Abstract
The trithorax (trxG) and Polycomb (PcG) group proteins recognize and propagate inheritable patterns of gene expression through a poorly understood epigenetic mechanism. A distinguishing feature of these proteins is the presence of a 130-amino-acid methyltransferase domain (SET), which catalyzes the methylation of histones. It is still not clear how SET proteins distinguish gene expression states, how they are targeted, or what regulates their substrate specificity. Many SET domain-containing proteins show robust activity on core histones but relatively weak activity on intact nucleosomes, their physiological substrate. Here, we examined the binding of two SET domain-containing proteins, ALL1 and SET7, to chromatin substrates. The SET domains from these proteins bind and methylate intact nucleosomes poorly but can recognize disrupted nucleosomal structures associated with transcribed chromatin. Interestingly, the remodeling of dinucleosomes by the ISWI class of ATP-dependent chromatin remodeling enzymes stimulated the binding of SET domains to chromatin and the methylation of H3 within the nucleosome. Unexpectedly, dinucleosomes remodeled by SWI/SNF were poor substrates. Thus, SET domains can distinguish nucleosomes altered by these two classes of remodeling enzymes. Our study reveals novel insights into the mechanism of how SET domains recognize different chromatin states and specify histone methylation at active loci.
The Polycomb (PcG) and trithorax (trxG) group proteins are evolutionarily conserved factors that maintain stable and inheritable states of gene expression (4). PcG proteins repress homeotic and other genes during development, while trxG proteins maintain active gene states (4, 36). Certain members of the trithorax and Polycomb group proteins contain a 130-amino-acid motif called the SET domain, which has histone methyltransferase (HMT) activity (34). In addition to the core 130-amino-acid HMT region, many SET domain proteins contain less-conserved 50- to 80-amino-acid pre- and post-SET regions, which are often included in the definition of the functional SET domain (34). All of the functions of the pre- and post-SET domains are not known; however, we found evidence they play a role in recognizing “open” nucleosome intermediates and single-stranded DNA or RNA structures in transcribed and supercoiled DNA (23). In principle, this may underlie the ability of SET domains to discriminate existing active and silent gene states.
SET domain proteins maintain gene activity by methylating chromatin to create “codes” of active and repressed gene states (17, 20, 34). For example, methylation of lysine 4 of histone H3 is associated with gene activation, while methylation of lysine 9 or 27 is associated with silencing (20, 34, 39). Although it has been shown that the SET domain of Drosophila melanogaster trithorax can bind and methylate the amino-terminal tail of histone H3, the H3 tail is inaccessible to the enzyme in intact nucleosomes (19). Furthermore, a feature of many SET domain HMTs is that they display robust activity on histones but relatively poor activity on intact nucleosomes (48). This has led to the speculation that SET domain proteins recognize chromatin that has been altered or remodeled by transcription or ATP-dependent chromatin remodeling complexes. However, this has not been shown directly.
Chromatin structure is regulated by the cooperation between histone-modifying enzymes and ATP-dependent chromatin remodeling factors (for a review see reference 31). There are multiple families of ATP-dependent chromatin remodeling enzyme complexes, which are classified by the structure and sequence similarity in the catalytic subunit (8, 13). The two best-characterized of these families are the SWI/SNF and ISWI classes. While the details on the mechanism of remodeling by these two groups are still being debated, it is generally accepted that the former remodels chromatin by disrupting the nucleosome and the latter slides nucleosomes while maintaining their canonical structure (8, 9, 13). Many ATP-dependent chromatin remodeling complexes contain subunits that recognize modified histone tails (3, 8, 10, 14-16, 41). There is ample evidence that histone modifications help recruit remodelers to genes through histone tail binding domains within subunits of the complex. For example, the bromodomain of Swi2/Snf2 anchors the SWI/SNF complex to acetylated chromatin templates in vitro and participates in its recruitment to genes in vivo (14, 15). Histone acetylation recruits RSC to templates in vitro and enhances it ability to stimulate RNA polymerase II (RNAPII) transcription through a nucleosome (6). The PhD finger in NURF specifically recognizes histone H3 trimethylated on lysine 4, and loss of this histone mark impaired NURF recruitment and function in vivo (49). While it has been shown that histone modifications can recruit and affect the activity of ATP-dependent chromatin remodeling enzymes, it is less clear if remodeling of nucleosomes by these factors directly enhances the activity of histone-modifying enzymes. Examples suggesting such regulation have been described in vivo; however, these effects cannot be attributed directly to changes in nucleosome structure.
Here we examined the recognition of nucleosomes by SET domain proteins and found that disruption of the canonical nucleosome structure by transcription or remodeling is required for binding and modification of the H3 tail. Furthermore, we examined the interaction of two SET proteins, ALL1 and Set7/9, with nucleosomes remodeled by distinct classes of chromatin remodeling complexes in a well-defined system. Interestingly, SET domains can bind and methylate nucleosomes remodeled by the ISWI class of chromatin remodeling enzymes, but surprisingly, those remodeled by SWI/SNF were not recognized or modified. Therefore, we have identified the requirements for SET domain interactions with chromatin and demonstrate a functional difference in the product of the remodeling reaction of these two different classes of ATP-dependent chromatin remodeling complexes.
MATERIALS AND METHODS
Construction of DNA templates.
DNA templates for in vitro assembly of mononucleosomes were constructed by cloning a NlaIII-NotI fragment containing the 147-bp minimal 601 positioning sequence (42) into the NotI-XbaI sites of pBluescript II(+), generating plasmid pBS601. Mononucleosome-length DNA templates containing variable amounts of linker DNA were generated from this construct by digesting with pairs of restriction endonucleases flanking the insertion sites. For dinucleosomal DNA templates, two minimal 601 fragments were cloned into different sites within the polylinker of pBluescript II(+) and liberated using appropriate pairs of restriction endonucleases (usually SacI/SacII and EcoRI/HindIII). DNA templates were end-labeled with [32P]dATP and Klenow fragment of DNA polymerase I. The “nucleosome B” template contained sequences −221 to +1 from the mouse mammary tumor virus promoter (46) and was cloned into the HindIII site of pBluescript II(+).
Histone isolation and chromatin reconstitution.
H1-depleted nucleosomes were prepared by the fractionation of micrococcal nuclease-digested chicken erythrocyte nuclei on CM-25 Sephadex columns (GE Healthcare) (21). Thiol-reactive HeLa nucleosomes were isolated by mercury affinity chromatography on Bio-Gel 501 (Bio-Rad) (1). Nucleosomes were formed by reconstituting purified histone octamers on to DNA by serial dilution as described in a previous publication, except that glycerol was omitted from the buffers (40). Reconstituted nucleosomes were analyzed on a 5 to 5.5% native polyacrylamide gel at a 50:1 acrylamide-to-bisacrylamide ratio and stained with ethidium bromide. In some cases, nucleosomes were purified on preparative native polyacrylamide gels.
Protein purification and GST pulldown assays.
Sequences corresponding to the SET domains, and mutant derivatives, of HMTs were amplified by PCR and cloned into the Ndel-EcoRI sites of the plasmid pGEX-2TKN (23). Recombinant glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli and purified by immobilization on glutathione-Sepharose (GE Healthcare) (23). The GST pulldown experiments were performed as described previously (23). The remodeling complexes were purified from Saccharomyces cerevisiae by affinity purification. Isw2 was purified through a triple FLAG tag from strain YTT966 (18, 43). SWI/SNF (Swi2-TAP), Isw1a (Ioc3-TAP), and Isw1b (Ioc2-TAP) were purified by the tandem affinity purification (TAP) method as described elsewhere (43, 45), except that the proteins were eluted from the column in a buffer containing 0.4 M salt. Quantities and purities of the remodelers were examined by silver staining of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. Amounts of remodeler used in each assay were based on the weight (mass) of the complex. The ATPase activity of the remodeling complexes was assayed in a 5-μl reaction mixture containing 0.1 μg of mono- or dinucleosomal template (or buffer only, as a control), 0.2 ng of ISWI complexes or 0.5 ng of SWI/SNF, 0.35 μCi of [γ-32P]ATP, and 0.1 mM cold ATP. These are approximately the same concentrations of remodeler and nucleosomes used in the remodeling assays (see below). The reactions were terminated by spotting 1 μl onto polyethyleneimine-cellulose F plates (EMD Chemicals) and resolved in a buffer containing 0.15 M LiCl-0.15 M formic acid. The plates were exposed to a phosphorimager screen and quantified on a Typhoon scanner (GE/Molecular Dynamics).
Histone methylation and remodeling assays.
Methylation of histones or nucleosomes was performed at room temperature for 1 to 2 h as described in a previous publication (48) in a buffer containing 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 2.5 mM MgCl2, 2.5 mM dithiothreitol, 0.05% NP-40, 50 μg/ml bovine serum albumin (fraction V; Sigma), 0.2 mM phenylmethylsulfonyl fluoride. Each reaction mixture contained 10 μg of histones (free or assembled into nucleosomes), 0.5 μg of GST-SET7 or 5 μg of ALL1 GST-SET polypeptide (amino acids 3745 to 3969), and of 0.5 μCi of S-[3H]adenosyl methionine ([3H]SAM; 5 to 15 Ci/mmol, 0.5 μCi/μl; Perkin-Elmer) per 30 μl of reaction mixture. Methylation reactions were stopped by the addition of SDS-PAGE sample buffer. If required, histones or nucleosomes were precipitated with 25% trichloroacetic acid or concentrated using Microcon 10K concentrators before loading onto SDS-PAGE or native gels. Gels were stained with Coomassie blue, destained, treated with En3Hance (Perkin-Elmer) or PPO-POPOP solutions (Sigma), dried, and exposed to X-ray film.
Nucleosome remodeling was carried out at room temperature for 1.5 h in nucleosome assembly buffer (15, 16, 29) supplemented with 1 mM ATP and an additional 2.5 mM MgCl2. The final concentrations of all components were 20 mM Tris-HCl (pH 7.6), 100 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 2.5 mM dithiothreitol, 0.05% NP-40, 50 μg/ml bovine serum albumin (fraction V; Sigma), 0.2 mM phenylmethylsulfonyl fluoride, and 1 mM ATP. The remodeling reactions were carried out in 50-μl volumes and contained 1.25 μg of nucleosomes and 2.5 ng of ISWI complexes or 5 ng of Swi-Snf complexes. Remodeling was terminated by the addition of 25 to 50 mU of apyrase (New England Biolabs) per reaction mixture for 20 min at room temperature. The remodeled products were analyzed by native gel electrophoresis or by probing nucleosome structures with micrococcal nuclease, DNase I, or restriction endonuclease (Worthington Biochemical and New England Biolabs) (15, 16, 29). For DNase I and MNase digestion assays, the reaction volume was scaled up to 100 μl, and 20-μl aliquots were digested with increasing amounts of nuclease. For restriction endonuclease (RE) accessibility assays, nucleosomes were remodeled by Swi/Snf for 40 min at room temperature and remodeling was terminated by apyrase treatment as described above. The remodeled products were digested with 5 U of RE enzyme for 30 min at room temperature. The DNA was purified by proteinase K digestion and phenol chloroform extraction and separated on ethidium bromide-containing gels.
RESULTS
The SET domain binds “unfolded” nucleosome intermediates.
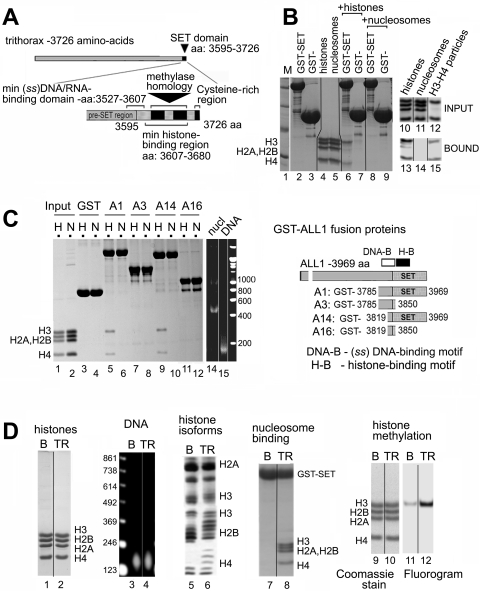
We studied the interaction between two prototypical SET domains and purified core histones (22), H1-depleted nucleosomes (21), and H3/H4 tetrasome particles (21). Immobilized GST-tagged polypeptides containing segments of trithorax or ALL1 were used in pulldown assays, and the bound material was examined on agarose- or SDS-PAGE gels, as indicated in the figure legends (Fig. 1A and B). It has been shown that the SET domain of trithorax binds tightly to the N-terminal tail of histone H3, and this binding can be modulated by the covalent modifications of histone tails (19). In our assays, GST-trithorax (Fig. 1B, lane 6) and GST-ALL1 (Fig. 1C, lane 5) interacted with core histones, pulling down predominantly histones H3 and H4. It is expected that H3 and H4 exist in tetramer form under the physiological ionic conditions used in the binding assay, which would explain the presence of both histones in the bound material. In contrast, the SET domains of trithorax and ALL1 bound very poorly to intact, canonical nucleosomes (Fig. 1B, lane 8, and C, lane 6). No histones or nucleosomes were retained on control GST polypeptides. Next, we examined the binding of GST-ALL1 polypeptides with deletions in different regions of the SET domain to histones and nucleosomes. The binding of the SET domain of ALL1 to histones was dependent upon a region previously described as a histone binding motif (H-B), and fragments of ALL1 lacking this region failed to bind to histones (Fig. 1C, compare lane 5 to lanes 7 and 11). The pre-SET domain that binds single-stranded nucleic acids (DNA-B) was dispensable for the interaction with histones, as the A14 construct retained its histone binding function (Fig. 1C, lane 9).
FIG. 1.
The SET domain of trithorax binds core histones and altered nucleosomal structures but not intact nucleosomes. (A) Schematic of the domain structure of trithorax. Positions of highly conserved blocks of homology with methyltransferases, a C-terminal cysteine-rich region, and positions of the histone- and single-stranded DNA binding motifs are labeled. (B, left) Binding of GST-trithorax to histones and nucleosomes. Immobilized SET domain of trithorax (GST-SET) or GST alone was incubated with histones (lanes 6 and 7) or nucleosomes (lanes 8 and 9), washed with 0.5 M NaCl wash buffer, and analyzed by SDS-PAGE. Input material is shown in lanes 4 and 5. (Right) GST-SET binding to nucleosomes and tetrasomes. Input and bound material are shown in lanes 10 to 12 and 13 to 15, respectively. (C) Map of the residues in the SET domain of ALL1 required for histone (H) and nuclosome (N) binding. Immobilized GST-ALL1 polypeptides (right) were assayed for binding to HeLa histones or nucleosomes as described for panel B. (D) Binding of GST-ALL1 to “unfolded” thiol-reactive nucleosomes isolated on Hg-agarose columns. Bulk (B) and thiol-reactive (TR) nucleosomes are indicated above each panel. The histone (lanes 1 and 2) and DNA (lanes 3 and 4) contents of the nucleosome fractions were analyzed by electrophoresis in SDS-PAGE or agarose gels, respectively. The degree of histone acetylation was analyzed in Triton-acid-urea gels (lanes 5 and 6). Binding of GST-ALL1 (construct A1 in panel C) to bulk and thiol-reactive nucleosomes was analyzed in pulldown assays (lanes 7 and 8). Bulk and thiol-reactive nucleosomes were incubated in solution with GST-ALL1 in the presence of [3H]SAM and then resolved by SDS-PAGE. Histone positions and amounts were determined by Coomassie blue staining (lanes 9 and 10), and methylation was detected by fluorography (lanes 11 and 12).
Since SET domains bound core histones, but not canonical nucleosomes, this suggested that the H3 tails are inaccessible in nucleosomes and that nucleosome remodeling or alteration is required for the recognition of the H3 tail. We first tested if removal of H2A/2B dimers from the nucleosome would stimulate SET domain binding. We formed “tetrasomes” by disassociating H2A/H2B dimers from nucleosomes using high salt. Under the same binding conditions, trithorax SET domain bound to the tetrasome nearly as well as to free histones (Fig. 1B, compare lanes 13 and 15). This suggests that the structure of intact nucleosomes is a major factor limiting the interaction between SET domains and the H3 tail and that disrupting this structure stimulates SET domain binding.
Removal of H2A/2B dimers can occur in vivo during transcription or from the actions of chromatin remodeling enzymes. Furthermore, it is a widely held belief that disruption of the nucleosome structure during transcription by RNAPII facilitates the methylation of chromatin within the open reading frames of active genes. The unfolding of nucleosomes during transcription uncovers a previously shielded cysteine residue in histone H3 located at the center of the nucleosome core (1), making it possible to separate active from inactive chromatin by chromatography on mercury affinity columns (7). The binding of GST-ALL1 to bulk nucleosomes and thiol-reactive species was compared. The amount of DNA and stoichiometry and amounts of the core histones were similar in the bulk and thio-reactive chromatin preparations (Fig. 1D, lanes 1 to 4). To confirm the efficacy of the Hg-agarose fractionation, we verified that the chromatin retained on the column was hyperacetylated (Fig. 1D, lane 5 versus 6), another hallmark of actively transcribed chromatin. Isolated thiol-reactive nucleosomes bound to the SET domain of ALL1 significantly better than bulk nucleosomes (Fig. 1D, lane 8 versus 7). Finally, the SET domain of ALL1 methylated these unfolded nucleosomes more efficiently than bulk nucleosomes (Fig. 1D, lane 11 versus 12). These initial studies suggest that the SET domain of histone methyltransferase enzymes selectively binds and modifies nucleosomes altered through the process of transcription and/or remodeling.
SET domains do not bind stably to remodeled mononucleosomes.
The studies described above suggest that disrupting the chromatin structure during the process of transcription enhances the binding of SET domains to chromatin. However, not all HMTs target and methylate histones at active genes, and some modify chromatin in regulatory regions and promoters that are not transcribed. Therefore, other mechanisms must exist to stimulate the HMT activity of SET domain proteins. We speculated that ATP-dependent chromatin remodeling complexes could fulfill such a role. There are several classes of ATP-dependent chromatin remodeling complexes (8, 13). Saccharomyces cerevisiae is commonly used as a source of remodeling complexes because it is easy to obtain large quantities of highly purified material. We focused on representatives of the imitation switch (ISWI) class, including Isw1a/Isw1b (43, 45) and Isw2 (9, 43), and the prototype of the Swi-Snf class, SWI/SNF (5, 33). The ISWI class modifies chromatin by sliding nucleosomes to new translational positions without disrupting the canonical structure of the nucleosome (35). The Swi-Snf class of enzymes can slide nucleosomes but also carries out other mechanisms of remodeling, such as loop formation, H2A/H2B dimer removal, and octamer transfer (12). Interestingly, work in yeast has shown that Isw1-containing complexes regulate the dynamics of H3 lysine 4 trimethylation (K4me3) during the induction of genes in vivo (28), and thus, they may regulate the activity of histone methyltransferase enzymes.
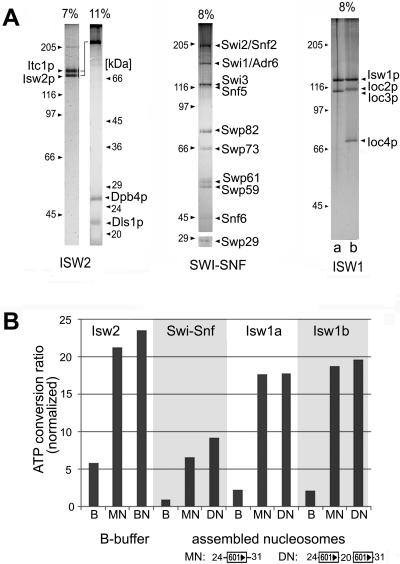
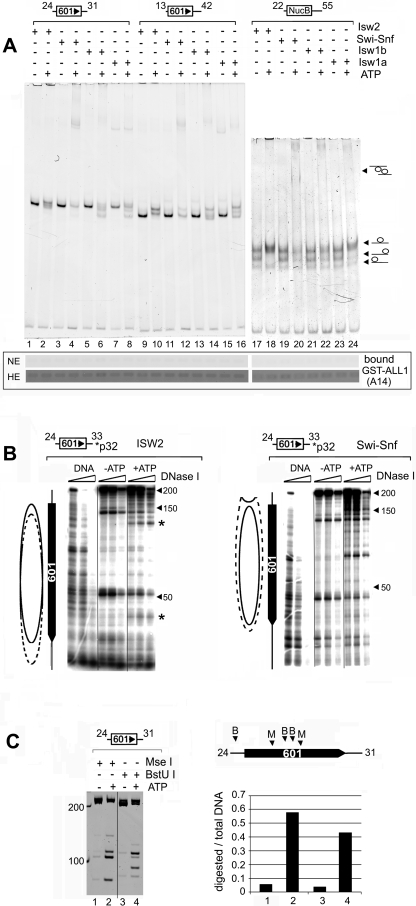
Remodeling complexes were purified (Fig. 2A), and their ATPase activities were compared in the presence of mono- and dinucleosomes. The ATPase activities of all four complexes were strongly stimulated by nucleosomes, with no significant differences observed when the complexes were incubated with either mono- or dinucleosomes (Fig. 2B). While the ATPase activities in the presence of nucleosomes varied somewhat between the complexes, the stimulation over the buffer control was approximately the same, four- to fivefold. The complexes were used to remodel nucleosomes reconstituted from purified histones and DNA templates containing one or two copies of the synthetic 601 nucleosome positioning sequence (42) or the nucleosome B DNA from the mouse mammary tumor virus promoter (46). Templates were also constructed that contained various lengths of linker DNA. The different combinations of templates and remodelers were used to generate distinct nucleosomal species in an attempt to correlate alterations in nucleosome structure to increased SET domain binding. First we tested whether ATP-dependent chromatin complexes could stimulate the binding of SET domains to mononucleosomes. Remodeling was assessed by gel mobility shift assays and was carried out using substoichiometric amounts of remodeling complex (see figure legends) to prevent the remodeling complex from interfering with the binding assay. This is particularly important for the reactions containing SWI/SNF. Excess SWI/SNF can lead to the generation of disassociated octamer, so we chose conditions under which the nucleosomes are remodeled but not disassembled. Furthermore, the removal of histones from the template was unlikely to occur under the reaction conditions used here because no histone chaperones or competitor DNA was included in the reaction mixture. Remodeling of mononucleosomes with any of the three ISWI complexes led to a noticeable shift in nucleosome mobilities, consistent with the sliding of nucleosomes to new translational positions along the DNA (Fig. 3A). The change in mobility was ATP dependent (Fig. 3A, compare odd- versus even-numbered lanes). However, incubation with SWI/SNF resulted in an ATP-dependent reduction in the nucleosomal species, with minimal evidence of sliding. A slight increase in free DNA was observed in some reaction mixtures, and a slower-migrating species appeared in the gel only in the presence of ATP (Fig. 3A). We interpret this species to be a dinucleosomal “bridge,” referred to as an altosome by others, that is formed by the remodeling of mononucleosomes by the human SWI/SNF complex or yeast RSC (26, 38, 44). Isw1b, but not Isw1a, also caused the appearance of a band migrating very close to that of the dinucleosomal bridge intermediate generated by SWI/SNF. The similar migration of the Isw1b-remodeled species suggests it is similar to the altosomes generated by SWI/SNF, but this will require additional confirmation. Remodeling was terminated by the depletion of ATP by apyrase treatment, and GST-ALL1 (A14 shown in Fig. 1C) was added. The binding of the nucleosomes to GST-ALL1 was assessed by analyzing the amount of nucleosomal DNA brought down on the beads. Surprisingly, we found that GST-ALL1 failed to bind to mononucleosomes remodeled by all four complexes (Fig. 3A, lower panel). Overexposure of the gel (high exposure) revealed the presence of background levels of DNA in all lanes, indicating that the failure to detect bound DNA was not due to a recovery problem. Furthermore, GST-ALL1 did not bind to remodeled mononucleosomes formed on templates containing different nucleosome positioning sequences (NucB) and lengths of linker DNA (Fig. 3A and data not shown).
FIG. 2.
Purification and analysis of yeast remodeling complexes. (A) Silver staining of SDS-PAGE gels, showing composition and purity of ATP-dependent chromatin remodeling complexes. The Isw1 panel, far right, shows the Isw1a (lane a) and Isw1b (lane b) complexes. The migrations of molecular mass (in kDa) standards are indicated on the sides of each panel. (B) ATPase activities of the complexes under the remodeling conditions used in this study (see Materials and Methods). Data are displayed as the Pi released/total ATP ratio and were normalized to the minimal ATPase activity observed in reaction mixtures containing Swi-Snf and buffer only.
FIG. 3.
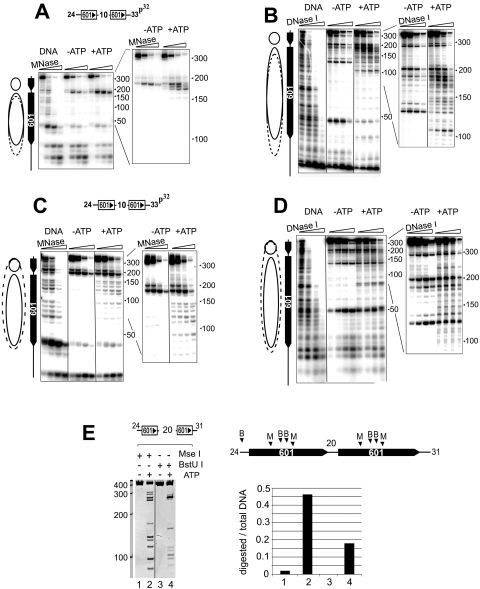
The SET domain of ALL1 does not bind remodeled mononucleosomes. (A, top) Analysis of remodeled mononucleosomes by native PAGE. Remodeling assay mixtures (50 μl) contained 1.2 μg of nucleosomes and 2.5 ng of ISWI or 5 ng of Swi-Snf complexes and were incubated at room temperature for 1.5 h. Details can be found in Materials and Methods. One-fifth of the reaction mixture was resolved on native 5% polyacrylamide gels and stained with ethidium bromide. The locations of nucleosomes positioned on the ends and middle of the DNA fragment are indicated on the left. (Bottom) The remainder of each reaction mixture was used in a GST pulldown assay with immobilized GST-ALL1 (A14 in Fig. 1C). The binding of nucleosomes was detected by analyzing nucleosomal DNA content on ethidium bromide-stained agarose gels. Two exposures of the gel are shown, normal exposure (NE) and high exposure (HE). The high exposure revealed the background retention of nucleosomes on the beads to demonstrate the recovery of DNA. This amount is similar to that typically retained on control beads (data not shown). (B) Verification of the remodeling of mononucleosomes by DNase I footprinting. Nucleosomes were assembled onto 32P-end-labeled DNA templates (depicted above each panel) and remodeled by ISW2 (left) or SWI/SNF (right) as described for panel A. Remodeling was terminated by incubating the reaction mixtures with apayrase and digestion with increasing amounts of DNase I. Products were purified and resolved on denaturing gels and visualized by autoradiography. The solid oval indicates the position of the nucleosome in the absence of ATP, while the dotted oval estimates its location after remodeling. The asterisks indicate the two hypersensitive sites shifted toward the unlabled end of the DNA fragment. (C) Restriction endonuclease analysis of SWI/SNF remodeled nucleosomes. Conditions are described in Materials and Methods. The locations of the MseI (M) and BstU I (B) sites on the template are indicated in the schematic diagram on the right. The graph on the right depicts the fraction of DNA cleaved as a ratio of total amount of DNA in each lane. The numbers under the bar graph correspond to the lanes in the gel.
To further verify that the mononucleosomes were remodeled, we examined the DNase I sensitivities of the ISW2 and SWI/SNF remodeled nucleosomes. The results showed that incubation of the template with ISW2 in the presence of ATP caused a change in the position of two hypersensitive sites toward the unlabeled end of the DNA fragment (Fig. 3B, left). However, the overall sensitivity of the nucleosomal DNA to DNase I was not changed. These changes are consistent with nucleosome sliding. The changes in the two hypersensitive sites suggest that Isw2 moves some of the nucleosomes in the population toward the end of the fragment. Isw2 is reported to preferentially move nucleosomes toward the center when mononucleosomes are assembled at the ends of DNA (13, 51). Examination of the remodeled species generated on this template on PAGE gels indicated that a significant fraction of the nucleosomes migrated faster, consistent with the movement of the nucleosome toward the end (Fig. 3A, lane 2). This may be due to the nearly symmetrical nature of the DNA fragment used in our study. Moreover, others have shown that Isw2 can move a nucleosome from the center of a nearly symmetrical DNA fragment toward the end (51). Since the linker DNA on each side of the nucleosome exceeds Isw2's minimal extranucleosomal DNA requirement of >20 to 22 bp (13, 51, 52), it could theoretically bind to either side of the nucleosome, generating a mixed population. In contrast, when Isw2 remodeled a mononucleosome with only 13 bp of DNA at one end, only slower-migrating species were observed, consistent with sliding of the nucleosome to the center (Fig. 3A, lane 10).
Incubation of the template with SWI/SNF caused an overall increase in DNase I sensitivity over the 601 positioning sequence, with less evidence for sliding (Fig. 3B, right). The increased DNase I sensitivity suggests that SWI/SNF disrupts the structure of nucleosome. The differences in DNase I digestion patterns of SWI/SNF- and ISW2-remodeled nucleosomes are consistent with published work on the activities of these two complexes (11, 12, 24). The failure of SWI/SNF to stimulate SET domain binding to nucleosomes was surprising, so we further verified the remodeling of the template by using a RE accessibility assay. Two RE enzymes, with sites located near the middle of the 601 sequence, were used (see figure legends). Here we observed an ATP-dependent increase in the digestion of these sites (Fig. 3C), providing a fourth line of evidence that SWI/SNF remodeled the mononucleosomes under the conditions used here. Even though the ATP-dependent complexes remodeled the mononucleosome templates, this failed to stimulate the binding of SET domains to the remodeled species.
Remodeling of dinucleosomes by the ISWI class promotes SET domain binding.
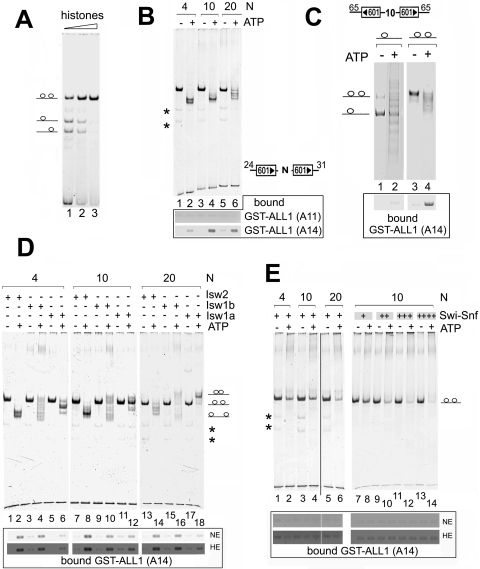
We considered that histone methyltransferases encounter multiple nucleosomes in vivo and may only target remodeled polynucleosome structures. In addition, EZH2, an H3-K27 HMT, displays greater activity on polynucleosome structures than mononucleosomes in vitro (27), further suggesting that some HMTs preferentially bind polynucleosomes. To test this, we examined the remodeling and binding of the SET domain of ALL1 to dinucleosomes formed on DNA templates containing two 601 positioning sequences separated by 4, 10, or 20 bp of DNA. The amount of histone octamers required to fill both nucleosome positioning sequences was determined empirically by titration, and an example is shown in Fig. 4A. We first examined the binding of GST-ALL1 derivatives to dinucleosomes remodeled by the Isw2 complex. Incubation of the templates with Isw2 caused the ATP-dependent repositioning of nucleosomes, evidenced by the shift of a single band into different, faster-migrating species (Fig. 4B). The change in migration suggests that Isw2 is sliding nucleosomes from the 601 sequences to the ends of the DNA. Interestingly, the number of bands and the migration of the remodeled species differed, depending upon the length of the sequence between the 601 positioning sequence. This too could be explained by the extranucleosomal DNA requirements of these enzymes (8, 13). We found that remodeling dinucleosomes by Isw2 significantly stimulated the binding of the SET domain of ALL1 to the remodeled templates (Fig. 4B, lower panel, A14). Importantly, a derivative lacking the histone binding region of the SET domain, but retaining the single-stranded nucleic acid binding domain (23) (DNA-B; A11), failed to interact with the remodeled dinucleosomes. This suggests that Isw2 is exposing features of the nucleosome masked in the unremodeled template, most likely the H3 tail, rather than simply exposing DNA. Furthermore, even though remodeling of the templates containing different lengths of spacer DNA generated distinct species, based on the different mobilities on the PAGE gel, GST-ALL1 bound to the different species (Fig. 4B). The material bound to GST-ALL1 must be nucleosomal DNA, rather than the small amount of free DNA in the reconstitution preparations, because we have shown that SET domains do not bind to linear double-stranded DNA (16) (data not shown), and the free DNA in the mononucleosomes preparations was not retained by GST-ALL1 (Fig. 3A).
FIG. 4.
The SET domain of ALL1 binds dinucleosomes remodeled by the ISWI class of chromatin remodeling enzymes. (A) Example of dinucleosome assembly. Dinucleosomes were reconstituted onto DNA containing two 601 minimal nucleosome positioning sequences (the orientation of the positioning sequences and the sizes of adjacent DNA are shown in each panel). (B) Isw2 remodeling and analysis of dinucleosomes were carried out as described for Fig. 3A. GST pulldown assays using ALL1 SET domain derivatives containing the minimal SET domain (A14) or only the nucleic acid binding domain (A11) (23) are shown on the bottom. Bound nucleosomes were detected by the DNA content in ethidium bromide-containing agarose gels. The numbers above each panel (4, 10, and 20) indicate the number of base pairs of DNA located between the two minimal 601 sequences. Asterisks mark locations of small amounts of contaminating mononucleosomes in the dinucleosome reconstitutions. (C) Binding of GST-ALL1 to remodeled dinucleosomes is not dependent on the length of extranucleosomal DNA and requires two nucleosomes on the template. Dinucleosomal templates were assembled with substoichiometric and saturating amounts of histones to obtain predominantly mono- and dinucleosomal species, respectively. The substrates were remodeled in ISW2 and GST pulldown assays performed as described for Fig. 3A. (D) The same experiment as shown in panel B, except that Isw2, Isw1a, and Isw1b were used to remodel dinucleosomes. (E) The same experiment as in panel B, except that 2.5 ng (left) or 2 to 10 ng (right) of SWI/SNF complex was used to remodel dinucleosomes. NE and HE indicate normal and high exposure of the gels, respectively, as described for Fig. 3A.
Nucleosomes formed on the 601 sequence exhibit a degree of asymmetry in biological processes. For example, a nucleosome assembled onto the 601 sequence in one orientation presents a stronger polar barrier to RNA polymerase II transcription than a nucleosome formed on the 601 sequence in the opposite orientation (2). Therefore, we also tested templates containing different orientations of the 601 positioning sequences to determine if this affected the remodeled product and the ability of SET domains to bind to the dinucleosome. Here too we found that GST-ALL1 only bound Isw2-remodeled dinucleosomes and that the binding was not dependent upon the orientation of the position sequences (data not shown.).
The ability of GST-ALL1 to recognize the remodeled dinucleosomal species could be explained by the longer length of DNA on these templates, versus the requirement for two nucleosomes. To address this directly, we compared the ability of GST-ALL1 to bind to remodeled mono- and dinucleosomes on the same DNA template. A fragment of DNA capable of accommodating two nucleosomes was assembled into chromatin using substoichiometric and saturating amounts of histones to generate predominately mononucleosomal and dinucleosomal species, respectively (Fig. 4C). Here we found that GST-ALL1 failed to bind the remodeled mononucleosomal species but could bind the dinucleosomal species (Fig. 4C, lane 2 versus 4). The small amount of nucleosomes retained by GST-ALL1 from the mononucleosomal sample likely originates from the small amount of dinucleosomal species contained in the reconstitutions (Fig. 4C, lane 2). This result further supports the idea that the binding of SET domains to dinucleosomal species is not a function of DNA length or sequence composition.
Next, we compared the abilities of different ATP-dependent chromatin remodeling complexes to stimulate the binding of SET domains to dinucleosomes. In addition to Isw2, we remodeled dinucleosomes with Isw1a, Isw1b, and SWI/SNF. Isw2, Isw1a, and Isw1b slide nucleosomes to new positions but display different biochemical properties and, thus, will generate different remodeled species (11, 13). Interestingly, remodeling by Isw1a and Isw1b generated different products, which was dependent upon the length of the spacer DNA. When templates containing 4 bp of spacer DNA were remodeled by the Isw1a and Isw1b enzymes, a faster-migrating species formed (Fig. 4D, lanes 4 and 6). However, when the intervening sequence was increased to 10 bp, slower-migrating species were detected and the amount of the slower-migrating species increased further when the length of intervening sequence was increased to 20 bp (Fig. 4D, lanes 16 and 18). The slower-migrating species suggest that the nucleosomes are moved to the center of the DNA fragment. Thus, incubating the various dinucleosomal templates with the three types of ISWI remodelers generated different species.
Similar to Isw2, remodeling by either Isw1a or Isw1b stimulated the binding of GST-ALL1 to the dinucleosome templates (Fig. 4D). While it is clear an ATP-dependent increase in GST-ALL1 binding to nucleosomes remodeled by Isw1a was observed, we repeatedly found that these remodeled dinucleosomes were not recognized as well as Isw2 or Isw1b remodeled nucleosomes. This suggests that Isw1a generates species that are functionally different from nucleosomes remodeled by the other two ISWI complexes, but this conclusion will require further investigation. Finally, we performed experiments where we remodeled nucleosomes using a mixture of two different ISWI remodeling complexes to see if binding could be stimulated further by the actions of two different remodeling enzymes, but we found that combining the remodelers did not increase the binding of GST-ALL1 to the templates (data not shown). Thus, even though Isw2, Isw1a, and Isw1b have different biochemical properties, all are capable of stimulating the binding of SET domains to dinucleosomal templates.
SWI/SNF and ISWI remodeling is considered to occur by different mechanisms, or at least through different intermediates (9, 11, 12). Therefore, we next compared the binding of GST-ALL1 to nucleosomes remodeled by SWI/SNF and the ISWI complexes. SWI/SNF caused an ATP-dependent reduction in the intensity of the band formed from the intact dinucleosome and the increase in the amount of a slower-migrating species (Fig. 4E). The slower-migrating species may result from the formation of bridges between the dinucleosome templates, similar to the altosomes formed between two remodeled mononucleosomes or aggregates. Despite evidence of remodeling, SWI/SNF failed to stimulate the binding of GST-ALL1 to the templates (Fig. 4E, left). So far, we have used amounts of the remodeling complexes based on estimates of protein levels, based on weight (see Materials and Methods). However, SWI/SNF has approximately twofold less ATPase activity than the ISWI complexes (Fig. 2B). We therefore repeated the experiment, titrating SWI/SNF into the remodeling reaction mixture. Increasing the amount of SWI/SNF reduced the amount of intact dinucleosomes but failed to stimulate the binding of GST-ALL1 to the remodeled template (Fig. 4E, right). This result indicates that the failure to stimulate binding of GST-ALL1 to the template was not due to limiting SWI/SNF amounts.
ISWI complexes slide nucleosomes to new translational positions and do not generate remodeled products with exposed DNA or histone core domain surfaces. It was unexpected that sliding of nucleosomes would have such a strong effect on SET domain binding to chromatin while remodeling by SWI/SNF did not. We considered that most studies detailing the products of ISW2 remodeling have used mononucleosome substrates, and it is possible that remodeling of dinucleosomes occurs through a different mechanism. We therefore characterized the dinucleosome products of ISW2 remodeling by nuclease digestion and compared them to those generated by SWI/SNF. Assembled dinucleosomes were incubated with ISW2 or SWI/SNF in the presence or absence of ATP, and the remodeled products were probed with MNase and DNase I. Examination of the MNase digestion pattern revealed that ISW2 did not increase digestion over the positioning sequence but led to a pattern consistent with the sliding of the distal nucleosome, defined as the one closest to the labeled end, away from the center of the DNA (Fig. 5A). This was evidenced by the appearance of two to four additional cleavage sites appearing within the linker region between the two 601 sequences at approximately 175 bp from the end (Fig. 5A, insert on right). In addition, a reduction in the intensity of the bands near the end of the 601 positioning sequence was observed (Fig. 5A, left). Likewise, DNase I mapping also suggested that ISW2 slides nucleosomes in the dinucleosome template. In the absence of ATP, the DNA underlying the nucleosome is protected from digestion, and two hypersensitive sites on the edges of the nucleosome were observed running at about 130 bp and 48 bp in the gel (Fig. 5B). The hypersensitive sites may result from the enhanced digestion of these bases due to the translational positioning of the nucleosomes over 601. In the ISW2 remodeled species, these two hypersensitive sites shifted downward in the gel, suggesting a movement of the nucleosome about 20 bp toward the end of the fragment (Fig. 5B). In addition, the sensitivity of the linker region was enhanced and spread downward in the gel (Fig. 5B, insert), which also suggests sliding to the ends of the fragment.
FIG. 5.
Analysis of ISW2 and SWI/SNF remodeled dinucleosomes. 32P-end-labeled dinucleosomes were remodeled by ISW2 (A and B) or SWI/SNF (C and D) as described in the legend for Fig. 3. Remodeling was terminated by apyrase treatment, and nucleosomes were digested with micrococcal nuclease (A and C) or DNase I (B and D). Digestion products were resolved on denaturing gels and visualized by autoradiography. (E) Restriction endonuclease analysis of SWI/SNF remodeled dinucleosomes. Conditions are described in Materials and Methods. The locations of the MseI (M) and BstU I (B) sites on the template are indicated in the schematic diagram on the right. The graph on the right depicts the fraction of DNA cleaved as a ratio of total amount of DNA in each lane. The numbers under the bar graph correspond to the lanes in the gel.
There was less evidence that the proximal nucleosome slid in the other direction, which would have been predicted based upon the gel mobility shift assay that suggested that ISW2 slid the DNA toward the ends of the DNA, as the MNase digestion pattern revealed an increase in digestion asymmetrically toward the labeled end of the fragment (Fig. 5A). This is unlikely to be caused by sequence bias of MNase, since DNase I footprinting also revealed a pattern whereby the linker region expanded toward the labeled end of the DNA (Fig. 5B). The reason for this asymmetry of nucleosome sliding may be the propensity of ISW2 to equally space polynucleosomes on a DNA fragment. Sliding of the distal nucleosome 15 to 20 bp toward the labeled end would achieve equal spacing of nucleosomes across this template.
The DNase I and MNase digestion patterns suggest that ISW2 is sliding nucleosomes on the template. This is in contrast to when the template was remodeled by SWI/SNF. Incubating the dinucleosome templates with SWI/SNF increased the accessibility of the DNA to MNase over the 601 positioning sequence, predominantly around the edge of the nucleosome near the linker region (Fig. 5C). Likewise, SWI/SNF increased the accessibility of the 601 sequence to DNase I, and no shift in the positions of the two hypersensitive sites was observed after remodeling (Fig. 5D). The changes in the MNase and DNase I footprints cannot be attributed to binding of the complexes to the nucleosomes, because a ratio of about 1 remodeler to 25 to 50 nucleosomes was used in these experiments. Finally, we verified that SWI/SNF remodeled the dinucleosomes by using the restriction endonuclease accessibility assay. As observed on mononucleosomes, an ATP-dependent increase in digestion by MseI and BstUI at sites within the 601 sequences was observed (Fig. 5E). Therefore, four different assays (PAGE, MNase, DNase I, and RE accessibility) indicate that SWI/SNF remodeled the dinucleosome-containing templates.
Our data suggest that ISW2 slides nucleosomes to the ends of the dinucleosome templates while SWI/SNF disrupts them; thus, these initial analyses have verified that the two remodeling complexes work by a mechanism similar to that described for mononucleosomes. Studies analyzing the details of Isw2 remodeling have used mononucleosome templates. Additional detailed studies will be needed to fully characterize the products of remodeling on dinucleosomal templates by these remodelers. Nonetheless, we provide strong evidence that the ISWI and Swi-Snf classes of enzymes remodel dinucleosomes and that the SET domains of HMTs can distinguish differences in the products of remodeling by these enzymes.
ISWI remodeling stimulates nucleosomal histone methylation.
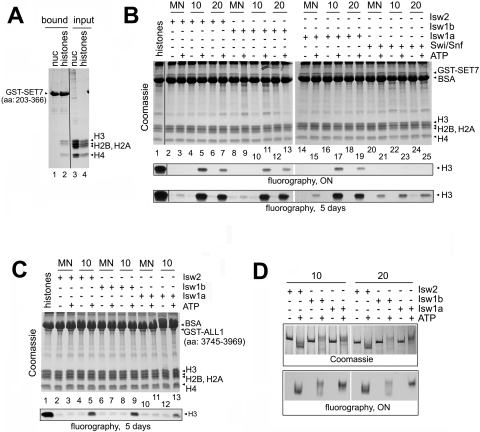
The results obtained thus far indicate that the binding of SET domains to dinucleosomes is stimulated by ISWI remodeling. Next, we evaluated the ability of the chromatin remodeling complexes to stimulate the nucleosomal HMT activity of two well-characterized HMTs, SET7/9 and ALL1. Human SET7/9, described as both SET7 (48) and SET9 (32), monomethylates lysine 4 of histone H3 in vitro and in vivo. It is referred to as SET7 from here on. We used SET7 in these experiments because it is highly active in recombinant form. First, we verified the histone and nucleosome binding properties of GST-SET7 and found that it bound histone H3 and H4 in histone preparations but not nucleosomes (Fig. 6A, lane 1 versus 2). A third band was observed in the SET7 bound material migrating below H3, but above H2A. This may be proteolyzed H3, which is observed in some histone preparations. Therefore, SET7 has the same histone and nucleosome binding properties as trithorax and ALL1. HMT assays were performed on free histones and mono- (MN) and dinucleosomes containing a 10- and 20-bp spacer between the positioning sequences, respectively. In agreement with a previous report (48), SET7 displayed robust activity on free histones but significantly less activity on intact mono- or dinucleosomes (Fig. 6B, compare lane 1 to lanes 2, 4, and 6). Thus, incorporation of H3 into nucleosomes prevented access of its N-terminal tail to SET7. However, incubating dinucleosomes with Isw2, Isw1a, or Isw1b in the presence of ATP greatly stimulated HMT activity (Fig. 6B, lanes 5, 7, 11, 13, 17, and 19). Importantly, incubating mononucleosomes with any of the ATP-dependent chromatin remodeling complexes stimulated their methylation by SET7 only very weakly compared to dinucleosomes; detection of this low level of activity required a fivefold overexposure of the autoradiograph (Fig. 6B, lower panel). This result correlates well with the nucleosome binding assays, which showed that SET domains do not bind to remodeled mononucleosomes (Fig. 3A). SWI/SNF was unable to stimulate the binding of SET domains to dinucleosomes (Fig. 4E) and, consistent with those observations, SWI/SNF only very weakly enhanced the HMT activity of SET7 (Fig. 6B, lanes 20 to 25). The low level of HMT activity detected on SWI/SNF remodeled nucleosomes was far below that observed on dinucleosomes remodeled by the ISWI complexes (Fig. 6B, compare lanes 5 and 6 versus 23 and 25, for example). In addition, the weak stimulation of HMT activity by SWI/SNF was observed on mono- and dinucleosomal templates equally well.
FIG. 6.
Remodeling of dinucleosomes by ISWI complexes stimulates histone methylation by SET domain proteins. (A) The SET domain of SET7 binds histones, but not nucleosomes. GST pulldown experiments were conducted with immobilized GST-SET7 polypeptides (residues 203 to 366) as described in the legend for Fig. 1B. (B) Remodeling of dinucleosomes facilitates their methylation by SET7. Twenty micrograms of mono- (MN) or dinucleosomes with a 10- or 20-bp spacer was incubated with 50 ng ISWI or 100 ng Swi/Snf remodeling enzymes with or without ATP for 1.5 h in 500 μl. After the depletion of ATP, 0.5 μg of GST-SET7 was added in the presence of [3H]SAM for 1 h. The samples were concentrated by trichloroacetic acid precipitation and resolved by SDS-PAGE. Histone recovery and content were examined by Coomassie blue staining, and [3H]SAM incorporation was determined by fluorography (lower panel). BSA, bovine serum albumin. (C) The same experiment as shown in panel B, except that 5 μg of GST-ALL1 (residues 3745 to 3969) was used in the HMT assay. (D) Resolution of remodeled and methylated nucleosomes on native PAGE gels. Amounts and locations of total nucleosomes were visualized by Coomassie blue staining (upper panel), and methylated histones were analyzed by fluorography (lower panel).
To show that these results were not specific to SET7, we verified our results by conducting HMT assays using the SET domain of ALL1 and nucleosomes remodeled by ISWI complexes (Fig. 6C). While the overall activity of ALL1 was lower on all substrates tested (note the differences in exposure), including free histones, remodeling of dinucleosomes by the three ISWI complexes significantly enhanced its ability to methylate H3 (Fig. 6C). Thus, our data indicate that remodeling of dinucleosome templates by the ISWI class of remodelers is required to stimulate SET domain binding and HMT activity.
While it was highly unlikely under the reaction conditions used here, it is a formal possibility that remodeling by the ISWI complexes generates free histones, which are then modified by the HMTs. So, to verify that the H3 methylated in these assays is within the remodeled nucleosomes, we separated the remodeled and methylated dinucleosome products on native polyacrylamide gels. The positions of the nucleosomes in the gel were visualized by Coomassie blue staining, and the incorporation of tritiated [3H]SAM into H3 was detected by fluorography. The results in Fig. 6D clearly show incorporation of [3H]SAM into a band comigrating with nucleosomes remodeled by the ISWI complexes. Furthermore, this was only observed in the presence of ATP, and the exact migrations of the bands containing tritium-labeled histones in each lane were superimposed onto the locations of the remodeled nucleosome products. This indicates that SET7 methylated histone H3 within the context of the nucleosome and that it recognizes features of a remodeled dinucleosome.
DISCUSSION
We have characterized the requirements for the recognition and modification of nucleosomal substrates by SET domain-containing HMTs. Most HMTs display a significantly higher level of enzymatic activity on free histones than on nucleosomal histones (reference 48 and references therein). In some cases, nucleosomal HMT activity is barely detectable. This suggests that the histone tails are shielded and not fully accessible within the context of the nucleosome, and alterations to the nucleosome must be made to allow the tails to be recognized by SET domain-containing proteins. Many HMTs modify nucleosomes at the promoter of genes and/or do so cotranscriptionally by associating with RNAPII (25). It has been proposed that the remodeling of nucleosomes by the passage of RNAPII alters the nucleosome structure to expose the tails and facilitate methylation; however, this has not been shown directly. Since RNAPII is also responsible for recruiting HMTs to genes, the effects of recruitment versus nucleosome alteration per se could not be distinguished in the previous studies. It may be difficult to show this directly in vitro, however, as RNAPII transcribes through mononucleosomes poorly in the absence of chromatin remodeling factors in vitro (6). If recognition and modification of nucleosomes by SET domains require polynucleosome structures, as observed here, it may be more difficult to establish an assay, as transcription though two or more nucleosomes may not occur without the aid of chromatin remodeling factors and histone chaperones. In an attempt to determine if transcribed chromatin is a better substrate for HMTs, we analyzed the binding of SET domains to chromatin isolated from cells on mercury affinity columns. Nucleosome intermediates associated with transcription, such as those with exposed thiol-reactive cysteine residues and “tetrasomes,” bind to SET domains better than unmodified nucleosomes. SET domains methylated these “altered” nucleosomes more efficiently as well. While this approach has its caveats, the results suggest that the alterations in nucleosome structure caused by RNAPII transcription may be important in the recognition and modification of H3 by HMTs and that RNAPII plays more than just a recruitment function during this process of cotranscriptional histone modification. In addition, the identifications of a histone binding and single-strand DNA/RNA binding motifs in SET domains suggest that both the exposure of the tails and the generation of single-stranded nucleic acid structures during transcription and remodeling may play a role in the targeting of these factors to chromatin also (19, 23).
The use of highly purified nucleosomal templates and remodeling complexes has allowed us to address if chromatin remodeling complexes can induce SET binding to nucleosomes. We used yeast complexes and the SET domains from human HMTs in our studies, and our conclusions are based on the assumption that remodeling by yeast complexes is equivalent to that of their respective human counterpart. It would be difficult to argue otherwise based upon sequence similarities between yeast and human subunits in the chromatin remodeling complexes and the biochemical analysis conducted thus far. Remodeling of dinucleosomes by ISWI complexes, but not SWI/SNF, stimulated HMT activity of both SET7 and ALL1. The ISWI class of enzymes reposition nucleosomes through a sliding mechanism and maintain the integrity of the nucleosome (12, 13, 29, 43). On the other hand, SWI/SNF generates intermediates that are more disruptive to nucleosome structure and under certain conditions can cause H2a/2b dimer release and octamer transfer (9, 11, 12). These characteristics might have predicted that SWI/SNF would be more effective at stimulating HMT activity. This was not observed under the conditions used here. The changes in nucleosome structure caused by ISWI remodeling responsible for stimulating SET domain binding and HMT activity may not be detectable by standard methods that focus on histone core-DNA interactions, i.e., nuclease accessibility. ISWI complexes show a distinct dependence on histone tails for remodeling, specifically the H4 tail (10, 13). ISWI remodeling may involve breaking the interactions between the histone tails and DNA or the nucleosome, which would free the tails to be recognized by HMTs. The N-terminal tail of H3 makes contact with DNA, which could shield it from the SET domain (50). The H3 tail should form the most thermodynamically stable interactions during histone deposition/assembly. It seems logical that the process of sliding would break these contacts, and after sliding, the tails may form new interactions that are less stable and more susceptible to SET domain binding. This model could explain the requirement for dinucleosome templates. ISWI remodeling only stimulated SET domain binding to dinucleosomes, not mononucleosomes, in the purified system. An adjacent nucleosome may be required to stabilize the remodeled product or intermediate, which is then recognized by the SET domain. HMTs encounter multiple nucleosomes in vivo and sequence-specific DNA binding proteins, which could stabilize the remodeled nucleosomes and allow SET domain binding to chromatin, so it is likely that what we observed here has physiological significance. Interestingly, comparison of the abilities of SET domains to recognize different templates remodeled by the three ISWI complexes suggests that the exact placement of the two nucleosomes along the length of the DNA and the amount of linker DNA are not major determinants. This is clear on the dinucleosome template containing 20 bp of spacer DNA between the 601 sequence (Fig. 4D). On this template, ISW2 generated a faster-migrating dinucleosome species, suggesting nucleosomes relocated near the ends, while ISW1a generated predominantly a slower-migrating species, suggesting nucleosomes are positioned in the middle, and both were recognized and methylated by the SET domain of ALL1.
ISW1 regulates the dynamics of histone H3 K4 methylation in yeast during transcription in vivo (28, 30). The mechanism is unclear, but it was suggested that it does so by modifying chromatin structure to allow for the release of RNAPII. Here we show that ISW1 can stimulate methylation of K4 in vitro, and our results suggest that the remodeling activities of Isw1 directly regulate K4me in vivo. Interestingly, ISWI complexes contain histone binding domains that selectively bind methylated H3 (20, 25, 37, 49). This presents an interesting scenario for establishing methylated chromatin domains, where the targeting of ISWI remodelers through adjacent histone methyl marks can reinforce or propagate the epigenetic mark by making the chromatin better substrates for HMT enzymes. This too could explain a mechanism for the inheritable patterns of histone methylation after DNA replication, where histones are distributed between parental and daughter strands and methylation patterns must be reestablished after the deposition of new histones onto the strands. Participation of ISWI remodelers in promoting histone methylation after replication is consistent with their known roles in chromatin assembly and spacing (24, 29, 35). Even Isw2, once thought to only regulate transcription, has recently been shown to participate in replication fork progression (47). Therefore, the ability of ISWI remodelers to stimulate the methylation of chromatin may be important for the propagation of the epigenetic mark.
Acknowledgments
This work was funded by grants from the Leukemia Research Foundation (Evanston, IL) and the Russian Foundation for Basic Research (08-04-01010) to W.A.K. and from the Association of International Cancer Researchers (06-466) to J.C.R. and W.A.K.
We thank Toshio Tsukiyama for strains expressing FLAG-tagged Isw2.
Footnotes
Published ahead of print on 14 September 2009.
REFERENCES
- 1.Allfrey, V. G., and T. A. Chen. 1991. Nucleosomes of transcriptionally active chromatin: isolation of template-active nucleosomes by affinity chromatography. Methods Cell Biol. 35:315-335. [DOI] [PubMed] [Google Scholar]
- 2.Bondarenko, V. A., L. M. Steele, A. Ujvari, D. A. Gaykalova, O. I. Kulaeva, Y. S. Polikanov, D. S. Luse, and V. M. Studitsky. 2006. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell 24:469-479. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, L. A., R. R. Latek, and C. L. Peterson. 2004. The SANT domain: a unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 5:158-163. [DOI] [PubMed] [Google Scholar]
- 4.Breiling, A., L. Sessa, and V. Orlando. 2007. Biology of polycomb and trithorax group proteins. Int. Rev. Cytol. 258:83-136. [DOI] [PubMed] [Google Scholar]
- 5.Cairns, B. R., Y. J. Kim, M. H. Sayre, B. C. Laurent, and R. D. Kornberg. 1994. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc. Natl. Acad. Sci. USA 91:1950-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carey, M., B. Li, and J. L. Workman. 2006. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol. Cell 24:481-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, T. A., and V. G. Allfrey. 1987. Rapid and reversible changes in nucleosome structure accompany the activation, repression, and superinduction of murine fibroblast protooncogenes c-fos and c-myc. Proc. Natl. Acad. Sci. USA 84:5252-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clapier, C. R., and B. R. Cairns. 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78:273-304. [DOI] [PubMed] [Google Scholar]
- 9.Dang, W., and B. Bartholomew. 2007. Domain architecture of the catalytic subunit in the ISW2-nucleosome complex. Mol. Cell. Biol. 27:8306-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dang, W., M. N. Kagalwala, and B. Bartholomew. 2006. Regulation of ISW2 by concerted action of histone H4 tail and extranucleosomal DNA. Mol. Cell. Biol. 26:7388-7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dechassa, M. L., B. Zhang, R. Horowitz-Scherer, J. Persinger, C. L. Woodcock, C. L. Peterson, and B. Bartholomew. 2008. Architecture of the SWI/SNF-nucleosome complex. Mol. Cell. Biol. 28:6010-6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan, H. Y., K. W. Trotter, T. K. Archer, and R. E. Kingston. 2005. Swapping function of two chromatin remodeling complexes. Mol. Cell 17:805-815. [DOI] [PubMed] [Google Scholar]
- 13.Gangaraju, V. K., and B. Bartholomew. 2007. Mechanisms of ATP dependent chromatin remodeling. Mutat. Res. 618:3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassan, A. H., K. E. Neely, M. Vignali, J. C. Reese, and J. L. Workman. 2001. Promoter targeting of chromatin-modifying complexes. Front Biosci. 6:D1054-D1064. [DOI] [PubMed] [Google Scholar]
- 15.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 16.Horn, P. J., and C. L. Peterson. 2001. The bromodomain: a regulator of ATP-dependent chromatin remodeling? Front Biosci. 6:D1019-D1023. [DOI] [PubMed] [Google Scholar]
- 17.Jenuwein, T. 2006. The epigenetic magic of histone lysine methylation. FEBS J. 273:3121-3135. [DOI] [PubMed] [Google Scholar]
- 18.Kassabov, S. R., N. M. Henry, M. Zofall, T. Tsukiyama, and B. Bartholomew. 2002. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol. Cell. Biol. 22:7524-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katsani, K. R., J. J. Arredondo, A. J. Kal, and C. P. Verrijzer. 2001. A homeotic mutation in the trithorax SET domain impedes histone binding. Genes Dev. 15:2197-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128:693-705. [DOI] [PubMed] [Google Scholar]
- 21.Krajewski, W. A. 2008. Evidence for the nucleosome-disruption process regulated by phosphorylation of 120 kDa protein complex in Drosophila embryo cell-free system. Biochimie 90:534-541. [DOI] [PubMed] [Google Scholar]
- 22.Krajewski, W. A., and P. B. Becker. 1999. Reconstitution and analysis of hyperacetylated chromatin. Methods Mol. Biol. 119:207-217. [DOI] [PubMed] [Google Scholar]
- 23.Krajewski, W. A., T. Nakamura, A. Mazo, and E. Canaani. 2005. A motif within SET-domain proteins binds single-stranded nucleic acids and transcribed and supercoiled DNAs and can interfere with assembly of nucleosomes. Mol. Cell. Biol. 25:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langst, G., and P. B. Becker. 2004. Nucleosome remodeling: one mechanism, many phenomena? Biochim. Biophys. Acta 1677:58-63. [DOI] [PubMed] [Google Scholar]
- 25.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128:707-719. [DOI] [PubMed] [Google Scholar]
- 26.Lorch, Y., B. R. Cairns, M. Zhang, and R. D. Kornberg. 1998. Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell 94:29-34. [DOI] [PubMed] [Google Scholar]
- 27.Martin, C., R. Cao, and Y. Zhang. 2006. Substrate preferences of the EZH2 histone methyltransferase complex. J. Biol. Chem. 281:8365-8370. [DOI] [PubMed] [Google Scholar]
- 28.Mellor, J. 2005. The dynamics of chromatin remodeling at promoters. Mol. Cell 19:147-157. [DOI] [PubMed] [Google Scholar]
- 29.Mellor, J., and A. Morillon. 2004. ISWI complexes in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1677:100-112. [DOI] [PubMed] [Google Scholar]
- 30.Morillon, A., N. Karabetsou, A. Nair, and J. Mellor. 2005. Dynamic lysine methylation on histone H3 defines the regulatory phase of gene transcription. Mol. Cell 18:723-734. [DOI] [PubMed] [Google Scholar]
- 31.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 32.Nishioka, K., S. Chuikov, K. Sarma, H. Erdjument-Bromage, C. D. Allis, P. Tempst, and D. Reinberg. 2002. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 16:479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson, C. L., A. Dingwall, and M. P. Scott. 1994. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc. Natl. Acad. Sci. USA 91:2905-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qian, C., and M. M. Zhou. 2006. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell Mol. Life Sci. 63:2755-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Racki, L. R., and G. J. Narlikar. 2008. ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Curr. Opin. Genet. Dev. 18:137-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ringrose, L., and R. Paro. 2007. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134:223-232. [DOI] [PubMed] [Google Scholar]
- 37.Santos-Rosa, H., R. Schneider, B. E. Bernstein, N. Karabetsou, A. Morillon, C. Weise, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2003. Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 12:1325-1332. [DOI] [PubMed] [Google Scholar]
- 38.Schnitzler, G., S. Sif, and R. E. Kingston. 1998. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell 94:17-27. [DOI] [PubMed] [Google Scholar]
- 39.Shilatifard, A. 2008. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 20:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steger, D. J., T. Owen-Hughes, S. John, and J. L. Workman. 1997. Analysis of transcription factor-mediated remodeling of nucleosomal arrays in a purified system. Methods 12:276-285. [DOI] [PubMed] [Google Scholar]
- 41.Suganuma, T., S. G. Pattenden, and J. L. Workman. 2008. Diverse functions of WD40 repeat proteins in histone recognition. Genes Dev. 22:1265-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thastrom, A., L. M. Bingham, and J. Widom. 2004. Nucleosomal locations of dominant DNA sequence motifs for histone-DNA interactions and nucleosome positioning. J. Mol. Biol. 338:695-709. [DOI] [PubMed] [Google Scholar]
- 43.Tsukiyama, T., J. Palmer, C. C. Landel, J. Shiloach, and C. Wu. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 13:686-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ulyanova, N. P., and G. R. Schnitzler. 2005. Human SWI/SNF generates abundant, structurally altered dinucleosomes on polynucleosomal templates. Mol. Cell. Biol. 25:11156-11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vary, J. C., Jr., V. K. Gangaraju, J. Qin, C. C. Landel, C. Kooperberg, B. Bartholomew, and T. Tsukiyama. 2003. Yeast Isw1p forms two separable complexes in vivo. Mol. Cell. Biol. 23:80-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vicent, G. P., A. S. Nacht, C. L. Smith, C. L. Peterson, S. Dimitrov, and M. Beato. 2004. DNA instructed displacement of histones H2A and H2B at an inducible promoter. Mol. Cell 16:439-452. [DOI] [PubMed] [Google Scholar]
- 47.Vincent, J. A., T. J. Kwong, and T. Tsukiyama. 2008. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat. Struct. Mol. Biol. 15:477-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, H., R. Cao, L. Xia, H. Erdjument-Bromage, C. Borchers, P. Tempst, and Y. Zhang. 2001. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell 8:1207-1217. [DOI] [PubMed] [Google Scholar]
- 49.Wysocka, J., T. Swigut, H. Xiao, T. A. Milne, S. Y. Kwon, J. Landry, M. Kauer, A. J. Tackett, B. T. Chait, P. Badenhorst, C. Wu, and C. D. Allis. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442:86-90. [DOI] [PubMed] [Google Scholar]
- 50.Zheng, C., and J. J. Hayes. 2003. Structures and interactions of the core histone tail domains. Biopolymers 68:539-546. [DOI] [PubMed] [Google Scholar]
- 51.Zofall, M., J. Persinger, and B. Bartholomew. 2004. Functional role of extranucleosomal DNA and the entry site of the nucleosome in chromatin remodeling by ISW2. Mol. Cell. Biol. 24:10047-10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zofall, M., J. Persinger, S. R. Kassabov, and B. Bartholomew. 2006. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat. Struct. Mol. Biol. 13:339-346. [DOI] [PubMed] [Google Scholar]