Lux expression is reported as light units/OD600.
Gel retardation assays.
E. coli BL21(DE3) cells containing a plasmid overexpressing C-terminal His-tagged AphA were grown at 37°C and induced with 1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) at 16°C. The AphA-His6 protein was purified through nickel columns according to the manufacturer's instructions (Qiagen). PCR products containing various lengths of the vpsT promoter region and the full tcpP promoter region were digested with EcoRI and end labeled by using [α-32P]dATP and the Klenow fragment of DNA polymerase I. Binding reaction mixtures contained 0.1 ng DNA and His6-AphA proteins in a buffer containing 10 mM Tris-HCl (pH 7.9), 1 mM EDTA, 1 mM dithiothreitol (DTT), 60 mM KCl, and 30 mg/ml calf thymus DNA. After 20 min of incubation at 37°C, samples were size fractionated by using 5% polyacrylamide gels in 1× TAE buffer (20 mM Tris-acetate, 1 mM EDTA [pH 8.5]). The radioactivity of free DNA and AphA-DNA complexes was visualized by using a Typhoon 9410 variable model imager (Molecular Dynamics).
Biofilm formation assays.
V. cholerae strains were grown on LB agar plates overnight and resuspended in LB at an OD600 of ∼0.6. A 1:100 dilution of this suspension was then inoculated into 1 ml of LB in glass test tubes. Biofilms were formed by allowing these cultures to stand at room temperature. At the time points indicated, culture supernatants were removed, and biofilms were washed in LB medium. Biofilm formation was quantified by use of crystal violet as described previously (44).
Infant mouse colonization assays.
The infant mouse colonization assay was performed as previously described (6), by inoculating approximately 105 V. cholerae cells per mouse into 6-day-old suckling mice. After a period of colonization, intestinal homogenates were collected, and the ratio of the two strains was determined by plating onto LB agar containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and normalized against the ratio of the two strains grown in vitro.
RESULTS AND DISCUSSION
Identification of AphA as a positive regulator of vpsT.
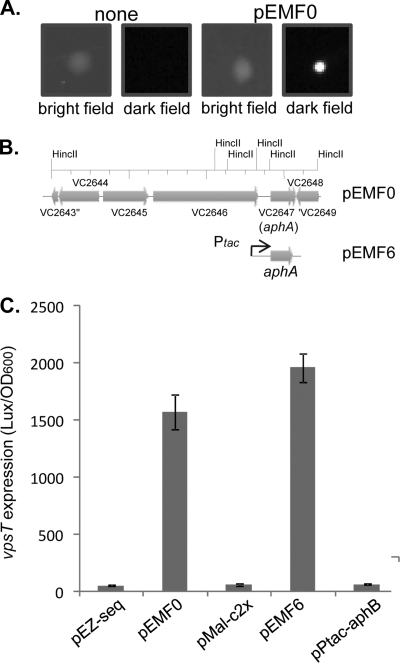
V. cholerae biofilm formation requires a number of transcriptional regulators. The expression of one of the key regulators, VpsT, is known to be controlled by a number of regulatory components, such as quorum sensing (42), c-di-GMP (21), the biofilm activator VpsR (40), and osmolarity (31). To further investigate whether additional regulation is involved in vpsT transcription, we constructed a PvpsT-luxCDABE transcriptional fusion on a plasmid. We found that V. cholerae strains containing this plasmid produced light (data not shown), but E. coli cells with the PvpsT-luxCDABE plasmid were dim (Fig. 1A, left two panels). We reasoned that a V. cholerae factor necessary for the activation of vpsT may not be present in E. coli to induce its expression. Thus, we introduced a V. cholerae genomic library into E. coli strains containing PvpsT-luxCDABE plasmids and screened for bright transformants. Of approximately 50,000 transformants screened, 11 bright colonies were isolated (Fig. 1A, right two panels). The plasmid from each isolate (pEMF0) was purified, and analysis by restriction enzyme digestion indicated that they all contained a similar 7.2-kb insertion and were possibly siblings of one original clone. DNA sequencing revealed that this fragment covered the V. cholerae genome region of VC2643 to VC2649 (Fig. 1B). Although most of the genes in this region encode products involved in bacterial metabolism, VC2647 encodes a key virulence activator, AphA. AphA is required for the induction of another virulence regulator, TcpP (32), and it also regulates the expression of a number of other genes, including a gene encoding penicillin amidase (15) and genes encoding enzymes involved in acetoin biosynthesis (14).
FIG. 1.
AphA is required to induce vpsT in E. coli cells. (A). Luminescence production of E. coli DH5α colonies containing the PvpsT-luxCDABE plasmid and V. cholerae genomic library plasmids. Pictures of colonies were taken either in light or in the dark. (B) Genetic organization of a V. cholerae fragment (in pEMF0) that activates vpsT expression. pEMF6 was constructed by cloning aphA into the pMal-c2x vector (New England Biolabs). (C) Luminescence of E. coli DH5α strains containing PvpsT-luxCDABE with vector controls, pEMF0, pEMF6, or a plasmid overexpressing aphB. Cells were grown in the presence of 0.1 mM IPTG in LB at 37°C for 8 h, and luminescence was measured by using a Bio-Tek plate reader and normalized against the OD600. The results are the averages of data from three experiments. Error bars indicate standard deviations.
To test whether AphA can activate vpsT expression in E. coli, we constructed a plasmid containing Ptac-aphA (pEMF6). In the absence of IPTG, the level of vpsT expression was low (data not shown). The overexpression of aphA in the presence of IPTG (100 μM, to induce the Ptac promoter) induced vpsT expression to levels similar to those when aphA was expressed from pEMF0 (Fig. 1C). This construct could also activate tcpP expression in E. coli (data not shown). These data suggest that AphA plays a role in the regulation of vpsT expression, at least in E. coli. In its previously described role of activating the tcpPH promoter, AphA acts cooperatively with its partner protein, AphB (16). To determine whether AphB could play a role in the activation of vpsT, we constitutively expressed aphB and found that AphB was unable to drive vpsT expression (Fig. 1C), indicating that AphB cannot promote vpsT activity and that AphA may be the primary activator.
AphA binds directly to the vpsT promoter region.
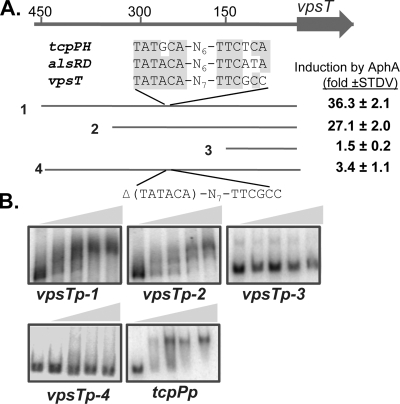
To investigate whether AphA activation of vpsT is direct or acts through another regulator present in E. coli, we purified AphA as a His6 fusion. Recombinant AphA is functional, as it could activate both tcpP and vpsT in E. coli (data not shown). We then performed electrophoretic mobility shift assays (EMSAs) using AphA-His6 and various lengths of vpsT promoter DNA (Fig. 2A). We also included tcpP promoter DNA as a control. Figure 2B shows that purified AphA-His6 was able to shift the two large vpsT promoter fragments as well as control tcpP DNA with similar affinities. All of these mobility shifts could be inhibited by the addition of unlabeled DNA, indicating that the binding of AphA to these DNA sequences is specific (data not shown). AphA was unable to shift the shortest vpsT promoter fragment encompassing 150 bp upstream of the vpsT translational start site, suggesting that the AphA binding site is located between 150 and 450 bp upstream of the vpsT gene. Consistent with the gel shift data, AphA could not induce vpsT expression when the 150-bp fragment was fused with the luxCDABE reporter in E. coli DH5α cells (Fig. 2A). Of note, we did not rule out the possibility that this fragment may be too small to include all the regulatory components of the vpsT promoter. However, by searching the vpsT promoter region for the AphA binding site identified previously (14), we found that a similar region is located around 240 bp upstream of vpsT. We deleted TATACA from the vpsT promoter sequences and found that AphA failed to activate vpsT expression (measured by the Lux reporter fusion) (Fig. 2A) and that AphA proteins could not bind to the mutated vpsT promoter region (vpsTp-4) (Fig. 2B). Further study is required to confirm that AphA binds to this sequence on the vpsT promoter. Taken together, these data suggest that AphA directly regulates vpsT expression.
FIG. 2.
AphA binds to the vpsT promoter region to regulate vpsT gene expression. (A) The vpsT locus and putative AphA-binding-site sequences in the promoter. Numbers are relative to the translational start of vpsT. The four promoter regions indicated were PCR amplified and cloned into pBBR-lux containing a transcriptional Lux reporter (9). The induction of vpsT by AphA was calculated by comparing Lux expression levels with pMal-c2x (vector control) or with pEMF6 (Ptac-aphA) in the presence of 0.1 mM IPTG. STDV, standard deviation. (B) Gel shift assays using purified AphA-His6 and DNA as shown above or the tcpP promoter as a control. Protein concentrations used in the gel shift assay (shown as shaded triangles) were 0, 10, 20, 40, and 80 ng/reaction mixture.
AphA regulates vpsT expression and biofilm formation in V. cholerae.
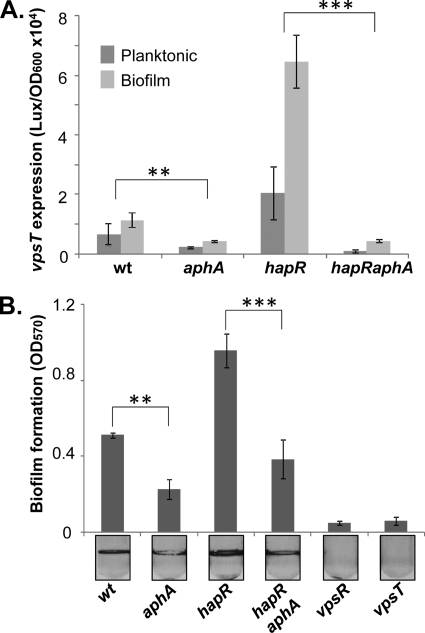
We next examined the effect of natively produced AphA on vpsT expression and biofilm formation in V. cholerae. We first compared vpsT expression by monitoring light production in strains containing wild-type aphA or an aphA in-frame deletion with the PvpsT-luxCDABE plasmid. The level of expression of vpsT was higher in biofilm-associated cells than in planktonic cells (Fig. 3A), consistent with previous findings that the vps gene expression level is elevated in biofilm cells compared to planktonic cells (1). The aphA mutants exhibited low vpsT expression levels in both planktonic and biofilm-associated cells (approximately one-third of the vpsT expression level in the wild type; Student t test P value of <0.05) (Fig. 3A and Table 1), and the overexpression of aphA in aphA mutants restored vpsT expression (Table 1), indicating that AphA is involved with vpsT regulation in V. cholerae. In addition, we tested the effect of AphA on vpsT expression in hapR mutants, as the expression of aphA itself is repressed by quorum sensing through the master quorum-sensing regulator HapR (18). We found that the effects of AphA on vpsT in both planktonic and biofilm cells became more prominent (Fig. 3A). We reason that since the aphA expression level is higher in hapR mutants, the deletion of aphA in the hapR mutant background produces a greater effect on vpsT expression.
FIG. 3.
AphA modulates vpsT expression and biofilm formation in V. cholerae. Different V. cholerae strains containing vpsT-luxCDABE plasmids were grown without shaking in LB for 24 h. Both planktonic and biofilm cells were withdrawn. (A) Luminescence was measured, and data were normalized against the OD600. (B) The biofilm mass was determined by staining surface-attached cells with crystal violet and dimethyl sulfoxide (DMSO) quantitation. The results are the averages of data from three experiments. Error bars indicate standard deviations. The Student t test for P value was performed (**, P value of <0.05; ***, P value of <0.01). wt, wild type.
TABLE 1.
Effects of AphA and other regulatory components on vpsT expression
| Genotypeb | Avg (SD) vpsT expression level (light units/OD600 [103])a |
|
|---|---|---|
| Vector | pBAD-aphA | |
| Wild type | 6.1 (0.4) | 15.7 (0.5) |
| aphA | 2.1 (0.03) | 7.1 (0.04) |
| vpsR | 0.092 (0.02) | 1.6 (0.02) |
| lrp | 2.3 (0.99) | 11.7 (2.5) |
| cdgC | 7.0 (1) | 13.9 (0.4) |
Values presented are the averages of three replicates. The numbers in parentheses indicate standard deviations. Cultures were grown at 22°C without shaking for 24 h. A total of 0.1% arabinose was added to the cultures. Luminescence was measured by using planktonic cells.
The wild type and the various in-frame deletion mutants indicated containing the PvpsT-luxCDABE reporter were introduced with either the vector control (pBAD24) or an aphA overexpression plasmid (pBAD-aphA).
It was previously reported that the expression of vpsT is regulated by a number of factors, including VpsR and c-di-GMP (42). Interestingly, the expression of aphA is activated by VpsR and the leucine-responsive regulatory protein (Lrp) (23). To determine whether VpsR and Lrp regulations of vpsT act through the activation of aphA, we compared PvpsT-luxCDABE expression levels in vpsR mutants in the presence or absence of overexpressed AphA (Table 1). The level of expression of vpsT in vpsR mutants was very low, and the overexpression of AphA only slightly increased vpsT expression levels. On the other hand, the overexpression of AphA could restore vpsT expression in lrp mutants. These data suggest that VpsR is a major positive regulator of vpsT expression. In addition, unlike data from a previous report (22), a deletion in cdgC, encoding a GGDEF-EAL domain protein, did not affect vpsT expression (Table 1). It is possible that the difference in strain background and rugose/smooth phenotypes results in these discrepancies.
Because VpsT is an essential activator for vps genes, a reduction of vpsT expression levels will lead to a decrease in biofilm formation. We thus measured the biofilm-forming capacities of different V. cholerae strains. The deletion of either vps activator (VpsR or VpsT) abolished biofilm formation in this V. cholerae strain (C6706 background) (Fig. 3B). The amount of biofilm formed by aphA mutants was significantly smaller than that formed by aphA+ strains. However, biofilm formation was not completely abolished in aphA mutants as it was in vps mutants (Fig. 3B and data not shown). This finding together with the observation that the overexpression of AphA in the vpsR mutant could not restore vpsT expression (Table 1) suggest that AphA plays a modulating role in V. cholerae biofilm formation.
Temporal requirements of AphA during V. cholerae biofilm formation.
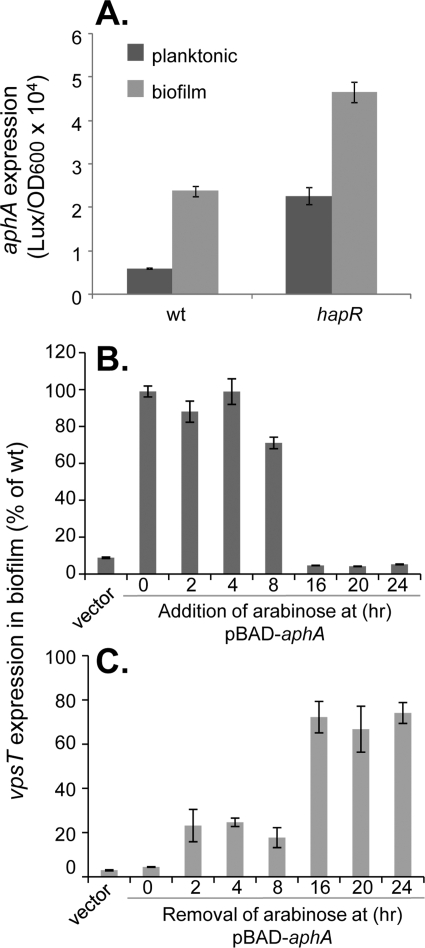
To further understand the physiological role of the AphA regulation of biofilm formation, we first examined the expression of aphA in planktonic and biofilm-associated cells using a plasmid containing PaphA-luxCDABE. Figure 4A shows that the aphA expression level in both the wild type and hapR mutants was significantly higher in biofilm-associated cells than in planktonic cells. Interestingly, since HapR represses aphA expression (24) and is present in higher levels in biofilm cells than in planktonic cells, the elevated level of expression of aphA in biofilms suggests that additional regulatory components are involved with aphA regulation in biofilms. Previous studies showed that environmental conditions such as temperature and pH do not alter aphA expression in LB medium (32). Throughout its natural cycle of infection, however, V. cholerae encounters many various environments that present unique challenges for bacterial survival and colonization. Therefore, it is possible that there is an unknown condition that may alter aphA expression and affect biofilm formation at some point in time during infection. We thus tested the temporal effect of AphA on vpsT expression by using V. cholerae aphA mutants containing an inducible PBAD-aphA plasmid. This strain formed biofilms similar to that of the wild type when 0.1% arabinose was included in the medium (data not shown). We then added or removed arabinose at different time points during biofilm development and measured vpsT expression in biofilms over a 24-h incubation period. When induced with arabinose up to 8 h after the beginning of incubation, we were able to restore vpsT expression levels to wild-type levels. In contrast, when pBAD-aphA was induced later during incubation, little expression of vpsT was observed (Fig. 4B). Similarly, the early removal of arabinose (up to 8 h postinoculation) resulted in lower vpsT expression levels (Fig. 4C). Taken together, these data suggest that AphA and, therefore, VpsT are required during the earlier stages of biofilm formation. aphA is expressed at a low cell density in LB, but how aphA is regulated temporally under certain environmental conditions and in host intestines is currently unclear and will be further investigated.
FIG. 4.
Temporal requirements of AphA in V. cholerae biofilm formation. (A) Expression of aphA in planktonic and biofilm-associated cells. The wild type and hapR mutants containing PaphA-luxCDABE plasmids were grown in LB at 22°C for 24 h, and the luminescence of planktonic and biofilm cells was measured and normalized against the OD600. (B and C) vpsT expression in biofilm-associated cells of aphA mutants containing pBAD-aphA plasmids. The aphA mutants containing pBAD24 were used as a vector control. The level of vpsT expression was compared to that of wild-type bacteria containing the PvpsT-luxCDABE plasmid. (B) The strains were inoculated in LB in the absence of arabinose, 0.1% arabinose was then added to the cultures at the time indicated, and growth continued for a total of 24 h at 22°C. (C) The strains were inoculated in LB in the presence of 0.1% arabinose. At the time points indicated, the culture supernatants were removed and replaced with equal volumes of fresh LB. After a total of 24 h of incubation, luminescence was measured and normalized against the OD600. The data were then compared with the vpsT expression level (Lux/OD600) of the wild-type strain in biofilms incubated for 24 h at 22°C and are presented as a percentage of wild-type vpsT expression levels. The results are averages of data from three experiments. Error bars indicate standard deviations.
Roles of AphA-regulated biofilm formation in the infant mouse colonization model.
Biofilm formation was previously shown to be important for V. cholerae survival in aquatic environments and resistance to gastric acids during passage through the host stomach (43). It is unclear, however, whether V. cholerae biofilm formation-related genes are important during infection. We used the infant mouse model to test the colonization abilities of aphA, vpsT, and vps mutants. The aphA mutant was profoundly defective in colonization (data not shown), consistent with a previous report showing that AphA is required to activate virulence genes and subsequently colonize the intestines (17). Both vpsT and vps mutants that abolished biofilm formation in vitro (Fig. 3B and data not shown) could colonize infant mice as well as the wild type (data not shown), suggesting that VPS-dependent biofilm formation may not be critical for the colonization of the infant mouse. Since V. cholerae infection of infant mice does not replicate some naturally occurring human cholera symptoms (e.g., watery diarrhea), this model may not be suitable for analyzing in vivo biofilm formation. Further studies using other animal models are required to elucidate the relationship between virulence gene induction and biofilm formation controlled by the shared regulator AphA.
In this study, we demonstrate that the indispensable virulence gene regulator AphA is also capable of promoting V. cholerae biofilm development through the direct stimulation of the vpsT promoter. This adds another regulatory component to the already complex regulatory pathway of biofilm formation. Throughout its natural cycle of infection, V. cholerae encounters several various environments that present challenges for bacterial survival and colonization. Endurance in hostile environments is facilitated by encapsulation within the matrix of a protective biofilm structure, and host colonization is aided by the expression of various virulence factors activated by a signal transduction cascade. AphA serves as a vital connection between these two processes. It is possible that during initial infection, V. cholerae cells utilize AphA to upregulate virulence factor production through the activation of tcpP expression, facilitating the bacterial colonization of intestinal surfaces. Simultaneously, AphA also enhances V. cholerae biofilm formation in the host, leading to resistance to various antimicrobial agents. Although biofilm formation apparently is not important for V. cholerae colonization in the infant mouse model, a recent report (4) indicated that stool samples collected from cholera patients contain a heterogeneous mixture of biofilm-like aggregates and free-swimming planktonic cells of V. cholerae. An estimation of the relative infectivity of these different forms of V. cholerae cells suggested that the enhanced infectivity of V. cholerae shed in human stools (26) is due largely to the presence of biofilm-like structures. aphA expression is repressed by quorum sensing at a high cell density (18). This repression presumably happens late in V. cholerae infection and may help bacteria exit the host (44). However, since AphA is required during the early stages of biofilm formation (Fig. 4), quorum-sensing repression of aphA expression may not affect biofilm structures. On the other hand, once V. cholerae bacteria come out of the host, the derepression of aphA in planktonic V. cholerae cells (becoming low-cell-density states) may help initiate biofilm formation in the environment. Intriguingly, the expression of aphA itself is also activated by the biofilm formation regulator VpsR (23). VpsR and VpsT also positively autoregulate their own and each other's expressions (1, 40). It may be interesting to further study how these positive regulatory loops contribute to the success of V. cholerae as a human pathogen and a colonizer of the aquatic environment.
Acknowledgments
We thank the laboratory members for discussion and reviewing the manuscript.
This study was supported by NIH/NIAID grant AI073419 (to J.Z.).
Editor: A. Camilli
Footnotes
Published ahead of print on 23 November 2009.
REFERENCES
- 1.Casper-Lindley, C., and F. H. Yildiz. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 186:1574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colwell, R. R., A. Huq, M. S. Islam, K. M. Aziz, M. Yunus, N. H. Khan, A. Mahmud, R. B. Sack, G. B. Nair, J. Chakraborty, D. A. Sack, and E. Russek-Cohen. 2003. Reduction of cholera in Bangladeshi villages by simple filtration. Proc. Natl. Acad. Sci. U. S. A. 100:1051-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 4.Faruque, S. M., K. Biswas, S. M. Udden, Q. S. Ahmad, D. A. Sack, G. B. Nair, and J. J. Mekalanos. 2006. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. U. S. A. 103:6350-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fong, J. C., and F. H. Yildiz. 2008. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J. Bacteriol. 190:6646-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gardel, C. L., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64:2246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammer, B. K., and B. L. Bassler. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101-104. [DOI] [PubMed] [Google Scholar]
- 9.Hammer, B. K., and B. L. Bassler. 2007. Regulatory small RNAs circumvent the conventional quorum sensing pathway in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 104:11145-11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hase, C. C., and J. J. Mekalanos. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 95:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haugo, A. J., and P. I. Watnick. 2002. Vibrio cholerae CytR is a repressor of biofilm development. Mol. Microbiol. 45:471-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houot, L., and P. I. Watnick. 2008. A novel role for enzyme I of the Vibrio cholerae phosphoenolpyruvate phosphotransferase system in regulation of growth in a biofilm. J. Bacteriol. 190:311-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kierek, K., and P. I. Watnick. 2003. Environmental determinants of Vibrio cholerae biofilm development. Appl. Environ. Microbiol. 69:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovacikova, G., W. Lin, and K. Skorupski. 2005. Dual regulation of genes involved in acetoin biosynthesis and motility/biofilm formation by the virulence activator AphA and the acetate-responsive LysR-type regulator AlsR in Vibrio cholerae. Mol. Microbiol. 57:420-433. [DOI] [PubMed] [Google Scholar]
- 15.Kovacikova, G., W. Lin, and K. Skorupski. 2003. The virulence activator AphA links quorum sensing to pathogenesis and physiology in Vibrio cholerae by repressing the expression of a penicillin amidase gene on the small chromosome. J. Bacteriol. 185:4825-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kovacikova, G., and K. Skorupski. 2000. Differential activation of the tcpPH promoter by AphB determines biotype specificity of virulence gene expression in Vibrio cholerae. J. Bacteriol. 182:3228-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovacikova, G., and K. Skorupski. 2001. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol. Microbiol. 41:393-407. [DOI] [PubMed] [Google Scholar]
- 18.Kovacikova, G., and K. Skorupski. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 46:1135-1147. [DOI] [PubMed] [Google Scholar]
- 19.Liang, W., A. Pascual-Montano, A. J. Silva, and J. A. Benitez. 2007. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology 153:2964-2975. [DOI] [PubMed] [Google Scholar]
- 20.Liang, W., A. J. Silva, and J. A. Benitez. 2007. The cyclic AMP receptor protein modulates colonial morphology in Vibrio cholerae. Appl. Environ. Microbiol. 73:7482-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim, B., S. Beyhan, J. Meir, and F. H. Yildiz. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60:331-348. [DOI] [PubMed] [Google Scholar]
- 22.Lim, B., S. Beyhan, and F. H. Yildiz. 2007. Regulation of Vibrio polysaccharide synthesis and virulence factor production by CdgC, a GGDEF-EAL domain protein, in Vibrio cholerae. J. Bacteriol. 189:717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, W., G. Kovacikova, and K. Skorupski. 2007. The quorum sensing regulator HapR downregulates the expression of the virulence gene transcription factor AphA in Vibrio cholerae by antagonizing Lrp- and VpsR-mediated activation. Mol. Microbiol. 64:953-967. [DOI] [PubMed] [Google Scholar]
- 24.Liu, Z., F. R. Stirling, and J. Zhu. 2007. Temporal quorum-sensing induction regulates Vibrio cholerae biofilm architecture. Infect. Immun. 75:122-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matson, J. S., J. H. Withey, and V. J. DiRita. 2007. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect. Immun. 75:5542-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metcalf, W. W., W. Jiang, L. L. Daniels, S. K. Kim, A. Haldimann, and B. L. Wanner. 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1-13. [DOI] [PubMed] [Google Scholar]
- 28.Moorthy, S., and P. I. Watnick. 2004. Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol. Microbiol. 52:573-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller, R. S., S. Beyhan, S. G. Saini, F. H. Yildiz, and D. H. Bartlett. 2009. Indole acts as an extracellular cue regulating gene expression in Vibrio cholerae. J. Bacteriol. 191:3504-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shikuma, N. J., J. C. Fong, L. S. Odell, B. S. Perchuk, M. T. Laub, and F. H. Yildiz. 2009. Overexpression of VpsS, a hybrid sensor kinase, enhances biofilm formation in Vibrio cholerae. J. Bacteriol. 191:5147-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shikuma, N. J., and F. H. Yildiz. 2009. Identification and characterization of OscR, a transcriptional regulator involved in osmolarity adaptation in Vibrio cholerae. J. Bacteriol. 191:4082-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skorupski, K., and R. K. Taylor. 1999. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol. Microbiol. 31:763-771. [DOI] [PubMed] [Google Scholar]
- 33.Thelin, K. H., and R. K. Taylor. 1996. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect. Immun. 64:2853-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsou, A. M., T. Cai, Z. Liu, J. Zhu, and R. V. Kulkarni. 2009. Regulatory targets of quorum sensing in Vibrio cholerae: evidence for two distinct HapR-binding motifs. Nucleic Acids Res. 37:2747-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waters, C. M., W. Lu, J. D. Rabinowitz, and B. L. Bassler. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190:2527-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yildiz, F. H., X. S. Liu, A. Heydorn, and G. K. Schoolnik. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497-515. [DOI] [PubMed] [Google Scholar]
- 41.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yildiz, F. H., and K. L. Visick. 2009. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17:109-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu, J., and J. J. Mekalanos. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647-656. [DOI] [PubMed] [Google Scholar]
- 44.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]