Abstract
Mast cells (MCs) are strongly implicated in immunity toward bacterial infection, but the molecular mechanisms by which MCs contribute to the host response are only partially understood. We addressed this issue by examining the direct effects of a Gram-positive pathogen, Streptococcus equi, on bone marrow-derived MCs (BMMCs). Ultrastructural analysis revealed extensive formation of dilated rough endoplasmic reticulum in response to bacterial infection, indicating strong induction of protein synthesis. However, the BMMCs did not show signs of extensive degranulation, and this was supported by only slow release of histamine in response to infection. Coculture of live bacteria with BMMCs caused a profound secretion of CCL2/MCP-1, CCL7/MCP-3, CXCL2/MIP-2, CCL5/RANTES, interleukin-4 (IL-4), IL-6, IL-12, IL-13, and tumor necrosis factor alpha, as shown by antibody-based cytokine/chemokine arrays and/or enzyme-linked immunosorbent assay. In contrast, heat-inactivated bacteria caused only minimal cytokine/chemokine release. The cytokine/chemokine responses were substantially attenuated in Toll-like receptor 2-deficient BMMCs and were strongly dependent on cell-cell contacts between bacteria and BMMCs. Gene chip microarray analysis confirmed a massively upregulated expression of the genes coding for the secreted cytokines and chemokines and also identified a pronounced upregulation of numerous additional genes, including transcription factors, signaling molecules, and proteases. Together, the present study outlines MC-dependent molecular events associated with Gram-positive infection and thus provides an advancement in our understanding of how MCs may contribute to host defense toward bacterial insults.
Mast cells (MCs) are widely implicated in allergic disorders, and it is now established that they also make deleterious contributions to various other types of disease, including arthritis, multiple sclerosis, cancer progression, atherosclerosis, and aneurysm formation (26, 48, 52-54). However, it is also clear that MCs have several functions that are protective to their host. In particular, there is now a wealth of evidence suggesting that MCs are important players in host defense toward parasitic and bacterial insults (reviewed in references 9, 13, and 33), and recent studies have shown that MCs may also have immunosuppressive functions in connection with allograft tolerance (29) and in suppressing contact dermatitis (18).
A role for MCs in combating bacterial disease was originally described in two reports published simultaneously, where it was shown that MC-deficient animals (W/Wv mice) were markedly more susceptible to infection in models of acute septic peritonitis (cecum ligation and puncture) than were corresponding wild-type (WT) mice (11, 30). Remarkably, when the MC-deficient mice were reconstituted with bone marrow-derived MCs (BMMCs), resistance to infection was regained. Since then, a large number of reports have shown similar findings, i.e., that MC deficiency results in a markedly elevated susceptibility to a host of different bacterial insults. For example, MC-deficient mice are much more sensitive to infection caused by Citrobacter rodentium (62), Helicobacter felis (63), Listeria monocytogenes (17), Pseudomonas aeruginosa (50), and invasive group A streptococci (10) than are corresponding WT animals.
MCs are highly multifaceted cells capable of releasing a wide panel of compounds when appropriately stimulated (7, 16, 37, 45). Potentially, MCs could thus influence the antibacterial response by acting at a multitude of levels. For example, MC degranulation results in the release of a panel of preformed mediators, e.g., histamine, cytokines, proteoglycans, and proteases, all of which could have an impact on the proinflammatory response after a bacterial infection. In addition, MC stimulation may result in de novo production and release of a large panel of additional proinflammatory compounds, including eicosanoids, as well as a variety of cytokines and chemokines. In addition to contributing by releasing proinflammatory substances, there are several reports suggesting that MCs can act as phagocytes (3, 31, 49). On the other hand, there are reports showing that MCs contribute to antibacterial defense without any signs of phagocytic activity (17). It has also been demonstrated that MCs can kill bacteria by formation of extracellular traps (65) and that MCs may express antimicrobial peptides (10).
Importantly, although a role for MCs in combating bacterial infection is now widely accepted, the mechanism by which MCs contribute to host defense is only partially resolved. In the original reports, it was suggested that MC-derived tumor necrosis factor alpha (TNF-α) was the key factor conferring resistance to infection, being crucial for recruiting neutrophils to the site of infection (11, 30). On the other hand, a subsequent report showed that also TNF-α−/− MCs improved the survival of mice in a sepsis model (34), thus suggesting that additional MC-derived factors contribute to survival.
MCs are located close to the host-environment interface of most tissues. Hence, they are ideally situated to provide first line defense toward pathogenic microbes, and direct interaction of invading pathogens with MCs is therefore likely to be an early and key event during a bacterial infection. Despite this, there is only limited information regarding the direct and global effects of live bacteria on MCs. Here we addressed this issue by studying the effects of a Gram-positive pathogen, Streptococcus equi subspecies equi (hereafter referred to as simply S. equi), on MCs by using a number of approaches, including nonbiased strategies. S. equi, a serological group C streptococcus, causes a severe upper respiratory tract infection in horses known as strangles (59) and can be used for experimental infection of rodents (15). Moreover, the closely related S. equi subsp. zooepidemicus is known to be infectious for various additional mammals, including humans (1). We show that live, but not heat-inactivated S. equi induces a powerful and defined cytokine/chemokine response accompanied by induction of a panel of transcription factors/signaling molecules and that this response is strongly dependent on Toll-like receptor 2 (TLR2) and on cell-cell contacts between bacteria and MCs.
MATERIALS AND METHODS
Bacteria.
The bacterial strain used Streptococcus equi subsp. equi strain Bd3221 has been obtained from the National Veterinary Institute (SVA), Uppsala, Sweden. This strain was originally isolated from an infected horse and has previously been used in several studies (27, 28).
BMMCs.
Bone marrow cells were collected from femura and tibia by flushing the bones with 2.5 ml of phosphate-buffered saline (PBS). BMMCs (47, 60) from WT, TLR2−/−, and TLR4−/− mice, all on a C57BL/6 genetic background, were obtained by culturing the bone marrow cells in Dulbecco modified Eagle medium (SVA, Uppsala, Sweden) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen, Carlsbad, CA), 60 μg of penicillin (SVA)/ml, 50 μg of streptomycin sulfate (SVA)/ml, 2 mM l-glutamine (SVA), 5 ng of mouse interleukin-3 (IL-3; Peprotech, Rocky Hill, NJ)/ml, and 25 ng of mouse stem cell factor (Peprotech)/ml for 3 weeks. The cells were kept at a concentration of 0.5 × 106 cells/ml, and the medium was changed every third day. BMMCs were analyzed for the presence of mouse MC protease 6 (mMCP-6) and β-actin by using Western blot analysis as previously described (8). Staining with May-Grünwald-Giemsa was performed as described previously (2).
In vitro coculture of BMMCs and bacteria.
BMMCs were washed two times in PBS and resuspended in antibiotic-free medium (otherwise as described above) in a density of 106 cells/ml and plated in 24-well tissue plates. S. equi (strain Bd 3221) were grown overnight, without shaking in Todd-Hewitt broth (THB; Oxoid, Basingstoke, United Kingdom) supplemented with 0.7% yeast extract, washed two times in PBS, and added to a final concentration of ∼2.5 × 107 cells/ml (multiplicity of infection [MOI] = 1:25). At various time points, cells were collected by centrifugation. Media and cell fractions were frozen and stored at −20°C. As a positive control for degranulation efficiency, cells were treated with the calcium ionophore A23187 (2 μM final concentration), after which the conditioned medium was collected at various time points and analyzed for the content of histamine. Heat-inactivated S. equi was obtained by incubating bacteria for 20 min at 60°C. They were then plated on blood agar plates to verify the lack of viability. To obtain S. equi-conditioned media, bacteria were grown in culture medium overnight, followed by sterile filtering of the conditioned medium. For transwell experiments, transwell polystyrene plates (polycarbonate membranes with a 0.4-μm pore size; Costar Coring, Inc., Schiphol-Rijk, The Netherlands) were used. BMMCs were seeded in the lower well, and bacteria were added to the top chamber of the transwell system.
Transmission electron microscopy (TEM).
Cells were fixed in 2% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2), supplemented with 0.1 M sucrose for 10 h. Next, cells were postfixed in 1% osmium tetroxide in the same buffer for 90 min, dehydrated in graded series of ethanol, and embedded with the epoxy plastic Agar 100 (Agar Aids, Stansted, United Kingdom). Ultrathin sections were placed on Formvar-coated copper grids, along with 2% uranyl acetate and Reynolds lead citrate. Analysis was performed with a Hitachi electron microscope at 75 kV.
Cytokine antibody array and ELISA.
Secretion of cytokines was determined by using a RayBio mouse cytokine antibody array I (RayBiotech, Inc., Norcross, GA) according to the manufacturer's instructions. TNF-α, MCP-1, IL-6, IL-13, MCP-3, MIP-2, IL-3 (Peprotech), and histamine (Oxford Biomedical Research, Oxford, MI) levels in BMMC-conditioned medium were quantified by using enzyme-linked immunosorbent assay (ELISA) kits according to the manufacturers' instructions.
RNA preparation, microarray expression analysis, and data analysis.
Total RNA from 2.5 × 106 cultured cells was isolated by using NucleoSpin RNA II (Macherey-Nagel, Düren, Germany). The RNA quality was evaluated with an Agilent 2100 Bioanalyzer system. The microarray analysis was performed at the Uppsala array platform (Uppsala, Sweden). From each sample of total RNA, 2 μg was used to prepare biotinylated fragmented cDNA according to the Gene-Chip expression analysis technical manual (revision 5; Affymetrix, Inc., Santa Clara, CA). Affymetrix GeneChip expression arrays (GeneChip Mouse Gene 1.0 ST array) were hybridized for 16 h in a 45°C incubator and rotated at 60 rpm, whereafter the arrays were washed and stained by using a Fluidics Station 450. The arrays were finally scanned with the GeneChip scanner 3000 7G. Gene expression data analysis was carried out in the statistical computing language R (http://www.r-project.org) using packages available from the Bioconductor project (www.bioconductor.org). The raw data were normalized by using the robust multi-array average (RMA) (22) and background-adjusted, normalized, and log-transformed summarized values. An empirical Bayes moderated t test was applied to search for differentially expressed genes (51). The P values were adjusted to avoid the problem with multiple testing (20). Assays were performed in duplicates and were confirmed in independent experiments.
Statistical analysis.
Data are shown as means ± the standard error of the mean (SEM). Statistical analyses were performed by using GraphPad Prism 4.0c (GraphPad Software) and an unpaired Student t test for two-tailed distributions (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
RESULTS
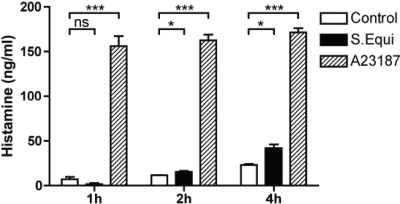
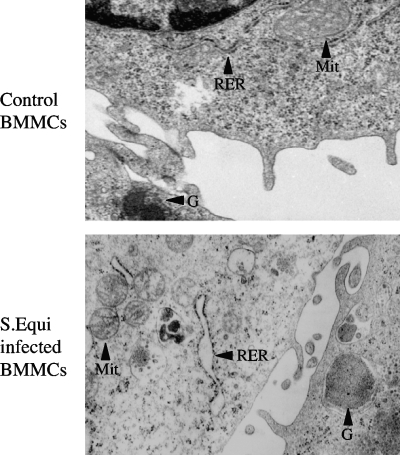
MC activation by a number of stimuli typically involves degranulation, causing the release of preformed granule constituents such as histamine, β-hexosaminidase, and proteases. However, it has been shown that MC stimulation, e.g., through TLR4, can result in substantial cytokine release without signs of degranulation (55, 61). To examine whether S. equi causes MC degranulation, we measured histamine release after the addition of live bacteria to BMMCs. As a control, calcium ionophore addition caused rapid and robust secretion of histamine (Fig. 1). Also, the addition of S. equi caused histamine release, but the release was much slower than the response induced by calcium ionophore, and the extent of histamine release was much lower (Fig. 1). To further examine the impact of S. equi on BMMCs, morphological examination using TEM was carried out. As shown in Fig. 2, nonstimulated BMMCs displayed normal morphological criteria, including presence of secretory granule, as well as nondilated rough endoplasmic reticulum (RER) and intact mitochondria. An examination of BMMCs in coculture with live S. equi revealed a striking presence of dilated RER, indicative of elevated translational activity. Further, and in line with the moderate histamine release caused by S. equi, extensive MC degranulation was not evident by morphological criteria (Fig. 2 and data not shown). Moreover, there were no signs of phagocytosis of the bacteria after inspection of >50 MCs by TEM (Fig. 2 and data not shown).
FIG. 1.
S. equi induces slow histamine release from BMMCs. BMMCs (106 cells/ml) were cultured either alone (control) or together with S. equi (MOI = 25). At the time points indicated, medium samples were analyzed for content of histamine. As a control, BMMCs were activated with a calcium ionophore (A23187), followed by measurement of histamine release (n = 3).
FIG. 2.
Effect of S. equi infection on BMMC ultrastructure. After coculture of BMMCs with S. equi for 4 h, cells were analyzed by transmission electron microscopy. Note the extensive formation of dilated RER induced by S. equi, a finding indicative of marked induction of RER-associated protein synthesis, whereas the RER in control BMMCs is not dilated. Note also the presence of secretory granule (G) and that S. equi infection does not induce extensive signs of degranulation. Intact mitochondria (Mit) are visible.
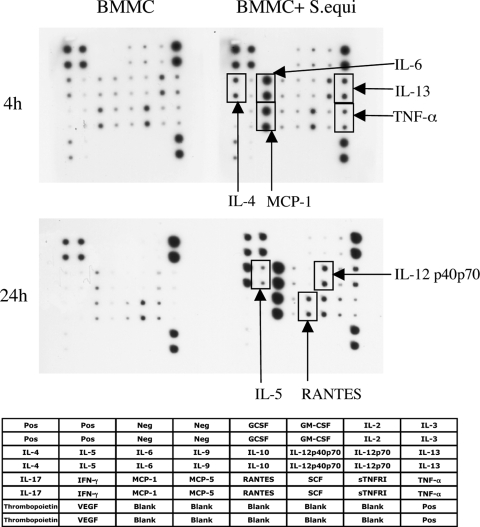
Considering the dramatic effect of S. equi on RER activity, we hypothesized that the infection induced the expression and release of proinflammatory compounds such as cytokines and chemokines. As an unbiased approach to investigate this, we used an antibody-based cytokine/chemokine array system. As shown in Fig. 3, infection of BMMCs with live bacteria caused a profound secretion of multiple cytokines/chemokines, in particular IL-6, MCP-1, IL-13, TNF-α, and IL-4. Clearly, upregulated secretion of these was seen at 4 h after S. equi addition. At 24 h after addition of bacteria, additional robust secretion of IL-12, RANTES, and IL-5 was also seen, suggesting that the latter compounds are released at a later stage of MC stimulation.
FIG. 3.
Array analysis demonstrating secretion of cytokines and chemokines after coculture of BMMCs with S. equi. BMMCs (106 cells/ml) were cultured either alone or cocultured with S. equi (MOI = 25). At 4 and 24 h, respectively, samples from the conditioned medium were analyzed for content of various cytokines and chemokines using antibody-based filter array analysis. The composition of the array is indicated. Compounds demonstrating an upregulated secretion are indicated.
As models for events associated with bacterial infection, isolated bacterial cell wall compounds such as LPS and peptidoglycan (PGN) are commonly used, with the concept being that triggering of TLR-dependent events by these substances will mimic those induced by viable bacteria. To investigate whether the effects seen after stimulation with live S. equi could be mimicked by bacterial cell wall components, we stimulated BMMCs with heat-inactivated bacteria and measured the cytokine/chemokine release. However, the addition of heat-inactivated S. equi caused only minimal cytokine/chemokine release, as judged by cytokine/chemokine array analysis (data not shown). Thus, an optimal effect on cytokine/chemokine responses requires live bacteria.
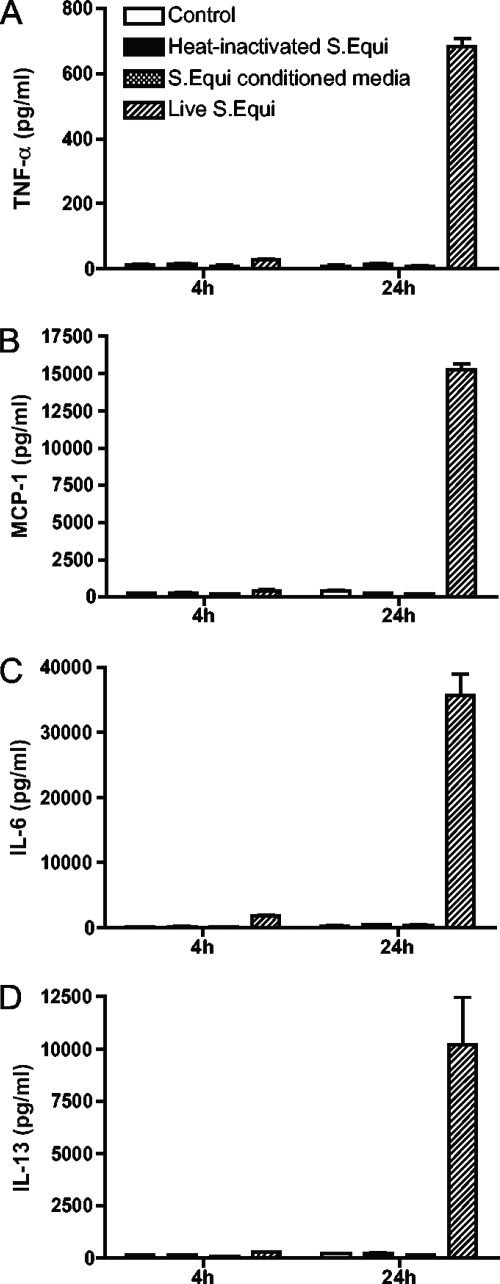
To verify and quantify the cytokine/chemokine responses induced by S. equi, we used specific ELISAs. Indeed, high levels of TNF-α, MCP-1, IL-6, and IL-13 secretion in response to live S. equi was verified (Fig. 4). All of these cytokines/chemokines were detectable at 4 h after the addition of S. equi, but their levels increased substantially at 24 h after stimulation. Again, heat-inactivated bacteria induced only marginal secretion of these cytokines/chemokines (Fig. 4), in agreement with the requirement for live bacteria in order to achieve a maximal cytokine/chemokine response.
FIG. 4.
Induction of cytokines and chemokines by live and heat-inactivated S. equi. BMMCs (106 cells/ml) were cultured either alone, in coculture with live or heat-inactivated S. equi (MOI = 25) or in the presence of S. equi-conditioned medium as indicated. At the time points indicated, samples from the conditioned media were analyzed for TNF (A), MCP-1 (B), IL-6 (C), or IL-13 (D) content by using specific ELISAs. The results are representative of three independent experiments (n = 4).
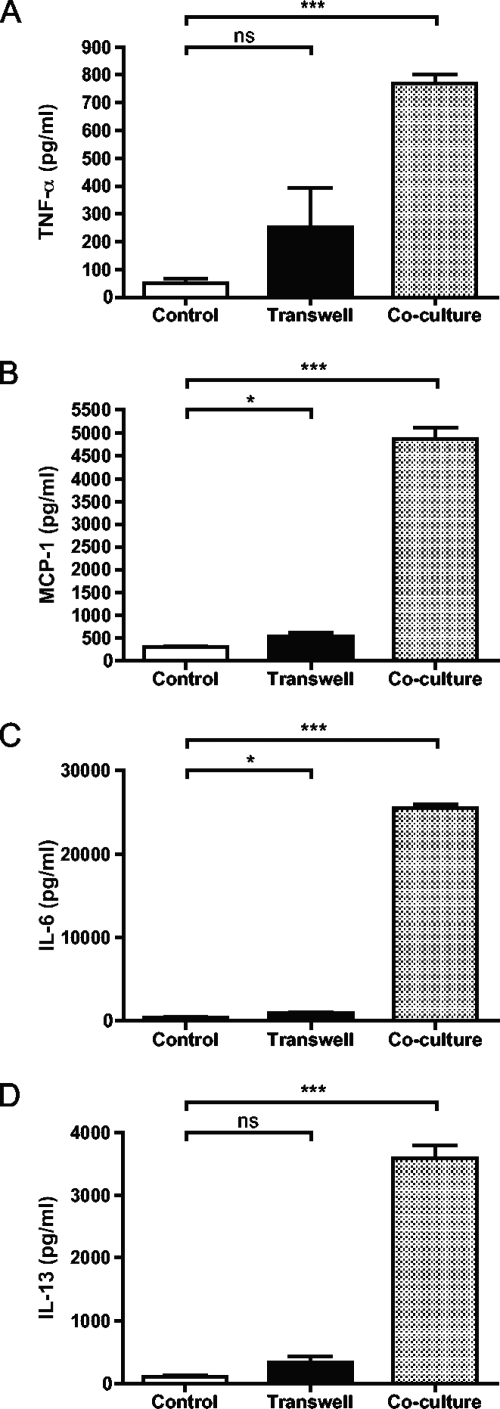
Next, the mechanism behind the effect of S. equi on BMMCs was investigated. A likely mode of BMMC activation is that pattern recognition receptors (PRRs) on the surface of BMMCs are engaged by S. equi-expressed pathogen-associated molecular patterns, with the most likely candidates being the various bacterial cell wall components. According to such a scenario, it would be expected that optimal BMMC activation would require cell-cell contact between the BMMCs and bacteria. As shown in Fig. 5, S. equi induced only low levels of TNF-α, MCP-1, IL-6, and IL-13 when bacteria and BMMCs were placed in separate chambers, whereas a strong response was seen when they were cultured together. Hence, optimal BMMC activation by the Gram-positive bacteria requires cell-cell contact. In further support for this notion, S. equi-conditioned medium was not able to induce measurable secretion of TNF-α, MCP-1, IL-13, or IL-6 (Fig. 4).
FIG. 5.
Dependence on cell-cell contact for induction of cytokines and chemokines. BMMCs (106 cells/ml) were cultured either alone (control) or in coculture with S. equi (MOI = 25). BMMCs and S. equi were either placed in separate chambers (Transwell) or in the same compartment (coculture). After 24 h, samples from the conditioned media were analyzed for TNF (A), MCP-1 (B), IL-6 (C), or IL-13 (D) using specific ELISAs (n = 4).
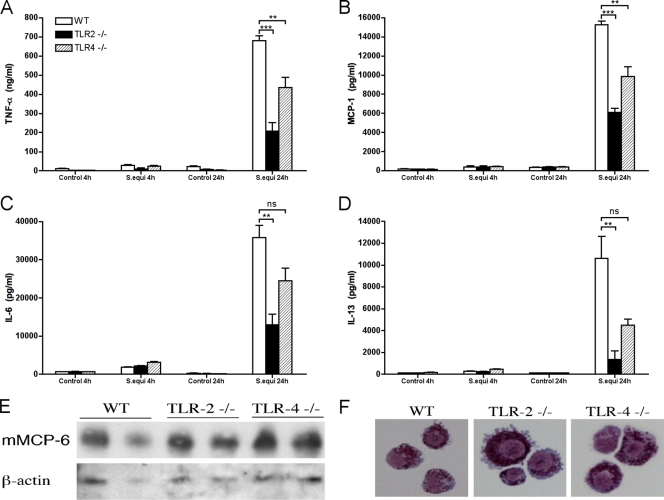
To further characterize the mechanism of MC activation, we sought to identify the cell surface receptor(s) responsible for BMMC activation and subsequent cytokine or chemokine release. Among the multiple PRRs that are known to mediate immune cell activation, we chose to focus on the TLRs (5, 12, 56), TLR2 and TLR4, since both of these TLRs have previously been shown to be expressed by MCs and shown to have a functional impact on MCs (36, 55). TLR2 is a receptor for PGN, i.e., a main component of the cell wall of Gram-positive bacteria, but it should be stressed that TLR2 also recognizes a number of additional ligands (64). TLR4, on the other hand, is mainly known to interact with LPS, the latter being a dominant component of the cell wall of Gram-negative bacteria. Considering that S. equi is Gram-positive, we therefore hypothesized that the cytokine or chemokine responses following S. equi stimulation may by blunted in particular in the absence of TLR2. To address these issues, we developed BMMCs from mice lacking TLR2 and TLR4, respectively, and measured the cytokine/chemokine responses after coculture with S. equi. Notably, the absence of either TLR2 or TLR4 did not affect MC maturation, as judged both by morphological criteria (Fig. 6E) and by the expression of a MC-specific marker, mouse mast cell protease 6 (Fig. 6F). In agreement with the data shown above, S. equi caused a robust secretion of TNF-α, MCP-1, IL-6, and IL-13 in WT BMMCs (Fig. 6). In contrast, the secretion of all of these cytokines and chemokines was markedly reduced in TLR2−/− BMMCs, indeed suggesting a major role for TLR2 (Fig. 6). Notably, however, residual cytokine/chemokine responses were seen also in the absence of TLR2, indicating that other, TLR2-independent activation mechanisms are required for an optimal response. Also, the TLR4−/− BMMCs responded less vividly to bacterial stimulation than did WT BMMCs, suggesting a contribution of TLR4 to the cytokine/chemokine response (Fig. 6). However, the effects of TLR4 deficiency were less pronounced and, in the case of IL-6 and IL-13, not statistically significant, suggesting that TLR2 has a higher impact on S. equi-induced activation of BMMCs than does TLR4.
FIG. 6.
Cytokine and chemokine secretion in response to S. equi is dependent on TLR2. WT, TLR2−/−, or TLR4−/− BMMCs (106 cells/ml) were cultured either alone (control) or in the presence of S. equi (MOI = 25). At the time points indicated, samples from the conditioned medium were analyzed for TNF (A), MCP-1 (B), IL-6 (C), or IL-13 (D) using specific ELISAs. The results are representative of three independent experiments (n = 4). (E) Western blot analysis for the MC-specific marker, mouse mast cell protease 6 (mMCP-6), in WT, TLR2−/−, and TLR4−/− BMMCs, with β-actin as a loading control. (F) Staining of cytospin slides from WT, TLR2−/−, and TLR4−/− BMMCs with May-Grünwald-Giemsa, showing that the absence of TLR2 or TLR4 did not cause altered morphology.
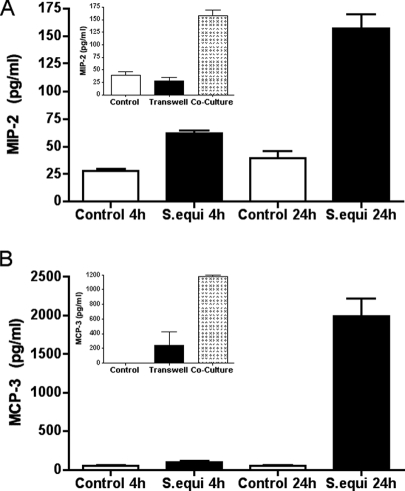
The results described above outline the chemokine/cytokine profile following Gram-positive bacterial infection of BMMCs. However, to get an even more comprehensive picture of the effect of the bacteria on BMMCs, we used Affymetrix gene chip microarray analysis. This analysis revealed a significant (P < 0.05) upregulation of 155 genes in response to S. equi infection, with the cutoff set at genes that were upregulated at least fourfold (Table 1 and see Table S1 in the supplemental material). In agreement with the ELISA and cytokine/chemokine array analysis (see Fig. 4 and 5), clearly upregulated expression of CCL2/MCP-1, IL-13, IL-6, IL-4, and TNF-α was apparent. However, a number of additional chemokines and cytokines showed an even more pronounced upregulation. Among the chemokines, particularly strong upregulation of CCL7/MCP-3, CCL4/MIP-1β, CCL1/I-309, CXCL-2/MIP-2, and CCL3/MIP-1α was seen. To verify their upregulated expression, specific ELISAs were used. Indeed, strong upregulation of CCL7/MCP-3 and CXCL2/MIP-2 in response to S. equi infection was seen (Fig. 7). Notably, secretion of CXCL-2/MIP-2 was seen already 4 h after BMMC stimulation. As shown in the insets in Fig. 7, the secretion of CCL7/MCP-3 and CXCL2/MIP-2 in response to S. equi infection was dependent on cell-cell contacts between bacteria and BMMCs.
TABLE 1.
Fifty genes showing the highest extent of significant (adjusted P < 0.05) upregulation after infection of BMMCs with S. equi
| mRNA description | Gene symbol | Probe set ID | Fold change | Adjusted P |
|---|---|---|---|---|
| Nuclear receptor subfamily 4, group A, member 3 (Nr4a3) | Nr4a3 | 10504838 | 110.0 | 0.00001 |
| Interleukin-3 (IL-3) | Il3 | 10385918 | 82.9 | 0.00001 |
| Colony-stimulating factor 2 (granulocyte-macrophage) (Csf2) | Csf2 | 10385912 | 59.4 | 0.00001 |
| Chemokine (C-C motif) ligand 7 (Ccl7) | Ccl7 | 10379518 | 46.5 | 0.00001 |
| Granzyme D (Gzmd) | Gzmd | 10420274 | 42.3 | 0.00001 |
| Chemokine (C-C motif) ligand 4 (Ccl4) | Ccl4 | 10379721 | 42.0 | 0.00001 |
| Nuclear receptor subfamily 4, group A, member 2 (Nr4a2) | Nr4a2 | 10482772 | 38.7 | 0.00001 |
| GTP binding protein (gene overexpressed in skeletal muscle) (Gem) | Gem | 10503334 | 36.8 | 0.00001 |
| RasGEF domain family, member 1B (Rasgef1b), transcript variant 1 | Rasgef1b | 10531610 | 34.1 | 0.00002 |
| Nuclear receptor subfamily 4, group A, member 1 (Nr4a1) | Nr4a1 | 10427035 | 31.4 | 0.00001 |
| MARCKS-like 1 (Marcksl1) | Marcksl1 | 10508465 | 30.6 | 0.00006 |
| Serine (or cysteine) peptidase inhibitor, clade E, member 1 (Serpine1) | Serpine1 | 10534667 | 29.6 | 0.00001 |
| Prostaglandin-endoperoxide synthase 2 (Ptgs2) | Ptgs2 | 10350516 | 29.3 | 0.00001 |
| Amphiregulin (Areg) | Areg | 10523182 | 26.3 | 0.00005 |
| Interleukin-2 (IL-2) | Il2 | 10497878 | 26.1 | 0.00001 |
| Activating transcription factor 3 (Atf3) | Atf3 | 10361091 | 23.1 | 0.00001 |
| A disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 9 (Adamts9) | Adamts9 | 10546450 | 22.8 | 0.00001 |
| Interleukin-13 (IL-13) | Il13 | 10385837 | 22.7 | 0.00001 |
| Oxidized low-density lipoprotein (lectinlike) receptor 1 (Olr1) | Olr1 | 10548385 | 20.0 | 0.00002 |
| Heparin-binding epidermal growth factor (EGF)-like growth factor (Hbegf) | Hbegf | 10458340 | 19.8 | 0.00001 |
| Interleukin-4 (IL-4) | Il4 | 10385832 | 19.4 | 0.00001 |
| Mitogen-activated protein kinase kinase kinase 8 (Map3k8) | Map3k8 | 10457225 | 18.9 | 0.00001 |
| Regulator of calcineurin 1 (Rcan1), transcript variant 1 | Rcan1 | 10440993 | 18.1 | 0.00002 |
| Chemokine (C-C motif) ligand 1 (Ccl1) | Ccl1 | 10389064 | 17.8 | 0.00002 |
| Sprouty homolog 1 (Drosophila) (Spry1) | Spry1 | 10491721 | 17.4 | 0.00010 |
| Follistatin (Fst) | Fst | 10412260 | 16.8 | 0.00001 |
| mRNA for mKIAA1726 protein | Zc3h12c | 10593492 | 16.5 | 0.00004 |
| EH-domain containing 4 (Ehd4) | Ehd4 | 10486396 | 15.9 | 0.00002 |
| Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta (Nfkbid) | Nfkbid | 10551891 | 15.9 | 0.00004 |
| Tumor necrosis factor (ligand) superfamily, member 9 (Tnfsf9) | Tnfsf9 | 10446229 | 15.2 | 0.00005 |
| Src-like adaptor (Sla), transcript variant 1 | Sla | 10429128 | 14.9 | 0.00004 |
| Tumor necrosis factor (TNF) | Tnf | 10450501 | 13.7 | 0.00002 |
| T-cell activation Rho GTPase-activating protein (Tagap) | Tagap | 10441601 | 13.3 | 0.00001 |
| Endothelin 1 (Edn1) | Edn1 | 10404783 | 13.0 | 0.00013 |
| Interleukin-6 (IL-6) | Il6 | 10520452 | 12.4 | 0.00012 |
| Sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, semaphorin 7A (Sema7a) | Sema7a | 10585778 | 12.3 | 0.00011 |
| RIKEN cDNA 1190002H23 gene (1190002H23Rik) | 1190002H23Rik | 10421810 | 11.9 | 0.00002 |
| Sphingosine kinase 1 (Sphk1), transcript variant 1 | Sphk1 | 10382802 | 11.7 | 0.00002 |
| Tribbles homolog 1 (Drosophila) (Trib1) | Trib1 | 10424370 | 11.1 | 0.00012 |
| Fatty acid amide hydrolase (Faah) | Faah | 10515220 | 11.1 | 0.00005 |
| Fermitin family homolog 2 (Drosophila) (Fermt2) | Fermt2 | 10419223 | 10.9 | 0.00002 |
| Early growth response 3 (Egr3) | Egr3 | 10416251 | 10.9 | 0.00007 |
| Tumor necrosis factor (ligand) superfamily, member 8 (Tnfsf8) | Tnfsf8 | 10513729 | 10.4 | 0.00005 |
| Nuclear factor, interleukin-3, regulated (Nfil3) | Nfil3 | 10409278 | 10.2 | 0.00004 |
| Protein tyrosine phosphatase, nonreceptor type 22 (lymphoid) (Ptpn22) | Ptpn22 | 10494978 | 10.0 | 0.00004 |
| Regulator of G-protein signaling 16 (Rgs16) | Rgs16 | 10350733 | 9.7 | 0.00005 |
| Polo-like kinase 3 (Drosophila) (Plk3) | Plk3 | 10515399 | 9.6 | 0.00005 |
| Coagulation factor III (F3) | F3 | 10495675 | 9.5 | 0.00068 |
| Chemokine (C-C motif) ligand 2 (Ccl2) | Ccl2 | 10379511 | 9.4 | 0.00012 |
| C-type lectin domain family 4, member e (Clec4e) | Clec4e | 10547664 | 8.9 | 0.00003 |
FIG. 7.
BMMCs secrete MIP-2 and MCP-3 in response to S. equi infection. BMMCs (106 cells/ml) were cultured either alone or in coculture with live S. equi (MOI = 25). At the time points indicated, samples from the conditioned media were analyzed for content of MIP-2 (A) and MCP-3 (B) by using specific ELISAs. Insets in panels A and B show that the induction of MIP-2 and MCP-3 is dependent on cell-cell contact between BMMCs and S. equi. The results are representative of two independent experiments (n = 4).
Several cytokines and growth factors were also strongly induced. In particular, a dramatic (>100-fold) upregulation of IL-3 was seen, and there was also a strong upregulation of the IL-2 and Tnfsf9/4-1BBL genes. Several growth factors were markedly upregulated, including CSF-2, amphiregulin, heparin-binding EGF-like growth factor (Hbegf), leukemia inhibitory factor (Lif), inhibin beta-A (Inhba), and CSF-1. In order to verify the upregulated expression of IL-3, we used an IL-3-specific ELISA. Since IL-3 is used in the culture medium as a MC growth factor, the background levels of IL-3 were therefore high (10 ng/ml). However, incubation of BMMCs alone resulted in gradual depletion of IL-3, whereas the presence of S. equi caused a small but significant increase in the medium content of IL-3 (∼120 ng/ml) after 24 h of incubation with bacteria (data not shown).
A large number of other genes were also profoundly upregulated in response to infection. Among these, transcription factors were highly represented. For example, three members of the nuclear receptor subfamily 4 (group A; Nr4a3, Nr4a2, and Nr4a1) were among the genes showing the highest extent of upregulation and additional upregulated transcription factors included Atf3, Nfil3, Tgif1, Axud1, Ets1, Erf, and Egr2. Also, a number of genes implicated in signaling processes were upregulated, as exemplified by Gem, Rasgef1b, Spry1, Rgs16, A630033H20Rik, Arhgef3, Rasl11b, Gpr171, Spry2, Arl5b, Rgl1, Htr1b, Arhgap5, Map3k8, Plk3, Tagap, Ptpn22, Pilra, and Rabgef1. Several proteolytic enzymes were also induced, most notably granzyme D, a disintegrinlike metallopeptidase, with thrombospondin type 1 motif 9 (ADAMTS9), ADAMTS6, and cathepsin L, as well as protease inhibitors—serpine 1 and tissue inhibitor of metalloproteinase 3 (TIMP-3) (Table 1 and see Table S1 in the supplemental material). We also noted a strong upregulation of follistatin, a protein implicated in sepsis (23), as well as upregulated expression of endothelin 1, a peptide with documented ability to cause MC degranulation (35). A number of other genes implicated in the regulation of MC degranulation were also induced, including sphingosine kinase 1 (Sphk1), regulator of calcineurin 1 (Rcan1) (66), src-like adaptor (Sla) (43), and Rabgef1 (57). Several genes related to lipid metabolism were also induced by the bacterial infection, as evidenced by the robust upregulation of oxidized low-density lipoprotein (lectinlike) receptor 1 (Olr1) and fatty acid amide hydrolase (Faah). Moreover, a dramatic upregulation of Ptgs2, i.e., the gene encoding cyclooxygenase 2 was seen, suggesting that MC activation by S. equi activates the de novo synthesis of prostaglandins. In agreement with this notion, lipoteichoic acid-stimulated MCs were recently shown to produce PGD2 (38).
The Affymetrix gene chip analysis also revealed a number of genes that were significantly downregulated more than fourfold after challenge pf BMMCs with S. equi. Of these, ganglioside-induced differentiation-associated-protein 10 (Gdap10) showed the highest extent of downregulation (∼24-fold), and it was also noteworthy that several zinc finger proteins and tRNAs were profoundly downregulated (see Table S2 in the supplemental material).
DISCUSSION
Despite the potential impact of direct interaction between MCs and invading bacteria on the immune response, the direct and global effects of live Gram-positive bacteria on MCs have only been partly outlined. Instead, previous attempts to outline the effect of Gram-positive bacteria on MCs have mainly utilized purified cell wall components (e.g., PGN and lipoteichoic acid) and have focused on a limited number of selected compounds, in particular proinflammatory cytokines. To obtain a more comprehensive picture of the global effects of Gram-positive bacteria on MCs, we instead used live bacteria and studied their impact by using unbiased approaches. By using an antibody-based cytokine array system, we show that BMMCs respond to live streptococci by early (at 4 h) secretion of substantial amounts of TNF-α, IL-6, IL-13, IL-4, and MCP-1, followed by a delayed onset of IL-5, RANTES, and IL-12 secretion. Interestingly, many of these cytokines and chemokines have previously been shown to be induced in MCs by various purified TLR2 agonists (38, 46, 55, 61). In accordance with the cytokine/chemokine array and ELISA analyses, the Affymetrix gene chip microarray analysis confirmed a strongly upregulated expression of the TNF-α, IL-6, IL-13, IL-4, and MCP-1 genes. Moreover, the microarray analysis revealed a strong upregulation of a number of additional cytokines and chemokines. Of these, the most striking induction was seen for IL-3, which was upregulated ∼80-fold in response to S. equi (Table 1). IL-3 is a potent growth factor for MCs, and its upregulation in response to S. equi may thus suggest an important autocrine function serving to limit toxic/proapoptotic effects of the bacteria and/or by promoting MC proliferation. Interestingly, it has been shown that IL-3 is of minor importance for maintaining the MC populations during physiological conditions, whereas it is vital for the expansion of the MC population during infection with Strongyloides venezuelensis (25). The dramatic upregulation of IL-3 is thus in accordance with an autocrine role for IL-3 in promoting MC proliferation in particular during conditions when the MCs are subjected to stress, such as during a bacterial infection.
The microarray analysis also indicated strong upregulation of CSF-2 (coding for GM-CSF), IL-2, and IL-4, suggesting that S. equi-challenged MCs respond by promoting the proliferation of a multitude of other immune cells, including macrophages and T and B lymphocytes. Another striking finding from the microarray analysis was the profound induction of a large number of chemokines, among which CCL7/MCP-3 and CCL4/MIP-1β showed the largest extent of upregulation, followed by CCL1/I-309, CCL2/MCP-1, CXCL2/MIP-2, and CCL3/MIP-1α (Table 1 and see Table S1 in the supplemental material). Hence, MCs respond directly to S. equi by mounting a powerful chemokine response, a notion that is clearly in line with the defective recruitment of inflammatory cells in response to bacterial infection that is seen in MC-deficient animals (11, 30). Previous reports have shown that MCs also can respond to other types of live streptococci. For example, MCs have been shown to inhibit the growth of Streptococcus pyogenes by forming extracellular traps (65), and it has also been demonstrated that MCs contribute to the defense toward invasive group A streptococci by secreting cathelicidin (10). Moreover, Streptococcus pneumoniae was previously shown to activate RBL-2H3 cells, a cell line with MC-like characteristics (4).
We also show that the Gram-positive challenge induces the expression of a large panel of additional genes, many of which have not previously been implicated in MC-mediated responses or in antibacterial defense. A striking example is the dramatic upregulation of three isoforms of the Nr4a transcription factor family, of which Nr4a3 showed the highest extent of upregulation of all genes, and with the two additional members also being among the 10 genes that were most highly induced. Clearly, their massive induction suggests a major role in the response toward Gram-positive infection. However, their exact contribution, for example, if signaling events leading to cytokine/chemokine release is Nr4a dependent, remains to be established. Notably, Nr4a transcription factors have previously been shown to promote LPS-induced inflammatory gene expression in macrophages (44), and the results presented here are thus compatible with a similar role also in MCs.
It is also evident that a large number of proteases, as well as protease inhibitors, are induced by Gram-positive infection. Interestingly, however, genes coding for the main proteases present in the secretory granule, i.e., chymases, tryptases, and MC-carboxypeptidase A, were not among these.
There is substantial evidence that MCs can be activated to secrete cytokines without an involvement of degranulation. For example, it has previously been shown that activation of BMMCs by LPS produces a TLR4-dependent robust secretion of TNF-α, IL-6, IL-13, and IL-1β without causing detectable degranulation (55). In the same study it was shown that stimulation of BMMCs with PGN resulted in a similar cytokine response but also rapid release of preformed granule components (55), both responses being TLR2 dependent, suggesting that TLR2 and -4 may have differential effects on MC degranulation. On the other hand, several subsequent studies have argued against these findings by showing that TLR2 engagement by PGN and other Gram-positive cell wall components can induce cytokine release without signs of degranulation (21, 38, 46). Here we show that challenge of BMMCs with live Gram-positive bacteria results in significant release of histamine, a sign of MC degranulation. However, the histamine release occurred with slow kinetics, and the ultrastructural analysis did not reveal extensive MC degranulation. Since MC degranulation is a rapid event, usually detected within a few minutes after MC activation, the slow histamine response in response to S. equi suggests that the bacterial infection does not induce massive degranulation of the BMMCs. In fact, we cannot exclude that histamine was released as a consequence of de novo synthesis rather than release from preformed pools, the latter notion being underscored by the robust upregulation of the histidine decarboxylase gene (hdc) in response to S. equi challenge (see Table S1 in the supplemental material). Our data thus conform to the notion that MC activation by bacterial cell wall products occurs without extensive release of preformed granule and extend this notion by showing that also challenge with live bacteria causes MC activation without signs of extensive degranulation. Interestingly, the microarray analysis revealed a strong upregulation of several genes involved in dampening mast cell degranulation, including regulator of calcineurin (Rcan) (66), Src-like adaptor (SLA) (43), and Rabgef1 (57). Hence, we may speculate that the slow release of granule mediators is associated with an upregulated expression of compounds involved in suppression of MC degranulation. In fact, we cannot exclude that the S. equi-mediated induction of genes involved in preventing MC degranulation may serve as a bacterial strategy to escape host defense mechanisms that are dependent on MC granule mediators. For example, the suppression of MC degranulation will lead to impaired secretion of mouse MC protease 6 (mMCP-6), a secretory granule protease that was recently implicated in antibacterial defense (58). On the other hand, the microarray analysis also revealed a strong upregulation of the genes coding for sphingosine kinase 1 (SphK1), an enzyme that recently has been implicated in FcɛRI-dependent degranulation of human MCs (41), suggesting that also compounds promoting MC degranulation are upregulated. Interestingly, though, in mice, SphK1 has been shown to be dispensable for FcɛRI-mediated degranulation, whereas the isoform, SphK2, has a major role (40). In support of activated SphK-dependent pathways, the microarray analysis also revealed an upregulated expression of myristoylated alanine-rich protein kinase C substrate (MARCKS), a protein that is recruited by sphingosine-1-phosphate (19), the latter being the enzymatic product of SphK.
Although TLR2 was shown to have a major role in S. equi-induced cytokine/chemokine production, it is clear that substantial cytokine/chemokine release occurs also in the absence of a functional TLR2, suggesting a contribution by TLR2-independent mechanisms. Interestingly, we also noted a clear reduction of the cytokine response in BMMCs lacking TLR4. Considering that LPS is regarded as the major stimulant for TLR4, this was somewhat unexpected since S. equi, being Gram-positive, does not express surface LPS. On the other hand, Gram-positive bacteria produce a number of pore-forming toxins denoted cholesterol-dependent cytolysins (6), and it has been documented that several of these toxins act as TLR4 agonists (32, 42). We may therefore speculate that the TLR4-dependent activation of BMMC by S. equi may be caused by toxins of this class, although the exact identity of the active TLR4 agonist produced by S. equi remains to be identified. Another explanation for the TLR2-independepnt cytokine/chemokine release could be that S. equi causes activation of NOD1/2, since both of these PRRs have been shown to be expressed by MCs (14, 39). However, since we did not see any uptake of bacteria by the BMMC, it appears less likely that these intracellular receptors have a major role in S. equi-induced cytokine/chemokine induction. A third possibility would be that PRRs belonging to the C-type lectin family account for the TLR2-independent cytokine/chemokine induction. In line with such a scenario, S. equi challenge resulted in a strong induction of C-type lectin domain family 4, member e (Clec4e) (Table 1).
Strikingly, heat-activated S. equi caused only minimal cytokine/chemokine induction compared to live counterparts. This was somewhat unexpected considering that TLR2 had a major impact on cytokine/chemokine secretion and that, presumably, the PGN component of the S. equi cell wall is a major agonist for the TLR2 expressed on the BMMC surface. Clearly, this indicates that optimal bacterium-driven effects on MCs requires viable bacteria and, therefore, that caution should be taken when attempting to mimic bacterial effects on MCs by using isolated cell wall components. However, the identity of the heat-labile factor(s) contributing to S. equi-mediated MC activation remains to be elucidated.
The effect of live bacteria on the global gene expression in MCs has not been extensively investigated previously. In one study, Kulka et al. studied the effect of E. coli on global gene expression in a human MC line (24). In agreement with the present study, E. coli was found to induce a number of CCL chemokines, TNF-α, transcription factors, and signaling molecules. However, the effects seen were considerably less pronounced compared to the present study, with a relatively low number of genes affected and with only few genes being upregulated more than fourfold (24). Likely explanations for the different effects seen in the present study and the study by Kulka et al. include the possibility that E. coli causes milder effects on MCs than doses S. equi, human/mouse species differences and that the present study used BMMCs, whereas the study by Kulka et al. was based on the use of a MC line (LAD-2).
In summary, the present study has revealed important insight into the direct effects of Gram-positive bacteria on MCs. We strongly believe that the molecular patterns identified here may provide important clues as to the mechanisms by which MCs operate during bacterial infection in vivo.
Supplementary Material
Acknowledgments
We are grateful to Hanna Göransson (Uppsala Array Platform) for helpful discussions and excellent support throughout this investigation.
This study was supported by grants from Formas (G.P. and B.G.), The Swedish Research Council (G.P.), Torsten and Ragnar Söderberg Foundation (G.P.), and the Swedish Horse Board (B.G.).
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 23 November 2009.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Abbott, Y., E. Acke, S. Khan, E. Muldoon, B. Markey, M. Pinilla, F. Leonard, K. Steward, and A. S. Waller. 2009. Zoonotic transmission of Streptococcus equi subsp. zooepidemicus from a dog to a handler. J. Med. Microbiol. 59:120-123. [DOI] [PubMed] [Google Scholar]
- 2.Abrink, M., M. Grujic, and G. Pejler. 2004. Serglycin is essential for maturation of mast cell secretory granule. J. Biol. Chem. 279:40897-40905. [DOI] [PubMed] [Google Scholar]
- 3.Arock, M., E. Ross, R. Lai-Kuen, G. Averlant, Z. Gao, and S. N. Abraham. 1998. Phagocytic and tumor necrosis factor alpha response of human mast cells following exposure to gram-negative and gram-positive bacteria. Infect. Immun. 66:6030-6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbuti, G., M. Moschioni, S. Censini, A. Covacci, C. Montecucco, and P. Montemurro. 2006. Streptococcus pneumoniae induces mast cell degranulation. Int. J. Med. Microbiol. 296:325-329. [DOI] [PubMed] [Google Scholar]
- 5.Barton, G. M., and J. C. Kagan. 2009. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat. Rev. 9:535-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billington, S. J., B. H. Jost, and J. G. Songer. 2000. Thiol-activated cytolysins: structure, function and role in pathogenesis. FEMS Microbiol. Lett. 182:197-205. [DOI] [PubMed] [Google Scholar]
- 7.Boyce, J. A. 2007. Mast cells and eicosanoid mediators: a system of reciprocal paracrine and autocrine regulation. Immunol. Rev. 217:168-185. [DOI] [PubMed] [Google Scholar]
- 8.Braga, T., M. Grujic, A. Lukinius, L. Hellman, M. Abrink, and G. Pejler. 2007. Serglycin proteoglycan is required for secretory granule integrity in mucosal mast cells. Biochem. J. 403:49-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawicki, W., and J. S. Marshall. 2007. New and emerging roles for mast cells in host defense. Curr. Opin. Immunol. 19:31-38. [DOI] [PubMed] [Google Scholar]
- 10.Di Nardo, A., K. Yamasaki, R. A. Dorschner, Y. Lai, and R. L. Gallo. 2008. Mast cell cathelicidin antimicrobial peptide prevents invasive group A Streptococcus infection of the skin. J. Immunol. 180:7565-7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Echtenacher, B., D. N. Mannel, and L. Hultner. 1996. Critical protective role of mast cells in a model of acute septic peritonitis. Nature 381:75-77. [DOI] [PubMed] [Google Scholar]
- 12.Elson, G., I. Dunn-Siegrist, B. Daubeuf, and J. Pugin. 2007. Contribution of Toll-like receptors to the innate immune response to Gram-negative and Gram-positive bacteria. Blood 109:1574-1583. [DOI] [PubMed] [Google Scholar]
- 13.Feger, F., S. Varadaradjalou, Z. Gao, S. N. Abraham, and M. Arock. 2002. The role of mast cells in host defense and their subversion by bacterial pathogens. Trends Immunol. 23:151-158. [DOI] [PubMed] [Google Scholar]
- 14.Feng, B. S., S. H. He, P. Y. Zheng, L. Wu, and P. C. Yang. 2007. Mast cells play a crucial role in Staphylococcus aureus peptidoglycan-induced diarrhea. Am. J. Pathol. 171:537-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flock, M., K. Jacobsson, L. Frykberg, T. R. Hirst, A. Franklin, B. Guss, and J. I. Flock. 2004. Recombinant Streptococcus equi proteins protect mice in challenge experiments and induce immune response in horses. Infect. Immun. 72:3228-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galli, S. J., S. Nakae, and M. Tsai. 2005. Mast cells in the development of adaptive immune responses. Nat. Immunol. 6:135-142. [DOI] [PubMed] [Google Scholar]
- 17.Gekara, N. O., and S. Weiss. 2008. Mast cells initiate early anti-Listeria host defenses. Cell. Microbiol. 10:225-236. [DOI] [PubMed] [Google Scholar]
- 18.Grimbaldeston, M. A., S. Nakae, J. Kalesnikoff, M. Tsai, and S. J. Galli. 2007. Mast cell-derived interleukin 10 limits skin pathology in contact dermatitis and chronic irradiation with ultraviolet B. Nat. Immunol. 8:1095-1104. [DOI] [PubMed] [Google Scholar]
- 19.Guo, Y., P. A. Singleton, A. Rowshan, M. Gucek, R. N. Cole, D. R. Graham, J. E. Van Eyk, and J. G. Garcia. 2007. Quantitative proteomics analysis of human endothelial cell membrane rafts: evidence of MARCKS and MRP regulation in the sphingosine 1-phosphate-induced barrier enhancement. Mol. Cell Proteomics 6:689-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hochberg, Y., and Y. Benjamini. 1990. More powerful procedures for multiple significance testing. Stat. Med. 9:811-818. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda, T., and M. Funaba. 2003. Altered function of murine mast cells in response to lipopolysaccharide and peptidoglycan. Immunol. Lett. 88:21-26. [DOI] [PubMed] [Google Scholar]
- 22.Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis, U. Scherf, and T. P. Speed. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249-264. [DOI] [PubMed] [Google Scholar]
- 23.Jones, K. L., D. M. de Kretser, S. Patella, and D. J. Phillips. 2004. Activin A and follistatin in systemic inflammation. Mol. Cell. Endocrinol. 225:119-125. [DOI] [PubMed] [Google Scholar]
- 24.Kulka, M., N. Fukuishi, M. Rottem, Y. A. Mekori, and D. D. Metcalfe. 2006. Mast cells, which interact with Escherichia coli, up-regulate genes associated with innate immunity and become less responsive to Fc(epsilon)RI-mediated activation. J. Leukoc. Biol. 79:339-350. [DOI] [PubMed] [Google Scholar]
- 25.Lantz, C. S., J. Boesiger, C. H. Song, N. Mach, T. Kobayashi, R. C. Mulligan, Y. Nawa, G. Dranoff, and S. J. Galli. 1998. Role for interleukin-3 in mast-cell and basophil development and in immunity to parasites. Nature 392:90-93. [DOI] [PubMed] [Google Scholar]
- 26.Lee, D. M., D. S. Friend, M. F. Gurish, C. Benoist, D. Mathis, and M. B. Brenner. 2002. Mast cells: a cellular link between autoantibodies and inflammatory arthritis. Science 297:1689-1692. [DOI] [PubMed] [Google Scholar]
- 27.Lindmark, H., and B. Guss. 1999. SFS, a novel fibronectin-binding protein from Streptococcus equi, inhibits the binding between fibronectin and collagen. Infect. Immun. 67:2383-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindmark, H., M. Nilsson, and B. Guss. 2001. Comparison of the fibronectin-binding protein FNE from Streptococcus equi subspecies equi with FNZ from S. equi subspecies zooepidemicus reveals a major and conserved difference. Infect. Immun. 69:3159-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu, L. F., E. F. Lind, D. C. Gondek, K. A. Bennett, M. W. Gleeson, K. Pino-Lagos, Z. A. Scott, A. J. Coyle, J. L. Reed, J. Van Snick, T. B. Strom, X. X. Zheng, and R. J. Noelle. 2006. Mast cells are essential intermediaries in regulatory T-cell tolerance. Nature 442:997-1002. [DOI] [PubMed] [Google Scholar]
- 30.Malaviya, R., T. Ikeda, E. Ross, and S. N. Abraham. 1996. Mast cell modulation of neutrophil influx and bacterial clearance at sites of infection through TNF-alpha. Nature 381:77-80. [DOI] [PubMed] [Google Scholar]
- 31.Malaviya, R., E. A. Ross, J. I. MacGregor, T. Ikeda, J. R. Little, B. A. Jakschik, and S. N. Abraham. 1994. Mast cell phagocytosis of FimH-expressing enterobacteria. J. Immunol. 152:1907-1914. [PubMed] [Google Scholar]
- 32.Malley, R., P. Henneke, S. C. Morse, M. J. Cieslewicz, M. Lipsitch, C. M. Thompson, E. Kurt-Jones, J. C. Paton, M. R. Wessels, and D. T. Golenbock. 2003. Recognition of pneumolysin by Toll-like receptor 4 confers resistance to pneumococcal infection. Proc. Natl. Acad. Sci. U. S. A. 100:1966-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall, J. S. 2004. Mast-cell responses to pathogens. Nat. Rev. 4:787-799. [DOI] [PubMed] [Google Scholar]
- 34.Maurer, M., B. Echtenacher, L. Hultner, G. Kollias, D. N. Mannel, K. E. Langley, and S. J. Galli. 1998. The c-kit ligand, stem cell factor, can enhance innate immunity through effects on mast cells. J. Exp. Med. 188:2343-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maurer, M., J. Wedemeyer, M. Metz, A. M. Piliponsky, K. Weller, D. Chatterjea, D. E. Clouthier, M. M. Yanagisawa, M. Tsai, and S. J. Galli. 2004. Mast cells promote homeostasis by limiting endothelin-1-induced toxicity. Nature 432:512-516. [DOI] [PubMed] [Google Scholar]
- 36.McCurdy, J. D., T. J. Lin, and J. S. Marshall. 2001. Toll-like receptor 4-mediated activation of murine mast cells. J. Leukoc. Biol. 70:977-984. [PubMed] [Google Scholar]
- 37.Metcalfe, D. D., D. Baram, and Y. A. Mekori. 1997. Mast cells. Physiol. Rev. 77:1033-1079. [DOI] [PubMed] [Google Scholar]
- 38.Mrabet-Dahbi, S., M. Metz, A. Dudeck, T. Zuberbier, and M. Maurer. 2009. Murine mast cells secrete a unique profile of cytokines and prostaglandins in response to distinct TLR2 ligands. Exp. Dermatol. 18:437-444. [DOI] [PubMed] [Google Scholar]
- 39.Okumura, S., K. Yuki, R. Kobayashi, S. Okamura, K. Ohmori, H. Saito, C. Ra, and Y. Okayama. 2009. Hyperexpression of NOD2 in intestinal mast cells of Crohn's disease patients: preferential expression of inflammatory cell-recruiting molecules via NOD2 in mast cells. Clin. Immunol. 130:175-185. [DOI] [PubMed] [Google Scholar]
- 40.Olivera, A., K. Mizugishi, A. Tikhonova, L. Ciaccia, S. Odom, R. L. Proia, and J. Rivera. 2007. The sphingosine kinase-sphingosine-1-phosphate axis is a determinant of mast cell function and anaphylaxis. Immunity 26:287-297. [DOI] [PubMed] [Google Scholar]
- 41.Oskeritzian, C. A., S. E. Alvarez, N. C. Hait, M. M. Price, S. Milstien, and S. Spiegel. 2008. Distinct roles of sphingosine kinases 1 and 2 in human mast-cell functions. Blood 111:4193-4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park, J. M., V. H. Ng, S. Maeda, R. F. Rest, and M. Karin. 2004. Anthrolysin O and other gram-positive cytolysins are Toll-like receptor 4 agonists. J. Exp. Med. 200:1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park, S. K., H. Qiao, and M. A. Beaven. 2009. Src-like adaptor protein (SLAP) is upregulated in antigen-stimulated mast cells and acts as a negative regulator. Mol. Immunol. 46:2133-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pei, L., A. Castrillo, and P. Tontonoz. 2006. Regulation of macrophage inflammatory gene expression by the orphan nuclear receptor Nur77. Mol. Endocrinol. 20:786-794. [DOI] [PubMed] [Google Scholar]
- 45.Pejler, G., M. Åbrink, M. Ringvall, and S. Wernersson. 2007. Mast cell proteases. Adv. Immunol. 95:167-255. [DOI] [PubMed] [Google Scholar]
- 46.Qiao, H., M. V. Andrade, F. A. Lisboa, K. Morgan, and M. A. Beaven. 2006. FcepsilonR1 and Toll-like receptors mediate synergistic signals to markedly augment production of inflammatory cytokines in murine mast cells. Blood 107:610-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Razin, E., C. Cordon-Cardo, and R. A. Good. 1981. Growth of a pure population of mouse mast cells in vitro with conditioned medium derived from concanavalin A-stimulated splenocytes. Proc. Natl. Acad. Sci. U. S. A. 78:2559-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Secor, V. H., W. E. Secor, C. A. Gutekunst, and M. A. Brown. 2000. Mast cells are essential for early onset and severe disease in a murine model of multiple sclerosis. J. Exp. Med. 191:813-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sher, A., A. Hein, G. Moser, and J. P. Caulfield. 1979. Complement receptors promote the phagocytosis of bacteria by rat peritoneal mast cells. Lab. Invest. 41:490-499. [PubMed] [Google Scholar]
- 50.Siebenhaar, F., W. Syska, K. Weller, M. Magerl, T. Zuberbier, M. Metz, and M. Maurer. 2007. Control of Pseudomonas aeruginosa skin infections in mice is mast cell-dependent. Am. J. Pathol. 170:1910-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smyth, G. K. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. [DOI] [PubMed]
- 52.Soucek, L., E. R. Lawlor, D. Soto, K. Shchors, L. B. Swigart, and G. I. Evan. 2007. Mast cells are required for angiogenesis and macroscopic expansion of Myc-induced pancreatic islet tumors. Nat. Med. 13:1211-1218. [DOI] [PubMed] [Google Scholar]
- 53.Sun, J., G. K. Sukhova, P. J. Wolters, M. Yang, S. Kitamoto, P. Libby, L. A. MacFarlane, J. Mallen-St Clair, and G. P. Shi. 2007. Mast cells promote atherosclerosis by releasing proinflammatory cytokines. Nat. Med. 13:719-724. [DOI] [PubMed] [Google Scholar]
- 54.Sun, J., G. K. Sukhova, M. Yang, P. J. Wolters, L. A. MacFarlane, P. Libby, C. Sun, Y. Zhang, J. Liu, T. L. Ennis, R. Knispel, W. Xiong, R. W. Thompson, B. T. Baxter, and G. P. Shi. 2007. Mast cells modulate the pathogenesis of elastase-induced abdominal aortic aneurysms in mice. J. Clin. Invest. 117:3359-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Supajatura, V., H. Ushio, A. Nakao, S. Akira, K. Okumura, C. Ra, and H. Ogawa. 2002. Differential responses of mast cell Toll-like receptors 2 and 4 in allergy and innate immunity. J. Clin. Invest. 109:1351-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takeda, K., T. Kaisho, and S. Akira. 2003. Toll-like receptors. Annu. Rev. Immunol. 21:335-376. [DOI] [PubMed] [Google Scholar]
- 57.Tam, S. Y., M. Tsai, J. N. Snouwaert, J. Kalesnikoff, D. Scherrer, S. Nakae, D. Chatterjea, D. M. Bouley, and S. J. Galli. 2004. RabGEF1 is a negative regulator of mast cell activation and skin inflammation. Nat. Immunol. 5:844-852. [DOI] [PubMed] [Google Scholar]
- 58.Thakurdas, S. M., E. Melicoff, L. Sansores-Garcia, D. C. Moreira, Y. Petrova, R. L. Stevens, and R. Adachi. 2007. The mast cell-restricted tryptase mMCP-6 has a critical immunoprotective role in bacterial infections. J. Biol. Chem. 82:20809-20815. [DOI] [PubMed] [Google Scholar]
- 59.Timoney, J. F. 2004. The pathogenic equine streptococci. Vet. Res. 35:397-409. [DOI] [PubMed] [Google Scholar]
- 60.Tsai, M., T. Takeishi, H. Thompson, K. E. Langley, K. M. Zsebo, D. D. Metcalfe, E. N. Geissler, and S. J. Galli. 1991. Induction of mast cell proliferation, maturation, and heparin synthesis by the rat c-kit ligand, stem cell factor. Proc. Natl. Acad. Sci. U. S. A. 88:6382-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Varadaradjalou, S., F. Feger, N. Thieblemont, N. B. Hamouda, J. M. Pleau, M. Dy, and M. Arock. 2003. Toll-like receptor 2 (TLR2) and TLR4 differentially activate human mast cells. Eur. J. Immunol. 33:899-906. [DOI] [PubMed] [Google Scholar]
- 62.Wei, O. L., A. Hilliard, D. Kalman, and M. Sherman. 2005. Mast cells limit systemic bacterial dissemination but not colitis in response to Citrobacter rodentium. Infect. Immun. 73:1978-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Velin, D., D. Bachmann, H. Bouzourene, and P. Michetti. 2005. Mast cells are critical mediators of vaccine-induced Helicobacter clearance in the mouse model. Gastroenterology 129:142-155. [DOI] [PubMed] [Google Scholar]
- 64.Wetzler, L. M. 2003. The role of Toll-like receptor 2 in microbial disease and immunity. Vaccine 21(Suppl. 2):S55-S60. [DOI] [PubMed] [Google Scholar]
- 65.von Kockritz-Blickwede, M., O. Goldmann, P. Thulin, K. Heinemann, A. Norrby-Teglund, M. Rohde, and E. Medina. 2008. Phagocytosis-independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood 111:3070-3080. [DOI] [PubMed] [Google Scholar]
- 66.Yang, Y. J., W. Chen, A. Edgar, B. Li, J. D. Molkentin, J. N. Berman, and T. J. Lin. 2009. Rcan1 negatively regulates FcɛRI-mediated signaling and mast cell function. J. Exp. Med. 206:195-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.