Abstract
Unrepaired DNA lesions can block the progression of the replication fork, leading to genomic instability and cancer in higher-order eukaryotes. In Saccharomyces cerevisiae, replication through DNA lesions can be mediated by translesion synthesis DNA polymerases, leading to error-free or error-prone damage bypass, or by Rad5-mediated template switching to the sister chromatid that is inherently error free. While translesion synthesis pathways are highly conserved from yeast to humans, very little is known of a Rad5-like pathway in human cells. Here we show that a human homologue of Rad5, HLTF, can facilitate fork regression and has a role in replication of damaged DNA. We found that HLTF is able to reverse model replication forks, a process which depends on its double-stranded DNA translocase activity. Furthermore, from analysis of isolated dually labeled chromosomal fibers, we demonstrate that in vivo, HLTF promotes the restart of replication forks blocked at DNA lesions. These findings suggest that HLTF can promote error-free replication of damaged DNA and support a role for HLTF in preventing mutagenesis and carcinogenesis, providing thereby for its potential tumor suppressor role.
Genomic instability underlies the development of various human diseases, including cancer. Cancer genomes are highly heterogeneous and can possess various instability phenotypes, including accumulation of gross chromosomal rearrangements (GCR) (9, 40). A plethora of evidence has indicated that defects in various DNA repair pathways promote genomic instability and trigger subsequent tumor formation (1). One example is the mutational or epigenetic inactivation of the mismatch repair gene MSH2 or MLH1 in a subset of colorectal cancers (4, 39). An apparently distinct subset of colon cancers, representing about 40% of malignant colorectal transformations, is characterized by epigenetic inactivation of the HLTF gene (22). While HLTF has been suggested to act as a transcription factor (31), recent studies indicated a role for HLTF in replication of damaged DNA, raising the possibility that it is this function of HLTF that can lead to suppression of genomic instability (23, 37).
In Saccharomyces cerevisiae, genetic data have indicated a crucial role for the Rad6-Rad18 protein complex, Rad5, and the Mms2-Ubc13 complex in the replication of damaged DNA (26). In contrast to nucleotide incorporation opposite the lesion by specialized translesion synthesis polymerases, which requires Rad6-Rad18-dependent monoubiquitylation of PCNA, template-switching-mediated bypass depends on Lys63 polyubiquitylation of PCNA by the Mms2-Ubc13 ubiquitin-conjugating enzyme complex and Rad5 ubiquitin ligase (10, 12, 36, 41). Yeast genetic data have shown that not only the ubiquitin ligase activity of Rad5 but also its ATPase activity is essential for its function in replication of damaged DNA (5). In agreement with the in vivo data, Rad5 is an ATP-hydrolysis-driven molecular motor which can facilitate template switching at stalled replication forks (2). While translesion synthesis can occur at the price of inserting wrong nucleotides during the synthesis process, template switching is inherently error-free. Consequently, in rad5Δ yeast cells, stalled replication forks could be resolved at the price of fork collapse and consequent genomic rearrangements (33).
Human HLTF has recently been shown to share several functional and structural similarities with yeast Rad5. Reduction of HLTF expression enhances DNA damage sensitivity and promotes GCR upon DNA damage, and HLTF is able to partially complement for yeast Rad5 function in a sensitized genetic background (23, 37). Moreover, HLTF has a yeast Rad5-like domain structure with a C3HC4 RING domain embedded into a SWI/SNF2 helicase motif. Similarly to other RING domain-containing proteins, HLTF is a ubiquitin ligase which, together with Rad6-Rad18 and Mms2-Ubc13 ubiquitin-conjugating complexes, carries out PCNA polyubiquitylation (17, 23, 37). However, the role of the ATPase/helicase domain of HLTF has not been addressed and direct evidence for the involvement of HLTF in replication of damaged DNA is missing.
We examined whether HLTF could facilitate the replication of damaged DNA. We found that HLTF is an ATP hydrolysis-driven double-stranded DNA translocase that can regress replication fork-like structures. In addition we show that the lack of HLTF, or mutational inactivation of its ATPase or RING domain in human cells, hinders fork movement upon DNA damage. We discuss the possible role of these activities of HLTF in cancer suppression.
MATERIALS AND METHODS
Proteins.
Wild-type and ATPase mutant DE557,558AA HLTFs were purified to apparent homogeneity, after being overexpressed as glutathione S-transferase (GST)-FLAG-fusion proteins in yeast using plasmids PIL1520 and PIL1734, respectively (37). Nicking endonucleases Nt.BbvCI and Nb.BbvCI, T4 gp32 (New England Biolabs), and Escherichia coli SSB (GE Healthcare)-purified proteins were purchased. The activity of single-stranded DNA (ssDNA) binding proteins under our experimental conditions was verified using gel-shift experiments (see Fig. S1 in the supplemental material).
DNA substrates.
To generate oligonucleotide-based DNA substrates, we annealed oligonucleotides listed in Table S1 in the supplemental material in various combinations, followed by purification on polyacrylamide gels as described previously (2). The term heterologous fork (HetF) refers to replication fork-like structures with noncomplementary leading and lagging arms, and homologous fork (HomF) indicates forks with complementary leading and lagging arms.
The plasmid-sized replication fork model substrate was created essentially as described previously, using pG46 and pG68 plasmids (2, 28). Briefly, the plasmids were gapped by digestion with nicking endonucleases Nt.BbvCI and Nb.BbvCI, respectively. Next, a pG46-containing sample was treated with shrimp alkaline phosphatase and subsequently labeled with T4 polynucleotide kinase while a pG68 derivative was linearized with XhoI digestion. The resulting plasmids containing complementary single-stranded gaps were then annealed at 53°C in 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 5 mM dithiothreitol (DTT) to form a joint molecule. Note that the joint molecule contains a structural mimic of a stalled replication fork in which the labeled lagging strand is longer by 14 nucleotides. pG68SapI(Het) was created from pG68 by inserting a small sequence heterology by cloning O1301/O1302 heteroduplex oligonucleotides into the SapI site of pG68. We note that in the original description of this experimental system the joint was formed by annealing the two gapped circular plasmids and converting the joint to a plectonemic joint by topoisomerase treatment; however, this conversion was not necessary for our purposes because HLTF cannot dissolve model forks to parental duplexes.
Plasmids for triple-helix displacement substrate preparation were made by cloning GAT CCT CGA TAT CTT TCT TTT TTC TTC TTT TCT TTC TTT TTC TCT CCT CAG CCT CAG CGT AG and GAT CCT CGA TAT CTT TCT TTT TTC TTC TTT TCT TTC TTT TTC TCT CCT CAG CAG CTG CCT CAG CGT AG sequences into the BamHI and SalI sites of pUC19 to yield plasmids pIL1828 and pIL1829, respectively. The double-stranded portion of the triplexes was amplified by PCR from pIL1828 or pIL1829 and O1107 and O1108 primers, and after purification the third strand, oligonucleotide O1377, was annealed in 45 mM morpholineethanesulfonic acid (MES)-NaOH (pH 5.5), 15 mM MgCl2; the mixture was then heated to 50°C and let cool down to room temperature. Substrates were purified from 10% native polyacrylamide gel electrophoresis (PAGE) gels developed in electrophoresis buffer containing 20 mM Tris-acetate (pH 5.5), 5 mM Na acetate, 1 mM MgCl2 by crushing and soaking the gel slices in a solution containing 15 mM Tris-HCl, pH 6.8, 10 mM MgCl2, and 1 mM DTT. To generate nicked or blunt-ended triple helices, the enzymatic manipulations were performed on the double-stranded base before the triplex-forming oligonucleotide was annealed. Briefly, the left arm of the product amplified from pIL1828 was removed by EcoRV and the right arm was nicked by using either Nt.BbvCI or Nb.BbvCI. For “blunt triplex,” the product amplified from pIL1829 was treated with EcoRV and PvuII, which leaves a 1-bp overhang and a 10-bp overhang on the two sides.
Fork reversal and triple-helix displacement assays.
Fork reversal and triple-helix displacement assays were carried out in buffer A containing 20 mM Tris-HCl, pH 7.0, 150 mM NaCl, 5 mM MgCl2, 5 mM ATP, 0.1 mg/ml bovine serum albumin, 1 mM DTT, and 10% glycerol with 0.5 nM 32P-labeled DNA and purified HLTF at the concentrations indicated in the figure legends. After the reaction mixtures were incubated at 37°C for 5 min or for the time indicated on the figure, equal volumes of helicase stop buffer containing 20 mM EDTA, 2 mg/ml proteinase K, 1% sodium dodecyl sulfate, 10% glycerol, and 0.02% bromphenol blue were added, followed by further incubation for 5 min before the DNA samples were loaded onto 10% native polyacrylamide gels and the products were separated by electrophoresis using 1× Tris-borate buffer containing no EDTA. Conditions for the branch migration assay were basically identical to those for helicase assays, except that either ATP or AMP-PNP was used at 1 mM with equimolar MgCl2.
Assays with the plasmid-sized forks were carried out essentially as described above but using 5 nM substrate DNA and 80 nM HLTF unless indicated otherwise. Reaction mixtures were quenched after incubation for 5 min at 37°C and analyzed by restriction enzyme digestion as described previously (2).
The reaction products of the triple-helix assays were separated on 10% native PAGE gels developed in electrophoresis buffer containing 20 mM Tris-acetate (pH 5.5), 5 mM Na acetate, and 1 mM MgCl2.
HLTF RNA interference in HeLa cells and immunolabeling of DNA fibers.
To obtain the HLTF DE557,558AA ATPase mutant expressing the construct, HLTF cDNA was taken from pIL1734 and cloned into the StuI site of pCS2+MT (pIL1267), a cytomegalovirus (CMV) promoter-driven vector for expressing proteins with an N-terminal fusion of 6MYC, resulting in pIL1792. The small interfering RNA (siRNA)-resistant wild-type and ATPase mutant HLTF-expressing plasmids were generated on plasmids pIL1370 (37) and pIL1792, respectively, by the QuikChange site-directed mutagenesis kit (Stratagene) using oligonucleotides O2088 (5′-GCT TGC AGG CGC CTT GGC GTA CAT CAT GGA CAA CAA ATT GGC-3′) and O2089 (5′-GCC AAT TTG TTG TCC ATG ATG TAC GCC AAG GCG CCT GCA AGC-3′), resulting in plasmids pIL1990 and pIL1991, respectively.
The C759S RING mutant was generated on pIL1990 by site-directed mutagenesis using the following primers: O2251 (5′-GTT CAG ATG AGG AAT CTG CAG TTT GCC TGG ATT CTT TAA C-3′) and O2252 (5′-GTT AAA GAA TCC AGG CAA ACT GCA GAT TCC TCA TCT GAA C-3′). The mutation was sequence verified and subcloned into pIL1990 as a NotI-SacI fragment, resulting in plasmid pIL2007. To obtain the RING-ATPase double mutant, the RING mutation containing the region from pIL2007 was subcloned as a NotI-SacI fragment into the pIL1991 plasmid, resulting in plasmid pIL2008.
Predesigned synthetic duplex siRNA for human HLTF, O1319 (GGUGCUUUGGCCUAUAUCAtt), and a nonspecific negative control siRNA (NC siRNA 1), O1359, were purchased from Ambion. In the presence of 100 nM siRNAs, HeLa cells were transfected using Lipofectamine 2000 (Invitrogen). For HLTF knockdown and complementation studies, cells were transfected first with siRNA and after 24 h with the siRNA-resistant wild-type and ATPase mutant HLTF expressing plasmids pIL1990 and pIL1991, respectively. DNA fibers were prepared 24 h after the plasmid transfection, when the knockdown efficiency and the overexpression of the siRNA-resistant HLTF were also confirmed by Western blot using anti-HLTF antibody on total cell extract.
Fiber assays were carried out as described earlier (14). Briefly, for pulse-labeling, HeLa cells were incubated with 25 μM iododeoxyuridine (IdU) for 15 min, washed, and then treated with 0.01% methyl methanesulfonate (MMS) for 20 min, which was followed by extensive washing before the second pulse-labeling with 250 μM BrdU for various times as indicated. Cells were then harvested, and DNA fiber spreads were prepared. BrdU-labeled tracks were detected with anti-BrdU antibody (OBT0030G; Oxford Biotechnology) and with Alexa Fluor-633-conjugated secondary antibody (A21094; Invitrogene). IdU-labeled tracks were detected using anti-BrdU/IdU monoclonal antibody (MD5100; Invitrogene) and Cy3-conjugated secondary antibody (C-2181; Sigma). Quality control for spreading DNA was performed by YOYO (Sigma) labeling (0.1 μM). Fibers were examined using an Olympus FV1000 confocal microscope and 63× lens.
RESULTS
HLTF is able to reverse model replication forks.
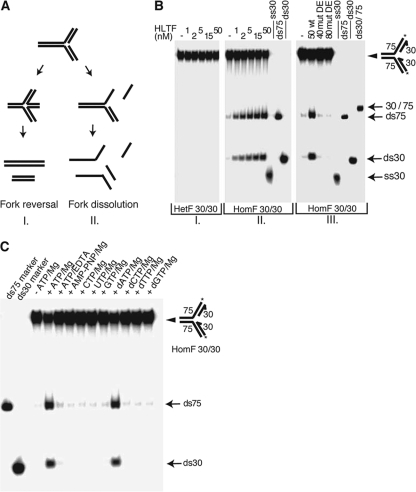
To test if HLTF is able to rearrange replication forks for template switching, we designed various model replication fork substrates from oligonucleotides. The logic of the experimental design is based on the assumption that coordinated fork reversion can occur only if the arms of the fork are homologous so that the nascent strands as well as the parental strands can anneal, and it should result only in double-stranded products (Fig. 1A, panel I). In contrast, a fork dissolution activity would act on both homologous and heterologous forks and result in various single-stranded products (Fig. 1A, panel II). To distinguish between these two possibilities, we tested if purified HLTF could process an oligonucleotide-based heterologous fork (HetF) or homologous fork (HomF). We found that HLTF left HetF unprocessed (Fig. 1B, panel I) but converted the HomF fork into distinct double-stranded products characteristic of annealed parental and nascent strands (Fig. 1B, panel II). The observed activity is intrinsic to HLTF, since an HLTF mutant protein carrying the DE557,558AA amino acid changes in its conserved DEQH motif involved in ATP hydrolysis showed no fork-processing activity (Fig. 1B, panel III). The fork reversal activity of HLTF was dependent on ATP (or dATP), and AMP-PNP, a nonhydrolyzable analogue of ATP, did not support the reaction, confirming the importance of ATP hydrolysis (Fig. 1C). Altogether, these results show that HLTF can carry out reversion of small replication fork-like model substrates in a reaction dependent on ATP hydrolysis, and it lacks fork dissolution activity.
FIG. 1.
HLTF reverses but does not dissolve oligonucleotide-based model replication-fork substrates. (A) Possible outcomes of enzymatic manipulation of model replication fork substrates. While fork reversal (I) can occur only on homologous forks which have complementary arms, fork dissolution (II) can happen on both homologous and heterologous forks. Whereas coordinated fork reversal requires specific enzymatic activity that leads only to two double-stranded oligonucleotides, fork dissolution by canonical helicase activity results in single-stranded oligonucleotides. (B) Fork regression activity of HLTF tested on heterologous fork (HetF) (I) and homologous fork (HomF) (II) model substrates. The DNA substrates in the gel are indicated by an arrowhead, while the positions of some of the possible products are shown by arrows. The 3′ ends of oligonucleotides are indicated by half arrows, and the positions of 5′ 32P labels are marked with asterisks. We note that formation of double-stranded products from HomF and not from HetF was used to indicate fork regression activity without dissolution of the fork. In panel III, the ATPase mutant HLTF DE557,558AA (DE) was examined with the HomF substrates. (C) HLTF activity requires ATP hydrolysis. Cofactor dependence of HLTF (15 nM) was tested on the oligonucleotide-based homologous fork model substrate, where one of the template strands and one of the opposite nascent strand were 5′ 32P labeled. The indicated ribonucleotides and deoxyribonucleotides were used at 5 mM concentrations in the presence of 5 mM MgCl2, except in lane 5, where 10 mM EDTA was used instead of MgCl2.
Mechanism of action of HLTF.
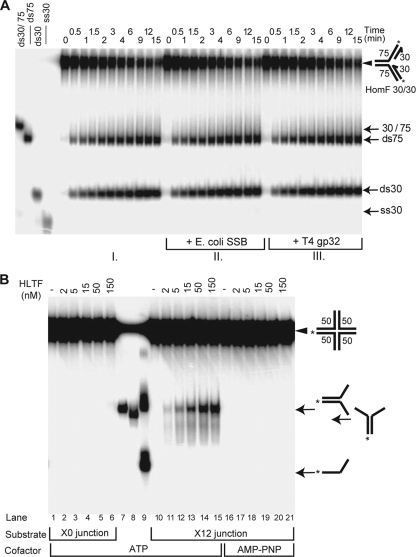
To rule out further any fork dissolution activity, we checked the kinetics of the product formation during HomF processing by HLTF and found that only the two double-stranded products appeared and they were formed at identical rates and without the appearance of any single-stranded intermediates (Fig. 2A, panel I). Moreover, single-stranded DNA binding proteins such as T4 gp32 and E. coli SSB, which are routinely used in conventional helicase assays to prevent product reannealing, had no effect in our time-course experiments (Fig. 2A, compare panel I to panels II and III; see Fig. S1 in the supplemental material). These data show that the outcome of the reaction is not the sum of kinetically separable events in which the nascent strands are released from the parental strands by a canonical helicase activity followed by the annealing of the nascent strands and of the parental strands together. Thus, HLTF concertedly unwinds and anneals the nascent and the parental strands without exposing extended single-stranded regions. The presence of such a highly specific fork reversal activity in HLTF suggested that HLTF would lack a conventional DNA helicase activity. Indeed, and in agreement with the fact that HLTF was not able to process HetF, we found that HLTF is unable to unwind double-stranded DNA present in partial heteroduplex, Y fork, or partial fork structures (see Fig. S2 in the supplemental material).
FIG. 2.
HLTF acts concertedly and possesses branch-migrating activity. (A) Kinetics of HLTF activity in the presence or absence of single-stranded DNA binding proteins. Processing of HomF substrate by HLTF (50 nM) was examined at the indicated time points in the absence (I) or presence of E. coli SSB (40 nM) (II) or T4 gp32 (90 nM) (III) proteins. (B) HLTF can migrate a moveable four-way junction. The X0 junction is static, while the core of the X12 junction, which was flanked with 19- to 20-nucleotide-long heterologies at the end of each arm, is movable. We note that product formation most likely requires spontaneous dissolution of terminal heterologies, which can explain the somewhat weaker activity of HLTF on X12 than on homologous fork substrates. Lane 7, marker Y fork containing O1114/O1115; lane 8, marker Y fork containing O1114/O1116; lane 9, boiled X12. Symbols are as described for Fig. 1.
The first intermediate of fork regression is a four-way junction, and to generate double-stranded products from a model replication fork substrate, HLTF should also be able to promote its branch migration. To test this possibility, we used synthetic four-way junctions that contained either the nonmovable heterologous arms (X0) or moveable arms (X12) containing a central homologous core (20). As expected, HLTF was not able to dissolve the X0 junction but promoted the conversion of the X12 junction into the Y fork in an ATP-hydrolysis-dependent manner, which indicates that HLTF exhibits branch-migrating activity (Fig. 2B).
HLTF can regress plasmid-sized replication fork model substrates.
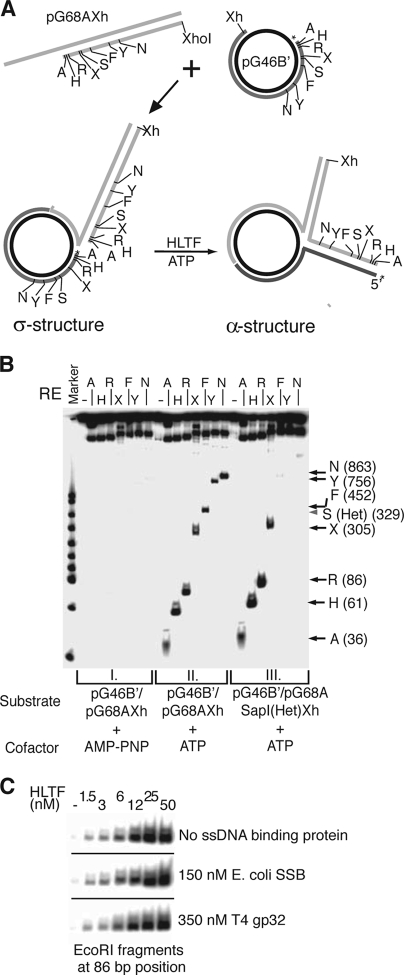
To approximate more closely the in vivo situation, where the leading and lagging strand DNA synthesis can become uncoupled and fork reversal can exceed hundreds of base pairs, we constructed a plasmid-sized model fork by annealing two nearly identical plasmids (3, 11, 35). At this configuration, the joint forms a σ-shaped molecule, where the labeled “lagging” strand is longer by 14 nucleotides (28) (Fig. 3A). Conversion of the σ structure to an α-shaped one by fork reversal can be conveniently followed by monitoring the transfer of the radioactive label from the circular lagging arm to the linear regressed arm context by using restriction endonucleases (RE) (Fig. 3A). We found that HLTF can revert the plasmid-sized fork in an ATP hydrolysis-dependent manner and can regress it through hundreds of bases, since at our longest checkpoint, at 863 base pairs long, a regressed arm appeared readily (Fig. 3B). In order to rule out that long single-stranded DNA regions are formed during the reaction which then anneal to recreate RE sites, we tested the effect of introducing a 30-nucleotide (nt)-long sequence heterology into the leading arm at 329-nt (Fig. 3B) or 452-nt (data not shown) positions and found that heterology prevented further processing of the fork, as revealed by the lack of appearance of RE site products downstream of the heterology. Moreover, we observed no inhibitory effect of single-stranded DNA binding proteins on the reaction, using either E. coli SSB or T4 gp32 (Fig. 3C; see Fig. S1 in the supplemental material). In conclusion, our results provide strong evidence that HLTF possesses fork reversal activity that proceeds without exposing extended single-stranded DNA intermediates.
FIG. 3.
HLTF can regress plasmid-sized model forks. (A) Schematic representation of the generation of joint DNA substrate (pG46B′/pG68AXh) (σ-structure) and the outcome of its HLTF-mediated regression named (α-structure). A, H, R, X, S, F, Y, N, and Xh refer to restriction endonuclease sites AvrII, BamHI, EcoRI, BsaXI, SapI, AflIII, BseYI, AlwNI, and XhoI, respectively. The positions of 5′-32P labels on the “lagging strand” are marked with asterisks. (B) Fork regression by HLTF is extensive and progressive. The transfer of restriction enzyme sites to the regressed arm by HLTF (50 nM) was followed in the presence of 5 mM ATP/Mg (panel II). The positions of the various restriction products generated by digestion of the regressed fork are indicated. The control set with 5 mM AMP-PNP/Mg shows the background level of spontaneous regression (panel I). Progressivity of the reaction is demonstrated by using pG46B′/pG68A SapI[Het]Xh substrate (panel III) in which a 30-bp sequence heterology was introduced at the SapI site (shown by arrowhead), which resulted in blocked regression beyond the heterology. (C) Fork reversal is not affected by single-stranded DNA-binding proteins. Regression through the EcoRI site was monitored at various HLTF concentrations in the presence or absence of E. coli SSB or T4 gp32 proteins.
HLTF is a double-stranded DNA translocase.
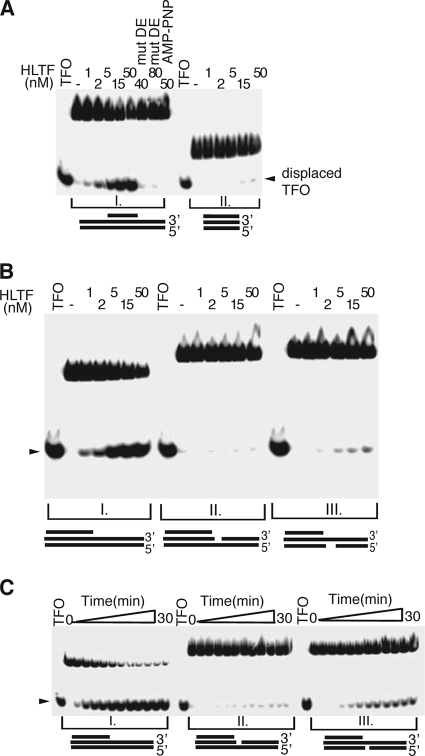
Having demonstrated a Rad5-like fork regression activity of HLTF, our next goal was to explore its mechanistic basis. The lack of fork dissolution activity and the inability of HLTF to process partial heteroduplex DNA substrates (see Fig. S2 in the supplemental material) suggest that instead of utilizing single-stranded DNA translocase activity like canonical helicases, HLTF may exploit a different mechanism during fork reversal, possibly translocating along double-stranded DNA (dsDNA). In order to test this possibility, we checked the activity of HLTF on a partial triple-stranded DNA made by annealing a third strand to the middle of a dsDNA by Hoogsteen bonding (29). Strikingly, HLTF was able to displace the third strand of this partial triple helix in an ATP-dependent manner (Fig. 4A, panel I). Removal of the double-stranded flanking arms of the partial triple helix abolished the displacement of the triple-helix-forming olignucleotide (Fig. 4A, panel II). Thus, we conclude that HLTF is a dsDNA translocase.
FIG. 4.
HLTF tracks along double-stranded DNA. (A) HLTF displaces radioactively labeled triplex-forming oligonucleotide (TFO) from partial triple-stranded DNA in an ATP hydrolysis-dependent manner. In the lanes indicated by mut DE, the ATPase mutant HLTF DE557,558AA was substituted for the wild-type HLTF, while in the lanes labeled AMP-PNP, ATP was replaced by AMP-PNP. We note that in the control reaction using isolated (blunted) triple helix, the TFO displacement was severely impaired, indicating that HLTF has to be loaded onto the dsDNA flanking the triplex forming region. (B) Triplex displacement activity of HLTF depends on the integrity of the double-stranded arm. HLTF at increasing concentration was incubated with intact (I), 3′-strand gapped (II), and 5′-strand gapped (III) DNA substrates which are represented schematically. We note that a 6-nucleotide-long gap on either the 3′ or the 5′ strand inhibited HLTF translocation and consequent TFO displacement. (C) Kinetics of triplex displacement by HLTF. The rate of displacement in the presence of 20 nM HLTF was examined at time points between 0 and 30 min (0, 0.5, 1.0, 1.5, 2.0, 4.0, 8.0, 12.0, 16.0, 20.0, 24.0, and 30.0 min) on intact (I), 3′-strand gapped (II), and 5′-strand gapped (III) DNA substrates as represented schematically. Note that some displacement occurred on the substrate with a gap on the 5′ strand but the 3′ gapped substrate was left unprocessed. These properties could classify HLTF as a 3′-5′ dsDNA translocase.
The known dsDNA translocases possess 3′-5′ polarity, and they cannot pass through gaps on the tracking strand but bypass small gaps on the other strand during translocation (29). We found that triple-helix displacement by HLTF is severely abolished by introducing gaps on either strand of the dsDNA flanking the triple helix (Fig. 4B). HLTF has some ability to translocate through a 6-nt-long gap on the 5′ strand but not through a 6-nt-long gap on the 3′ strand of the protruding double-stranded arm, which shows that HLTF exhibits some 3′-5′ polarity during translocation (Fig. 4B); however, kinetic analysis provided strong support that the preferred substrate for HLTF translocation is continuous intact dsDNA (Fig. 4C). This enzymatic property distinguishes HLTF from “canonical” dsDNA translocases.
HLTF has a role in replication of damaged DNA.
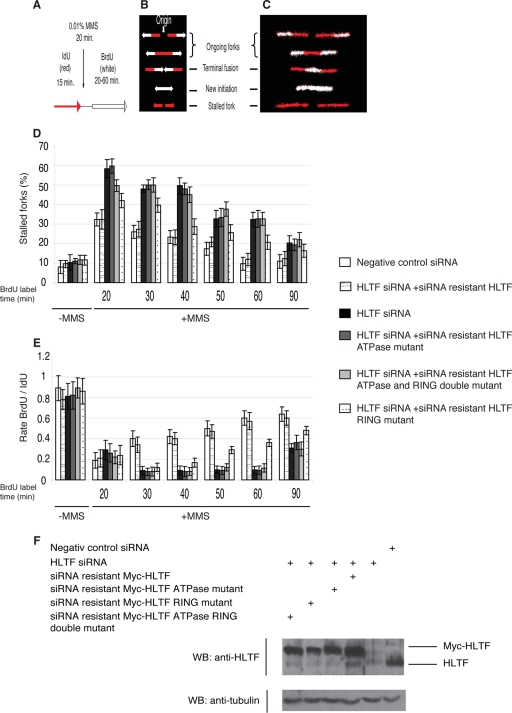
Our in vitro findings prompted us to investigate whether HLTF promotes the replication of damaged DNA in vivo and whether its ATPase domain has a role in this function. We examined the effect of knocking down the expression of HLTF by siRNA (Fig. 5F) on the progress of replication of damaged DNA using the chromosomal fiber technique (14). To track individual replication forks on damaged DNA, human cells were pulse-labeled with the nucleoside analog iododeoxyuridine (IdU), followed by treatment with the alkylating agent methyl methanesulfonate (MMS) before a second pulse-labeling with bromodeoxyuridine (BrdU) (Fig. 5A). The periods of DNA synthesis before and after MMS treatment are marked by the incorporation of different halogenated nucleotides, IdU (red) and BrdU (white), respectively. The newly synthesized DNA that has incorporated these bases can be visualized by fluorescence microscopy of prepared DNA fibers stained with specific antibodies against these halogenated nucleotides. Based on the length of individual labeled tracks and the labeling pattern, the rate of replication fork movement before and after DNA damage can be tracked as well as stalled and moving forks can be clearly distinguished (Fig. 5B and C; see Fig. S3 in the supplemental material).
FIG. 5.
HLTF is required for efficient replication fork restart on damaged DNA. (A) Protocol for defining sites and speeds of replication fork before and after treatment by MMS. (B) Schematic representation of possible fiber-labeling patterns. Arrows indicate the direction of fork movements. (C) Images of various types of labeled chromosome fibers. We note that ongoing forks are dually labeled by IdU and BrdU while forks that have stalled upon encountering a DNA lesion during MMS treatment are labeled only by IdU. (D) Effects of HLTF knockdown on MMS-induced stalling of replication forks in siRNA-transfected HeLa cell lines. For HLTF knockdown, HeLa cells were transfected with negative control or HLTF-specific siRNA. For complementation, the HLTF siRNA knocked-down cells were subsequently transfected by siRNA-resistant wild-type, ATPase mutant, RING mutant, or ATPase and RING double-mutant HLTF-expressing plasmids. The percentage of stalled forks was calculated on the basis of the sum of stalled and ongoing forks being 100%. (E) Effect of MMS treatment on replication movement. Apparent average fork rates were calculated by dividing the length of labeled fiber with the labeling time for both halogenated nucleotides. The ratio of apparent average fork rates after and before MMS treatment was followed in a 20- to 90-min period following MMS treatment. Data in panels D and E are means of three independent determinations, in which at least 100 fiber measurements were performed. Error bars represent standard deviations. (F) Efficacy of HLTF knockdown by siRNA and HLTF expression. Total cell extracts were prepared from aliquots of samples used for preparing DNA fibers. The efficiency of HLTF knockdown and expression of wild-type, ATPase, RING, and RING/ATPase double-mutant 6MYC-HLTF proteins were confirmed by Western blotting using anti-HLTF antibody. As a loading control, antitubulin antibody was used.
First, we determined the effect of MMS treatment and HLTF knockdown on inhibiting the progress of replication by calculating the percentage of stalled forks. As expected, MMS treatment resulted in the stalling of replication forks, which recovered gradually over a 20- to 90-min period in wild-type cells (Fig. 5D). Strikingly, we found that the recovery of the progression of replication forks was impaired in cells in which the HLTF level was depleted compared to control cells (Fig. 5D). For example, 60 min after MMS treatment, in control cells the forks had recovered the ability to continue synthesis, indicated by the fact that the percentage of stalled forks was similar to the level for the untreated control (∼10%), whereas in HLTF-knocked-down cells 3-fold more stalled forks could be detected (∼30%) (Fig. 5D). With an siRNA-resistant wild-type HLTF cDNA we were able to rescue this defect, which rules out an off-target effect of siRNA toward an unrelated protein. Importantly, however, an siRNA-resistant DE557,558AA ATPase mutant HLTF cDNA could not complement the impairment of resumption of replication on damaged DNA in HLTF knockdown cells, indicating that the ATPase domain of HLTF is indispensable for this function. We also tested if the C759S RING mutant and the RING-ATPase double-mutant HLTF show replication fork restart impairment. We found that the RING mutant HLTF also showed a defect in fork restart, though it appeared less severe than that of the ATPase mutant. Importantly, however, the level of the defect detected for the RING-ATPase double-mutant HLTF did not exceed the level found in the single ATPase mutant (Fig. 5D). These results imply that both the ATPase and the RING domain-mediated ubiquitin ligase activities support the same function of HLTF in promoting replication through damaged DNA. To show the effect of HLTF knockdown on replication forks that were not permanently blocked and resumed during the second pulse-labeling, we calculated the ratio of average fork rates in the first (IdU) and second (BrdU) labeling periods. Dividing the length of the labeled tracks by the time of the labeling period results in a measure of apparent average fork rate, which reflects the actual fork speed as well as fork stalling. We monitored the ratio of apparent average fork rates after and before MMS treatment at various times (20 to 90 min) following MMS treatment. This analysis revealed that the progression of replication forks on damaged DNA is slowed down to a much greater extent in HLTF-knocked-down cells than in control cells (Fig. 5E). For example, after 40 min of MMS treatment the apparent average rate of forks in HLTF-knocked-down cells was slowed down by about 9-fold, whereas in control cells the fork rate was only 2-fold slower. While the siRNA-resistant wild-type HLTF complemented this phenotype, the ATPase, the RING, and the double-mutant HLTF failed to do so. At 90 min after MMS treatment, the replication rate appeared to be almost recovered in the control cells. In the absence of HLTF, recovery also occurred, although it was still delayed and was not rescued by the mutants (Fig. 5E).
Altogether, these results indicate that HLTF strongly affects the resumption of DNA synthesis at replication forks stalled at DNA lesion sites and that both the ATPase and the RING domains of HLTF have a critical role in this function.
DISCUSSION
Here we show that HLTF exhibits an ATP-dependent double-stranded DNA translocase activity and that it can carry out replication fork reversal by concerted unwinding of the leading and lagging strand arms of the fork and then annealing together the nascent strands and the parental strands, the same biochemical activity that Rad5 possesses (2). We also show using a chromosomal fiber technique that restart of replication forks blocked at DNA lesions becomes impaired in the absence of HLTF. Moreover, we provide evidence that mutational inactivation of the ATPase domain of HLTF impairs its fork reversal activity as well as the movement of replication forks on damaged DNA. These results, combined with the high degree of structural homology between HLTF and yeast Rad5, and also with the functional homology as ubiquitin ligases in Mms2-Ubc13-Rad6-Rad18-dependent PCNA polyubiquitylation (37), support that HLTF plays a yeast Rad5-like role in replication of damaged DNA.
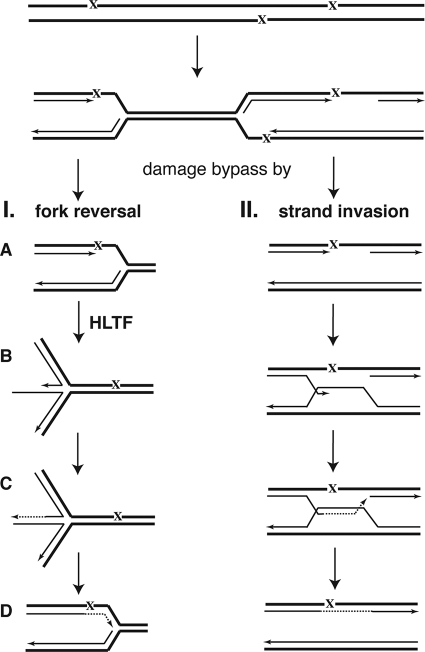
Replication fork reversal has been suggested to promote replication restart through several mechanisms. Remodeling of a replication fork stalled at a DNA lesion can facilitate the displacement of the replicative helicase and polymerase from the junction point, thereby making it accessible for repair or lesion bypass enzymes, or it can promote the recombination-dependent restart of replication (18, 34). After fork reversal the lesion could be relocated to its original double-stranded context, where it could be removed by nucleotide excision repair. Alternatively, fork reversal could promote template switching, where instead of the damaged strand the newly synthesized strand of the sister chromatid can be used as a template for replication. Although all these mechanisms can rescue the stalled replication fork, as for HLTF action we regard template switching as the most likely possibility. Considering that HLTF is a yeast Rad5 ortholog, our suggestion is supported by yeast genetic data showing that neither nucleotide excision repair enzymes nor translesion synthesis polymerases play a role in the Rad5-dependent pathway (36) and that Rad5 and Rad52 promote two alternate pathways (6). We suggest that in the context of a replication fork HLTF translocates along the parental dsDNA and remodels the nascent strands which are obstacles for further translocation. Our results predict that HLTF translocation for fork reversal can occur if and only if the parental strands at the junction point become reannealed. The suggested mechanism for HLTF not only reveals the mechanism of concerted fork reversal but clarifies the reason for the lack of any effect of ssDNA binding proteins on the reaction and also explains why HLTF cannot invade sequence heterology to act as a canonical DNA helicase. Processing stalled replication forks in a manner resembling the action of canonical helicases has been shown to promote template switching in trans that occurs between similar but not identical sequences, promoting thereby gross chromosomal rearrangements (16, 25). While the enzymatic basis of transtemplate switching is unknown, the requirement of DNA helix integrity for HLTF translocation could provide a means of eliminating such a risk. The properties of HLTF action can ensure the integrity of the DNA during fork reversal and are obligatory for a high-fidelity damage bypass by a template-switching mechanism in cis, as we propose in our model (Fig. 6).
FIG. 6.
Model for template switch damage bypass by HLTF-dependent fork reversal and strand invasion. Stalling of replication at an unrepaired DNA lesion (X) can lead to unresolved fork structures as well as gaps in the newly synthesized daughter strand opposite the lesion. Unresolved fork structures can particularly be formed if replication cannot be reinitiated downstream of a fork-blocking lesion and cryptic origins of replication cannot be activated at intradamage DNA fragments. The filling in of gaps and resolving fork structures can require postreplicative repair mechanisms. (I) Damage bypass by HLTF-dependent fork reversal. (A) When the lesion is located on the leading strand template, the leading and lagging strand synthesis can become uncoupled, and synthesis on the lagging strand can continue way past the blocked nascent leading strand. (B) The nascent strands then unwind from their respective templates and anneal with one another, and the parental strands also reanneal, which can be catalyzed by HLTF. The overall outcome of these reactions is the regression of the replication fork to form a four-way Holliday junction. (C) Following that, the sequences complementary to the damaged region are synthesized on the nascent leading strand using the nascent lagging strand as the template. (D) The regressed fork is then reversed, and synthesis resumes beyond the point of the lesion. (II) Damage bypass by strand invasion. The 3′ end of the newly synthesized gapped DNA invades the homologous region of the sister chromatid and uses its newly synthesized strand as a template for DNA synthesis.
Our in vivo results show that stalled replication can be rescued even in the absence of HLTF, although recovery of DNA synthesis is far slower compared to that of control cells. This indicates that in the absence of HLTF other pathways, like translesion synthesis or recombination, become operative in rescuing stalled replication forks, or it can denote functional redundancy. We note that similar to HLTF, human SHPRH, another potential tumor suppressor, also has a Rad5-like domain structure with a C3HC4 RING domain embedded into a SWI/SNF2 helicase motif. SHPRH can interact with Rad6-Rad18 and facilitate Mms2-Ubc13-dependent PCNA polyubiquitylation (24, 38). Although HLTF more closely resembles Rad5 structurally, it would be interesting to investigate if SHPRH can also carry out fork reversal and promote replication of damaged DNA.
Recently, RecQ helicases and FANCM protein have also been shown to have a biochemical activity for replication fork reversal (7, 8, 15, 28, 34). While yeast genetic data have provided strong support for the role of Rad5 in promoting error-free damage bypass via template switching, Sgs1, the only Saccharomyces cerevisiae RecQ helicase, has been found to have a role in suppressing crossovers during double-strand break repair (13). The MPH1 gene, a relative of FANCM in Saccharomyces cerevisiae, functions in an error-free pathway of DNA damage repair/tolerance that involves homologous recombination in which it dissociates Rad51-made D-loops (27, 30). In fission yeast, double deletion of FML1 and RAD8, the FANCM and HLTF orthologs, respectively, causes an increase in MMS and UV sensitivity relative to single mutants, which may indicate that they promote alternative ways of damage tolerance (34). Indeed, it has recently been suggested that FANCM may antagonize the movement of replication forks advancing toward sites of damage, which might result in fork reversal once the replication fork is stalled. Remodeling of forks by FANCM would stabilize the stalled replication fork and provide time and space for the lesion site to be repaired (7, 8). Both HLTF and FANCM are dsDNA translocases; thus, the underlying mechanisms of their fork processing activity may be similar (21). Interestingly, even though RecG helicase, the prokaryotic fork reversal enzyme, has also been suggested to utilize double-stranded DNA translocase activity for fork reversal, in contrast to HLTF, it can also catalyze the removal of nascent strands from heterologous model forks (19, 32). Likewise, the fork-processing activity of RecQ helicases is mechanistically distinguishable from that of HLTF. RecQ helicases exhibit single-stranded DNA translocase activity, and in contrast to HLTF, they can also dissolve heterologous model forks. In fact, during the action of RecQ helicases on replication fork-like substrates, single-stranded products also appear and they become annealed into double-stranded products only later, which is quite distinct from HLTF-catalyzed concerted fork reversal (15).
The rate of gross chromosomal rearrangements in yeasts increases as much as 200-fold in the absence of Rad5 (33). That could result if in the absence of coordinated fork reversal, aberrant DNA structures are formed and resolved at the price of genomic stability. As increased genomic instability and consequent complex genomic rearrangements are hallmarks of malignantly transformed cells in higher eukaryotes, we note that our data suggesting a role for HLTF in promoting error-free damage bypass by template switching are in concert with its previously proposed tumor suppressor function (22).
Supplementary Material
Acknowledgments
We thank Katalin Kovács and Ildikó Kravjar for the technical assistance, Jung-Hoon Yoon for optimizing siRNA knockdown of HLTF, Barnabas Szakal for helping in siRNA experiments and generating the GST-FLAG-HLTF-overexpressing plasmid, and Dean A. Jackson for sharing his expertise on fiber assays.
This work was supported by the Howard Hughes Medical Institute grant 55005612, the Wellcome Trust International Senior Research Fellowship, Hungarian Science Foundation grants OTKA 77495 and TÁMOP-4.2.2/08/1, the János Bolyai Research Scholarship of the Hungarian Academy of Sciences, the Hungarian National Office for Research and Technology grant KFKT-1-2006-0010, and the FEBS Collaborative Experimental Scholarships for Central & Eastern Europe.
We declare no competing financial interests.
Footnotes
Published ahead of print on 30 November 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bielas, J. H., K. R. Loeb, B. P. Rubin, L. D. True, and L. A. Loeb. 2006. Human cancers express a mutator phenotype. Proc. Natl. Acad. Sci. U. S. A. 103:18238-18242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blastyak, A., L. Pinter, I. Unk, L. Prakash, S. Prakash, and L. Haracska. 2007. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell 28:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cordeiro-Stone, M., A. M. Makhov, L. S. Zaritskaya, and J. D. Griffith. 1999. Analysis of DNA replication forks encountering a pyrimidine dimer in the template to the leading strand. J. Mol. Biol. 289:1207-1218. [DOI] [PubMed] [Google Scholar]
- 4.Fishel, R., M. K. Lescoe, M. R. Rao, N. G. Copeland, N. A. Jenkins, J. Garber, M. Kane, and R. Kolodner. 1993. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell 75:1027-1038. [DOI] [PubMed] [Google Scholar]
- 5.Gangavarapu, V., L. Haracska, I. Unk, R. E. Johnson, S. Prakash, and L. Prakash. 2006. Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:7783-7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gangavarapu, V., S. Prakash, and L. Prakash. 2007. Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 27:7758-7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gari, K., C. Decaillet, M. Delannoy, L. Wu, and A. Constantinou. 2008. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc. Natl. Acad. Sci. U. S. A. 105:16107-16112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gari, K., C. Decaillet, A. Z. Stasiak, A. Stasiak, and A. Constantinou. 2008. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell 29:141-148. [DOI] [PubMed] [Google Scholar]
- 9.Grady, W. M., and S. Markowitz. 2000. Genomic instability and colorectal cancer. Curr. Opin. Gastroenterol. 16:62-67. [DOI] [PubMed] [Google Scholar]
- 10.Haracska, L., C. A. Torres-Ramos, R. E. Johnson, S. Prakash, and L. Prakash. 2004. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:4267-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins, N. P., K. Kato, and B. Strauss. 1976. A model for replication repair in mammalian cells. J. Mol. Biol. 101:417-425. [DOI] [PubMed] [Google Scholar]
- 12.Hoege, C., B. Pfander, G. L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135-141. [DOI] [PubMed] [Google Scholar]
- 13.Ira, G., A. Malkova, G. Liberi, M. Foiani, and J. E. Haber. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson, D. A., and A. Pombo. 1998. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol. 140:1285-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanagaraj, R., N. Saydam, P. L. Garcia, L. Zheng, and P. Janscak. 2006. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 34:5217-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, J. A., C. M. Carvalho, and J. R. Lupski. 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131:1235-1247. [DOI] [PubMed] [Google Scholar]
- 17.Lee, K. Y., and K. Myung. 2008. PCNA modifications for regulation of post-replication repair pathways. Mol. Cells 26:5-11. [PMC free article] [PubMed] [Google Scholar]
- 18.McGlynn, P., and R. G. Lloyd. 2002. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 3:859-870. [DOI] [PubMed] [Google Scholar]
- 19.McGlynn, P., and R. G. Lloyd. 2001. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl. Acad. Sci. U. S. A. 98:8227-8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGlynn, P., A. A. Mahdi, and R. G. Lloyd. 2000. Characterisation of the catalytically active form of RecG helicase. Nucleic Acids Res. 28:2324-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meetei, A. R., A. L. Medhurst, C. Ling, Y. Xue, T. R. Singh, P. Bier, J. Steltenpool, S. Stone, I. Dokal, C. G. Mathew, M. Hoatlin, H. Joenje, J. P. de Winter, and W. Wang. 2005. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 37:958-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moinova, H. R., W. D. Chen, L. Shen, D. Smiraglia, J. Olechnowicz, L. Ravi, L. Kasturi, L. Myeroff, C. Plass, R. Parsons, J. Minna, J. K. Willson, S. B. Green, J. P. Issa, and S. D. Markowitz. 2002. HLTF gene silencing in human colon cancer. Proc. Natl. Acad. Sci. U. S. A. 99:4562-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Motegi, A., H. J. Liaw, K. Y. Lee, H. P. Roest, A. Maas, X. Wu, H. Moinova, S. D. Markowitz, H. Ding, J. H. Hoeijmakers, and K. Myung. 2008. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc. Natl. Acad. Sci. U. S. A. 105:12411-12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Motegi, A., R. Sood, H. Moinova, S. D. Markowitz, P. P. Liu, and K. Myung. 2006. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J. Cell Biol. 175:703-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Payen, C., R. Koszul, B. Dujon, and G. Fischer. 2008. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 4:e1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prakash, L. 1981. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. 184:471-478. [DOI] [PubMed] [Google Scholar]
- 27.Prakash, R., D. Satory, E. Dray, A. Papusha, J. Scheller, W. Kramer, L. Krejci, H. Klein, J. E. Haber, P. Sung, and G. Ira. 2009. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 23:67-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ralf, C., I. D. Hickson, and L. Wu. 2006. The Bloom's syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 281:22839-22846. [DOI] [PubMed] [Google Scholar]
- 29.Saha, A., J. Wittmeyer, and B. R. Cairns. 2002. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 16:2120-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schurer, K. A., C. Rudolph, H. D. Ulrich, and W. Kramer. 2004. Yeast MPH1 gene functions in an error-free DNA damage bypass pathway that requires genes from homologous recombination, but not from postreplicative repair. Genetics 166:1673-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheridan, P. L., M. Schorpp, M. L. Voz, and K. A. Jones. 1995. Cloning of an SNF2/SWI2-related protein that binds specifically to the SPH motifs of the SV40 enhancer and to the HIV-1 promoter. J. Biol. Chem. 270:4575-4587. [DOI] [PubMed] [Google Scholar]
- 32.Singleton, M. R., S. Scaife, and D. B. Wigley. 2001. Structural analysis of DNA replication fork reversal by RecG. Cell 107:79-89. [DOI] [PubMed] [Google Scholar]
- 33.Smith, S., J. Y. Hwang, S. Banerjee, A. Majeed, A. Gupta, and K. Myung. 2004. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 101:9039-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun, W., S. Nandi, F. Osman, J. S. Ahn, J. Jakovleska, A. Lorenz, and M. C. Whitby. 2008. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol. Cell 32:118-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svoboda, D. L., and J. M. Vos. 1995. Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: fork uncoupling or gap formation. Proc. Natl. Acad. Sci. U. S. A. 92:11975-11979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torres-Ramos, C. A., S. Prakash, and L. Prakash. 2002. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:2419-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Unk, I., I. Hajdu, K. Fatyol, J. Hurwitz, J. H. Yoon, L. Prakash, S. Prakash, and L. Haracska. 2008. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc. Natl. Acad. Sci. U. S. A. 105:3768-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unk, I., I. Hajdu, K. Fatyol, B. Szakal, A. Blastyak, V. Bermudez, J. Hurwitz, L. Prakash, S. Prakash, and L. Haracska. 2006. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc. Natl. Acad. Sci. U. S. A. 103:18107-18112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veigl, M. L., L. Kasturi, J. Olechnowicz, A. H. Ma, J. D. Lutterbaugh, S. Periyasamy, G. M. Li, J. Drummond, P. L. Modrich, W. D. Sedwick, and S. D. Markowitz. 1998. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc. Natl. Acad. Sci. U. S. A. 95:8698-8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wood, L. D., D. W. Parsons, S. Jones, J. Lin, T. Sjoblom, R. J. Leary, D. Shen, S. M. Boca, T. Barber, J. Ptak, N. Silliman, S. Szabo, Z. Dezso, V. Ustyanksky, T. Nikolskaya, Y. Nikolsky, R. Karchin, P. A. Wilson, J. S. Kaminker, Z. Zhang, R. Croshaw, J. Willis, D. Dawson, M. Shipitsin, J. K. Willson, S. Sukumar, K. Polyak, B. H. Park, C. L. Pethiyagoda, P. V. Pant, D. G. Ballinger, A. B. Sparks, J. Hartigan, D. R. Smith, E. Suh, N. Papadopoulos, P. Buckhaults, S. D. Markowitz, G. Parmigiani, K. W. Kinzler, V. E. Velculescu, and B. Vogelstein. 2007. The genomic landscapes of human breast and colorectal cancers. Science 318:1108-1113. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, H., and C. W. Lawrence. 2005. The error-free component of the RAD6/RAD18 DNA damage tolerance pathway of budding yeast employs sister-strand recombination. Proc. Natl. Acad. Sci. U. S. A. 102:15954-15959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.