Abstract
Mutations in either of two human presenilin genes (PS1 and PS2) cause Alzheimer’s disease. Here we describe genetic and physical interactions between Caenorhabditis elegans SEL-10, a member of the Cdc4p family of proteins, and SEL-12, a C. elegans presenilin. We show that loss of sel-10 activity can suppress the egg-laying defective phenotype associated with reducing sel-12 activity, and that SEL-10 can physically complex with SEL-12. Proteins of the Cdc4p family have been shown to target proteins for ubiquitin-mediated turnover. The functional and physical interaction between sel-10 and sel-12 therefore offers an approach to understanding how presenilin levels are normally regulated.
Alzheimer’s disease (AD) is a major cause of dementia among elderly people. The brains of affected individuals have a characteristic pathology, including hallmark amyloid plaques and neurofibrillary tangles. The main constituent of amyloid plaques is Aβ42(43), a β-amyloid precursor protein processing product. The “amyloid hypothesis” posits that deposition of this peptide causes AD; however, the mechanism by which this peptide exerts its toxic effect is unclear (1–3).
Familial forms of AD have enabled the identification of genes that may illuminate the underlying molecular basis of the disease. In addition to the gene encoding the β-amyloid precursor protein itself, familial AD mutations have defined two additional genes, which encode the proteins presenilin 1 (PS1) and presenilin 2 (PS2). Mutations in the presenilins account for a significant portion (up to 60%) of early-onset familial AD cases. Presenilins appear to have eight transmembrane domains (see ref. 4), and most studies have localized presenilins to endoplasmic reticulum/Golgi subcellular compartments (5–8). There is a growing body of evidence that presenilins are involved in the trafficking or processing of a selective group of proteins that includes β-amyloid precursor protein and LIN-12/Notch proteins, but the mechanism of presenilin function remains unknown.
Individuals carrying mutations in PS1 or PS2 have elevated levels of Aβ42(43) (9, 10), and elimination of PS1 activity reduces the proteolysis of the β-amyloid precursor protein (11). It has therefore been suggested that one approach to antiamyloidogenic therapy for AD is reducing presenilin activity (11). Here, we report the use of a simple model organism, Caenorhabditis elegans, to identify genes that may serve as the basis for such therapies.
In C. elegans, there are two functional presenilin genes, sel-12 and hop-1. The sel-12 gene was found in a genetic screen for mutations that suppress the phenotypic effects of a constitutively active transmembrane receptor of the LIN-12/Notch family (12). The SEL-12 protein appears to be a bona fide presenilin, since human PS1 or PS2 can substitute for SEL-12 in C. elegans (13). Another C. elegans gene, hop-1, encodes a somewhat more divergent protein that also appears to be a bona fide presenilin, since HOP-1 can substitute for SEL-12 (14). Furthermore, genetic studies suggest that sel-12 and hop-1 are functionally redundant in facilitating the activity of LIN-12/Notch proteins in several cell fate decisions (14).
Genetic screens for suppressors of mutations in the C. elegans sel-12 presenilin constitute one approach to identifying factors that may regulate presenilin level or activity. sel-12 mutations cause a highly penetrant egg-laying defective (Egl) phenotype (12). Suppressors can be easily identified by looking for the presence of eggs among the progeny of mutagenized sel-12 mutant hermaphrodites or by constructing double mutants carrying mutations in candidate genes and in sel-12. In principle, loss-of-function mutations that suppress the Egl defect of sel-12 mutants might augment or stabilize mutant SEL-12 proteins or HOP-1(+), or reduce or bypass the need for presenilin activity.
Here, we show that the sel-10 gene has properties of a candidate factor for regulating presenilin level or activity. The sel-10 gene was originally defined by suppressors of a subset of defects caused by reduced lin-12 activity and was found to encode a member of the Cdc4p family of F-box/WD40 repeat containing proteins (15, 16). Based on biochemical and functional studies, F-box/WD40 proteins have been proposed to recruit substrates for ubiquitination (17, 18). The F-box is believed to interact with core ubiquitination complexes, and the WD40 repeats are believed to interact with substrates (17–19). Ubiquitination generally leads to rapid protein degradation, although this is not necessarily its sole consequence (18), and there is evidence that presenilins are degraded by the ubiquitin–proteasome pathway (20–22). Different Cdc4p-like proteins have been implicated in the targeting of a variety of substrates. The resemblance of SEL-10 to Cdc4p led us to explore the possibility that sel-10 is a potential regulator of presenilin level or activity. The genetic and functional interactions we observed suggest that sel-10 is indeed a candidate for such a regulator.
MATERIALS AND METHODS
Genetic Methods.
Standard procedures and conditions for the maintenance and manipulation of C. elegans are described in ref. 23. All experiments were performed at 20°C, unless otherwise indicated. Mutations used in this study were: lin-12(e2621) III (gift of Jonathan Hodgkin), sel-10(ar41) V (15, 16), him-5(e1490) V (24), sel-12(ar131) and sel-12(ar171) X (12), unc-1(e538) X (23). Hermaphrodites carrying sel-12 mutations were scored as egg-laying proficient (non-Egl) if they displayed robust egg-laying comparable to a wild-type hermaphrodite.
Plasmids for Cell Culture Experiments.
Plasmids used in the transient transfection experiments were constructed in pQNCX (Qingyou Yan and J.K., unpublished observations), vectors that drive gene expression under the control of a CMV promoter.
pQNClacZ contains the bacterial lacZ gene.
pQNCsel-10myc encodes a protein with six myc epitope tags (25) fused in frame to the N terminus of SEL-10 at amino acid 13.
pQNCsel-10 hemagglutinin (HA) encodes a protein with an HA epitope tag fused in frame to the N terminus of SEL-10 at amino acid 13.
pQNCsel-12myc encodes a protein with six myc epitope tags fused in frame to the SEL-12 loop region between amino acids 336 and 337.
pQNCsel-12HA encodes a protein with an HA epitope fused in frame to the C terminus of SEL-12 at amino acid 441.
Transfections, Immunoprecipitations, and Western Blot Analysis.
293T (Bosc23) cells (26) were maintained in DMEM with 10% fetal bovine serum. A confluent plate of cells was split 1:3 the day before transfection. For one 60-mm plate of cells, 4 μg of each plasmid DNA was transfected by using the calcium phosphate precipitation method. The total amount of DNA was kept constant by supplementation with lacZ-containing plasmids.
Two days after transfection, cells were harvested and lysed in TENT buffer (50 mM Tris⋅Cl (pH 8.0)/2 mM EDTA/150 mM NaCl/1% Triton-X 100) containing protease inhibitors (2 μg/ml aprotinin/2 μg/ml leupeptin/2 μg/ml pepstatin/0.5 mM phenylmethylsulfonyl fluoride). Lysates were clarified by centrifugation at 10,000 × g for 10 min, protein content was determined by using the Bio-Rad Protein Assay Kit, and samples were normalized for protein content. Extracts were precleared with Sepharose CL-4B beads, incubated with antibodies (50 μl of 12CA5 anti-HA supernatant or 200 μl of 9E10 anti-myc supernatant) for 6 hours at 4°C, then incubated with 40 μl of 50% slurry of protein A-Sepharose for 1 hour at 4°C. The protein A-Sepharose beads were washed with TENT buffer three times by vortexing for 5 min, beads were boiled in 30 μl of 1× protein loading buffer and then electrophoresed on a 10% SDS–polyacrylamide gel and transferred onto nitrocellulose membrane.
A Western blot was first blocked overnight at 4°C with TBST (10 mM Tris, pH 8.0/150 mM NaCl/0.2% Tween 20) containing 1% BSA (TBST–BSA). The blot was then incubated with primary antibody diluted (1:50 for 12CA5; 1:10 for 9E10) in TBST–BSA for 1 hour, washed three times for 5 min each with TBST, and incubated with secondary antibody in TBST–BSA for 1 hour. After three washes, the signal was visualized by enhanced chemiluminescence (ECL, Amersham).
12CA5 anti-HA antibody was obtained from Berkeley Antibody, Richmond, CA. 9E10 anti-myc antibody was prepared from culture supernatants of the 9E10 hybridoma (27).
RESULTS AND DISCUSSION
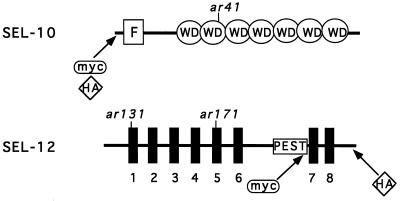
A schematic drawing showing relevant features of the SEL-10 and SEL-12 proteins is shown in Fig. 1. The positions of relevant mutations and of epitope tag insertions are also indicated.
Figure 1.
Schematic view of the SEL-10 and SEL-12 proteins. For SEL-10, the F-box and WD40 repeats, as well as the position of ar41 (W323STOP), are indicated. For SEL-12, thick vertical lines represent the eight transmembrane domains, and the sel-12(ar131) (C60S) and sel-12(ar171) (W225STOP) mutations are indicated. Information about the SEL-10 sequence is from ref. 16 and information about the SEL-12 sequence is from ref. 12, with a correction as in ref. 13.
Genetic Interactions Between sel-10 and sel-12.
A genetic suppressor or enhancer approach may enable the detection of a functionally relevant interaction between genes even when biochemical methods are not sensitive enough to detect differences in activity or level. Genes encoding products that normally promote the turnover of C. elegans presenilins or of components that act downstream of presenilin or in parallel pathways might be defined as suppressors of the Egl phenotype of sel-12 loss-of-function mutations.
If a ubiquitin-targeting factor is mutated, the target protein should be stabilized. F-box proteins like SEL-10 are candidates for ubiquitin-targeting factors (17, 18). We therefore explored the possibility that sel-10 might be involved in targeting SEL-12 or HOP-1 for degradation by looking for the ability of sel-10(ar41), a strong loss-of-function mutation in sel-10 (16), to suppress the Egl phenotype caused by sel-12 alleles. We examined the effects on two different sel-12 alleles, sel-12(ar131) (C60S) and sel-12(ar171) (W225STOP) (12). sel-10(ar41) displays striking suppression of the Egl phenotype of both of these alleles (Table 1). Suppression is recessive (data not shown). The suppression of the Egl phenotype implies a functional interaction between sel-10 and sel-12, but the biochemical nature of the interaction cannot be inferred from this simple genetic test.
Table 1.
Genetic interactions involving sel-10
| Relevant genotype | # Egl/total, % |
|---|---|
| sel-10 interactions with sel-12* | |
| sel-10(+); sel-12(ar131) | 97/99 (98%) |
| sel-10(ar41); sel-12(ar131) | 30/118 (25%) |
| sel-10(+); sel-12(ar171) | 107/107 (100%) |
| sel-10(ar41); sel-12(ar171) | 101/126 (80%) |
| sel-10 interactions with lin-12 | |
| lin-12(n676n930); sel-10(+) | (95%)‡ |
| lin-12(n676n930); sel-10(ar41) | (97%)‡ |
| lin-12(e2621); sel-10(+)† | 88/136 (65%) |
| lin-12(e2621); sel-10(ar41)† | 52/110 (47%) |
All strains also contained the markers him-5(e1490) and unc-1(e538) to facilitate genetic manipulations.
All strains also contained the marker him-5(e1490).
The data for the egg-laying defect of lin-12(n676n930) in a sel-10(+) and sel-10(ar41) background is presented in ref. 15. lin-12(e2621) does not affect the number of anchor cells (E.J.A.H., unpublished observations).
lin-12 partial loss-of-function mutants, like sel-12 mutants, are Egl (15). The cellular basis of these Egl phenotypes of sel-12 and lin-12 mutants is not known with certainty, but the phenotypes appear similar and possibly share a common underlying cause (12). However, genetic data suggest that sel-10 does not suppress the Egl phenotypes of sel-12 mutants simply by increasing lin-12 activity. First, sel-10 does not efficiently suppress the Egl phenotype associated with partial loss of lin-12 activity. Previously it was shown that the Egl phenotype of lin-12(n676n930), a partial loss-of-function allele, is not suppressed by sel-10 (ref. 15; see also Table 1). Here we show that sel-10(ar41) does not efficiently suppress the Egl phenotype of lin-12(e2621), the weakest partial loss-of-function allele of lin-12 available, in contrast to its efficient suppression of the Egl phenotype of sel-12(ar131) (Table 1). Second, there are mutations in other genes that efficiently suppress the Egl phenotype caused by lin-12(n676n930) but not by sel-12 mutations (B. Grant, C. Wen, and I.G., unpublished observations).
The genetic data suggested that the functional interaction between sel-10 and sel-12 is likely to reflect an effect on presenilin activity independent of an effect on lin-12 activity. The functional interaction between sel-10 and sel-12 might reflect a direct physical interaction between SEL-10 and presenilins or between SEL-10 and another factor or factors that influence presenilin level or activity. We explored this issue by coimmunoprecipitation experiments, as described below.
Expression of Epitope-Tagged SEL-10 and SEL-12 in Mammalian Cells.
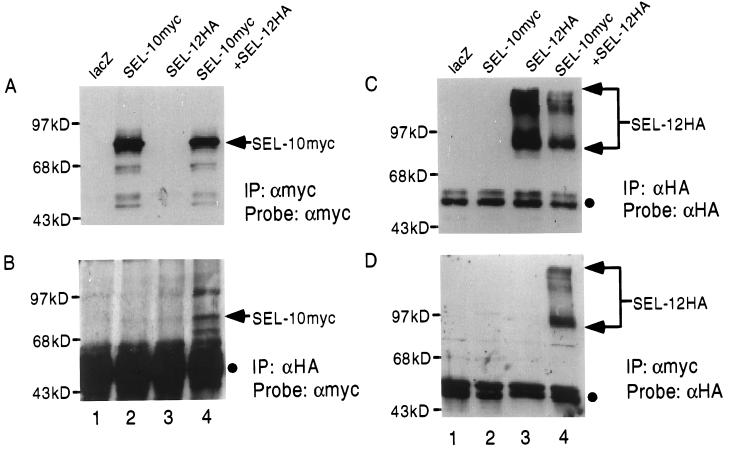
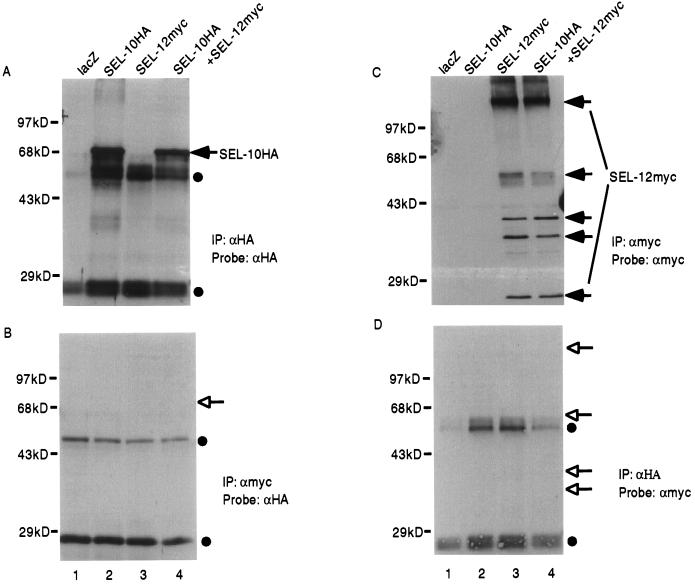
We tagged SEL-10 with either a six-myc epitope or an HA epitope at the amino terminus (Fig. 1), as described in Materials and Methods. 293T cells were transfected with expression plasmids encoding these epitope-tagged proteins. Protein detection was conducted by immunoprecipitation from transfected cell lysates followed by Western blotting of the immunoprecipitates. When this procedure was performed with anti-myc antibodies to detect SEL-10myc, we detected a prominent species of 80 kDa, the expected molecular mass of SEL-10myc (Fig. 2A, lanes 2 and 4). When it was performed with anti-HA antibodies to detect SEL-10HA, we detected a major species of 68 kDa, the expected molecular mass of SEL-10HA (Fig. 3A, lanes 2 and 4).
Figure 2.
Coimmunoprecipitation of SEL-10myc with SEL-12HA. Arrowheads indicate the positions of SEL-10myc or SEL-12HA, and filled circles indicate the position of the Ig heavy chain. (A) Samples were immunoprecipitated with anti-myc antibody and the Western blot was probed with anti-myc to visualize SEL-10myc. (B) Samples were immunoprecipitated with anti-HA antibody and the Western blot was probed with anti-myc to visualize SEL-10myc associated with SEL-12HA. The Ig heavy chain is more prominent because it is a longer exposure than in other panels. (C) Samples were immunoprecipitated with anti-HA antibody and the Western blot was probed with anti-HA to visualize SEL-12HA. (D) Samples were immunoprecipitated with anti-myc and the Western blot was probed with anti-HA to visualize SEL-12HA associated with SEL-10myc. Lane 1, lacZ expression plasmid. Lane 2, sel-10myc expression plasmid. Lane 3, sel-12HA expression plasmid. Lane 4, sel-10myc + sel-12HA expression plasmids. All transfections were normalized for DNA content with lacZ expression plasmid.
Figure 3.
SEL-12 epitope-tagged in the loop region does not associate with SEL-10. Arrowheads indicate the positions of SEL-10HA or SEL-12myc, open arrowheads indicate the expected positions of SEL-10HA or SEL-12myc, and filled circles indicate the positions of the Ig heavy and light chains. (A) Samples were immunoprecipitated with anti-HA antibody and the Western blot was probed with anti-HA to visualize SEL-10HA. In A, B, and D, the Ig heavy and light chains are visible at approximately 50 kDa and 28 kDa, respectively. (B) Samples were immunoprecipitated with anti-myc antibody and the Western blot was probed with anti-HA to determine whether SEL-10HA is present. (C) Samples were immunoprecipitated with anti-myc antibody and the Western blot was probed with anti-myc to visualize SEL-12myc. Arrows denote the multiple apparent SEL-12myc species, which are reminiscent of the multiple human presenilin species that have been reported (20). (D) Samples were immunoprecipitated with anti-HA and the Western blot was probed with anti-myc to determine whether SEL-12myc is present. Lane 1, lacZ expression plasmid. Lane 2, sel-10HA expression plasmid. Lane 3, sel-12myc expression plasmid. Lane 4, sel-10HA + sel-12myc expression plasmids. All transfections were normalized for DNA content with lacZ expression plasmid.
We tagged SEL-12 with an HA epitope at the C terminus or a six-myc epitope in the intracellular loop (Fig. 1; Materials and Methods). When immunoprecipitation and blotting were performed, we detected the SEL-12HA or SEL-12myc proteins as multiple species, including several of higher molecular weight than predicted (Fig. 2C, lanes 3 and 4; Fig. 3C, lanes 3 and 4). (Differences in the patterns may be attributed to the location of the epitope tags and sensitivity of the detecting antibodies.) The multiplicity and pattern of epitope-tagged SEL-12 species strongly resemble the multiplicity and pattern of similarly epitope-tagged human presenilin forms (20). Biochemical analysis of human presenilins has revealed that higher molecular-weight species also contain ubiquitin and/or increase on treatment with proteasomal inhibitors (20–22). It is not clear why high molecular-weight polyubiquitinated forms of presenilins are evident in the absence of inhibitors of the proteasome, and it will be interesting to determine whether there are other factors that regulate the rate or extent of polyubiquitinated-presenilin turnover.
Physical Interactions Between SEL-10 and SEL-12.
If SEL-10 does indeed target presenilins for ubiquitination then SEL-10 would be expected to associate physically with presenilins. We assessed potential physical interactions between SEL-10 and SEL-12 by coimmunoprecipitation using transiently transfected mammalian cells. When lysates of transfected cells containing SEL-10myc and SEL-12HA (C-terminal tag) were precipitated with either anti-myc antibodies (Fig. 2 A and D) or anti-HA antibodies (Fig. 2 B and C), the immunoprecipitates were found to contain both SEL-10myc and SEL-12HA. These results demonstrate that SEL-10 and SEL-12 presenilin are able to form a complex, suggesting that presenilins may be targeted by SEL-10.
However, when lysates of transfected cells containing SEL-10HA and SEL-12myc (loop tag) were analyzed for coimmunoprecipitation, we were unable to detect a physical interaction (Fig. 3 B and D). This lack of association raises two interesting points. First, the lack of coimmunoprecipitation of loop-tagged SEL-12myc with SEL-10HA supports the interpretation that the interaction we observed between SEL-12HA and SEL-10myc is specific and is not simply the result of nonspecific aggregation of SEL-12 and SEL-10 because of the experimental conditions. Second, the cytosolic loop of SEL-12 has a region with features of a PEST sequence (28) and potential phosphorylation sites. The WD40 region of Cdc4p has been shown to bind to phosphorylated target proteins to promote their degradation (18). The failure of coimmunoprecipitation when the epitope tag is near the PEST region may indicate that this region is mediating the interaction between SEL-10 and SEL-12.
We note that coimmunoprecipitation experiments similar to those described here as well as yeast two-hybrid experiments have identified other proteins that interact physically with the presenilin intracellular loop region. Two studies have provided evidence that the presenilin loop region interacts with β-catenin, a family of proteins that are components of cell junctions and of the Wnt signal transduction pathway, and δ-catenin, a novel β-catenin-related protein (29, 30). Another study has provided evidence that the presenilin loop region interacts with the microtubule-associated protein τ and with glycogen synthase kinase-3β, which can phosphorylate τ as well as intermediates in the Wnt signal transduction pathway (31). However, no evidence for the functional relevance of these interactions has been reported.
Concluding Remarks.
Loss of sel-10 activity leads to dramatic suppression of the Egl phenotype caused by reducing sel-12 activity. At this time, we do not know whether reducing sel-10 activity leads to suppression by stabilizing mutant SEL-12 proteins, stabilizing HOP-1(+), or lowering the need for presenilin activity by a bypass mechanism (perhaps by stabilizing other proteins). These possibilities are not mutually exclusive, and it is possible that the functional interaction we have observed is the sum of several relatively small biochemical effects. Further work will be directed toward understanding the details of how reducing sel-10 activity leads to suppression.
Proteins highly related to SEL-10 exist in mammals (16, 18) (our unpublished observations; M.E. Gurney, personal communication). The observations we have described here concerning the physical and functional interactions between SEL-10 and SEL-12 in C. elegans raise the intriguing possibility that SEL-10 is part of a conserved mechanism that regulates presenilin level or activity in mammals. Functional analysis of SEL-10-related proteins in mammalian cell culture and genetic systems will be necessary to assess this possibility.
Acknowledgments
We are grateful to Xiajun Li for valuable advice and assistance. We also thank Xiajun Li, Ron Liem, and Chenhui Wen for valuable comments on the manuscript and Jonathan Hodgkin for providing lin-12(e2621). This work was supported by Department of Defense DAMD 17-94-J-4410 (J.K.), DAMD 17-97-1-7291 (G.W.), and National Institutes of Health NS35556 (I.G.). I.G. is an Associate Investigator and E.J.A.H. was a Postdoctoral Associate of the Howard Hughes Medical Institute.
ABBREVIATIONS
- AD
Alzheimer’s disease
- PS1
presenilin 1
- PS2
presenilin 2
- Egl
egg-laying defective
- HA
hemagglutinin
References
- 1.Hardy J. Trends Neurosci. 1997;20:154–159. doi: 10.1016/s0166-2236(96)01030-2. [DOI] [PubMed] [Google Scholar]
- 2.Neve R L, Robakis N K. Trends Neurosci. 1998;21:15–19. doi: 10.1016/s0166-2236(97)01168-5. [DOI] [PubMed] [Google Scholar]
- 3.Hardy J, Duff K, Hardy K G, Perez-Tur J, Hutton M. Nat Neurosci. 1998;1:355–358. doi: 10.1038/1565. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Greenwald I. Proc Natl Acad Sci. 1998;95:7109–7114. doi: 10.1073/pnas.95.12.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kovacs D M, Fausett H J, Page K J, Kim T W, Moir R D, Merriam D E, Hollister R D, Hallmark O G, Mancini R, Felsenstein K M, et al. Nat Med. 1996;2:224–229. doi: 10.1038/nm0296-224. [DOI] [PubMed] [Google Scholar]
- 6.De Strooper B, Beullens M, Contreras B, Levesque L, Craessaerts K, Cordell B, Moechars D, Bollen M, Fraser P, St. George-Hyslop P, et al. J Biol Chem. 1997;272:3590–3598. doi: 10.1074/jbc.272.6.3590. [DOI] [PubMed] [Google Scholar]
- 7.Lah J J, Heilman C J, Nash N R, Rees H D, Yi H, Counts S E, Levey A I. J Neurosci. 1997;17:1971–1980. doi: 10.1523/JNEUROSCI.17-06-01971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levitan D, Greenwald I. Development (Cambridge, UK) 1998;125:3599–3606. doi: 10.1242/dev.125.18.3599. [DOI] [PubMed] [Google Scholar]
- 9.Duff K, Eckman C, Zehr C, Yu X, Prada C, Perez-Tur J, Hutton M, Buee L, Harigaya Y, Yager D, et al. Nature (London) 1996;383:710–713. doi: 10.1038/383710a0. [DOI] [PubMed] [Google Scholar]
- 10.Scheuner D, Eckman C, Jensen M, Younkin S. Nat Med. 1996;2:864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 11.De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Nature (London) 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 12.Levitan D, Greenwald I. Nature (London) 1995;377:351–354. doi: 10.1038/377351a0. [DOI] [PubMed] [Google Scholar]
- 13.Levitan D, Doyle T G, Brousseau D, Lee M K, Thinakaran G, Slunt H H, Sisodia S S, Greenwald I. Proc Natl Acad Sci USA. 1996;93:14940–14944. doi: 10.1073/pnas.93.25.14940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Greenwald I. Proc Natl Acad Sci USA. 1997;94:12204–12209. doi: 10.1073/pnas.94.22.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sundaram M, Greenwald I. Genetics. 1993;135:765–783. doi: 10.1093/genetics/135.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hubbard E J A, Wu G, Kitajewski J K, Greenwald I. Genes Dev. 1997;11:3182–3193. doi: 10.1101/gad.11.23.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 18.Patton E E, Willems A R, Tyers M. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 19.Neer E J, Schmidt C J, Nambudripad R, Smith T F. Nature (London) 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- 20.Kim T-W, Pettingell W H, Hallmark O G, Moir R D, Wasco W, Tanzi R E. J Biol Chem. 1997;272:11006–11010. doi: 10.1074/jbc.272.17.11006. [DOI] [PubMed] [Google Scholar]
- 21.Fraser P E, Levesque G, Yu G, Mills L R, Thirlwell J, Frantseva M, Gandy S E, Seeger M, Carlen P L, St. George-Hyslop P. Neurobiol Aging. 1998;19:S19–S21. doi: 10.1016/s0197-4580(98)00029-3. [DOI] [PubMed] [Google Scholar]
- 22.Marambaud P, Ancolio K, Lopez-Perez E, Checler F. Mol Med. 1998;4:147–157. [PMC free article] [PubMed] [Google Scholar]
- 23.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodgkin J, Horvitz H R, Brenner S. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth M B, Zahler A, Stolk J H. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pear W S, Nolan G P, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evan G I, Lewis G K, Ramsay G, Bishop J M. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogers S, Wells R, Rechsteiner M. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 29.Zhou J, Liyanage U, Medina M, Ho C, Simmons A D, Lovett M, Kosik K S. NeuroReport. 1997;8:2085–2090. doi: 10.1097/00001756-199705260-00054. [DOI] [PubMed] [Google Scholar]
- 30.Yu G, Chen F, Levesque G, Nishimura M, Zhang D M, Levesque L, Rogaeva E, Xu D, Liang Y, Duthie M, et al. J Biol Chem. 1998;273:16470–16475. doi: 10.1074/jbc.273.26.16470. [DOI] [PubMed] [Google Scholar]
- 31.Takashima A, Murayama M, Murayama O, Kohno T, Honda T, Yasutake K, Nihonmatsu N, Mercken M, Yamaguchi H, Sugihara S, et al. Proc Natl Acad Sci USA. 1998;95:9637–9641. doi: 10.1073/pnas.95.16.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]