Abstract
The two mitotic centrosomes direct spindle bipolarity to maintain euploidy. Centrosome amplification—the acquisition of ≥3 centrosomes—generates multipolar mitoses, aneuploidy, and chromosome instability to promote cancer biogenesis. While much evidence suggests that Cdk2 is the major conductor of the centrosome cycle and that it mediates centrosome amplification induced by various altered tumor suppressors, the role played by Cdk4 in a normal or deregulated centrosome cycle is unknown. Using a gene knockout approach, we report that Cdk2 and Cdk4 are critical to the centrosome cycle, since centrosome separation and duplication are premature in Cdk2−/− mouse embryonic fibroblasts (MEFs) and are compromised in Cdk4−/− MEFs. Additionally, ablation of Cdk4 or Cdk2 abrogates centrosome amplification and chromosome instability in p53-null MEFs. Absence of Cdk2 or Cdk4 prevents centrosome amplification by abrogating excessive centriole duplication. Furthermore, hyperactive Cdk2 and Cdk4 deregulate the licensing of the centrosome duplication cycle in p53-null cells by hyperphosphorylating nucleophosmin (NPM) at Thr199, as evidenced by observations that ablation of Cdk2, Cdk4, or both Cdk2 and Cdk4 abrogates that excessive phosphorylation. Since a mutant form of NPM lacking the G1 Cdk phosphorylation site (NPMT199A) prevents centrosome amplification to the same extent as ablation of Cdk2 or Cdk4, we conclude that the Cdk2/Cdk4/NPM pathway is a major guardian of centrosome dysfunction and genomic integrity.
The centrosome maintains genomic integrity by enforcing euploidy (20). A centrosome consists of two centrioles, containing proteins such as α-tubulin; structural proteins including pericentrin, γ-tubulin, and centrin-2; and cell cycle-regulatory proteins, which include p53 and cyclin E/Cdk2. Normal cells have one mature centrosome during early G1 (20). At late G1, each of the centrioles composing the mature centrosome separates and duplicates to form a new (or daughter) centriole between late G1 and late S phase, culminating in two fully mature centrosomes at G2. The two mitotic centrosomes associate with spindle fibers and migrate toward opposite sides of the spindle pole to establish bipolarity. This ensures that sister chromatids segregate toward each spindle pole. Following cytokinesis, each daughter cell receives one centrosome and an equal complement of chromatids. Normal centrosome duplication must be strictly controlled and strictly coordinated with S-phase initiation and progression (68). When this control fails, centrosome amplification occurs, leading to aberrant and multipolar mitotic spindles, increased frequency of chromosome segregation errors, aneuploidy, and chromosome instability (12, 20). Centrosome amplification, aneuploidy, and chromosome instability contribute to cancer biogenesis and progression by triggering reduced expression of tumor suppressors and overexpression of proto-oncogenes.
One of the pathways contributing to centrosome amplification is deregulated centrosome duplication triggered by the G1 cyclin-dependent kinases (Cdks) (27, 51, 59). The Cdks, a family of serine/threonine protein kinases, control the onset of the major cell cycle events, such as DNA synthesis and mitosis (65). Cdk activities are positively regulated by association with different cyclins, which are temporally expressed at specific phases of the cell cycle; they are negatively regulated by a variety of Cdk inhibitors (CKIs) (65). Individual and combinatorial gene knockouts of the cyclins and Cdks have uncovered redundancy in the regulation of DNA synthesis and specificity in their abilities to control development and tumorigenesis (2, 5, 6, 24, 25, 38, 52, 55, 61, 72, 77). However, how the cyclins and Cdks individually or cooperatively impinge on centrosome duplication is poorly understood. Biochemical and pharmacological evidence pointed to Cdk2 as the only Cdk coordinating the centrosome duplication and cell cycles (32, 33, 36, 40, 44). Cdk2 was proposed to coordinate the cell and centrosome duplication cycles by phosphorylating Rb to promote S phase (28) and by phosphorylating various centrosomal proteins to regulate the centrosome duplication cycle (10, 19, 51). Cyclin E/Cdk2 phosphorylates nucleophosmin (NPM)/B23 at T199 to regulate centrosome licensing; this phosphorylation allows centrioles to separate and centriole duplication to commence (70). Cdk2 directly promotes centrosome duplication by phosphorylating Mps-1 and CP110 and by modulating the activity of Plk4 (10, 19, 26). However, gene knockout approaches dethroned Cdk2 as the sole Cdk coordinating the cell and centrosome duplication cycles, since mouse embryonic fibroblasts (MEFs) from which Cdk2 (14) or cyclins E1 and E2 (25) had been deleted showed only a minor deviation from normal centrosome ratios and proliferated. These results implied that, as with the cell cycle, there is redundancy among the Cdks regulating the centrosome duplication cycle. These results were unexpected, given the involvement of Cdk2 in the regulation of two central steps in the centrosome duplication cycle: licensing and duplication. To date, the identity of the Cdks supporting Cdk2 in regulating normal centrosome duplication is unknown.
As the cyclins, Cdks, and CKIs control centrosome duplication, altered tumor suppressors and oncogenes deregulate those cell cycle-regulatory molecules, leading to centrosome amplification (12, 21). Ablated genes that result in elevated Cdk2 activity and elevated frequencies of centrosome amplification include E2F3, p53, Skp2, and p21Waf1; similarly, ectopically expressed cyclins E and A result in elevated Cdk2 activity and centrosome amplification in p53−/− MEFs (13, 27, 44, 49, 59, 68). Likewise, oncogenes and altered tumor suppressors that hyperactivate Cdk4 and result in high frequencies of centrosome amplification include ectopically expressed Her2 (47), H-RasV12, v-Mos (57), MEK1Glu217/Glu221 (58), cyclin D1 (50), and silenced MEK2 (73). Conversely, p16 restricts excessive centriole reduplication (42, 44). However, the relationships between altered genes, ectopic activities of specific Cdks, and centrosome amplification are correlational, as they deregulate cyclin/Cdk activities as well as complex signal transduction cascades that control a plethora of transcripts.
The abilities of the cell cycle and centrosomal checkpoints—signaling pathways that monitor the integrity and replication status of the genome and the centrosome—to inhibit entry into S phase and centrosome duplication are closely associated with the function of the p53 tumor suppressor (45, 74). The p53 transcription factor is inactivated in approximately 50% of human cancers (71). p53 regulates the transcription of a large number of genes to prevent entry into S phase in the presence of DNA or centrosome damage (45, 74). Indeed, ablation of p53 allows centrosome amplification, aneuploidy, and chromosome instability (22). A gene product central to centrosome duplication control is p21Waf1, expressed at low levels in a p53-dependent manner (48) to inhibit the cyclin E/Cdk2 complex (65). In addition, p21Waf1 has been implicated in the assembly of the cyclin D1/Cdk4 complex, and its overexpression inhibits the activity of Cdk4 at higher concentrations (30, 35, 76). The continual presence of p21Waf1 guards against premature activation of cyclin E/Cdk2 and perhaps against that of cyclin D/Cdk4, ensuring the coordinated initiation of centrosome and DNA duplication. In p21Waf1−/− MEFs, initiation of centrosome and DNA duplication is uncoupled, much like that in cells with constitutively active cyclin E/Cdk2 (13, 48, 68). Importantly, observations that the reintroduction of wild-type p53 into p53−/− cells resulted in nearly complete restoration of the centrosome duplication cycle while ectopic expression of p21Waf1 in p53−/− cells only partially restored that cycle (68) suggest that p53 controls centrosome duplication through multiple pathways, one of which is mediated by the negative regulation of Cdk2 by p21Waf1.
Since direct evidence linking Cdk2 or Cdk4 to centrosome amplification in p53−/− MEFs was lacking, we used a genetic approach to test whether Cdk4 and Cdk2 mediate that abnormal process. Our results revealed that p53 knockout does not signal centrosome amplification and chromosome instability exclusively through Cdk2, as suggested previously (21). We propose a new paradigm: ablation of p53 requires the presence of both Cdk2 and Cdk4 activities in order to induce high frequencies of centrosome amplification and chromosome instability.
MATERIALS AND METHODS
Generation of mouse embryonic fibroblasts.
Mice were crossed as Cdk2+/− × Cdk2+/−, Cdk4+/− × Cdk4+/−, Cdk2+/− Cdk4+/− × Cdk2+/− Cdk4+/− (meiotically recombined [6]), p53+/− Cdk2+/− × p53+/− Cdk2+/−, p53+/− Cdk4+/− × p53+/− Cdk4+/−, and p53+/− Cdk2+/− Cdk4+/− × p53+/− Cdk2+/− Cdk4+/−. After mating, embryos were isolated from females 13.5 days after detection of seminal plugs. Embryos were collected under sterile conditions and their livers extirpated for extraction of DNA and PCR genotyping. MEFs were generated using established methods (59). The individual genotypes were assessed by PCR genotyping with primers specific for the wild-type and knockout p53, Cdk2, and Cdk4 alleles (6). All experiments were performed on passage 2 (p2) MEFs.
Cell culture.
MEFs were maintained under proliferating conditions with 10% fetal bovine serum (FBS)-Dulbecco's modified Eagle medium (DMEM). For serum arrest experiments, cells were cultured in 0.2% FBS-DMEM for 60 h.
Centriole reduplication assay.
Three independent, proliferating MEFs of the genotypes indicated in Fig. 4E and F, plated in two-well chamber slides, were either left untreated or treated with 2 mM hydroxyurea (HU) for 48 h. For coimmunostaining of α- and γ-tubulins in order to examine centrioles, cells were first incubated on ice for 30 min, to destabilize microtubules nucleated at the centrosomes, and then briefly extracted (∼1 min) with cold extraction buffer [0.75% Triton X-100, 5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 2 mM EGTA (pH 6.7)]. Cells were then briefly washed in cold phosphate-buffered saline (PBS) and were fixed as previously described (59). Cells were immunostained with anti-α-tubulin monoclonal (DMA1; Sigma) and anti-γ-tubulin polyclonal (ab11317; Abcam) antibodies. The antibody-antigen complexes were detected with the appropriate Alexa Fluor-conjugated antibodies (Molecular Probes), and the frequencies of centrosome amplification were calculated by counting 200 cells per group.
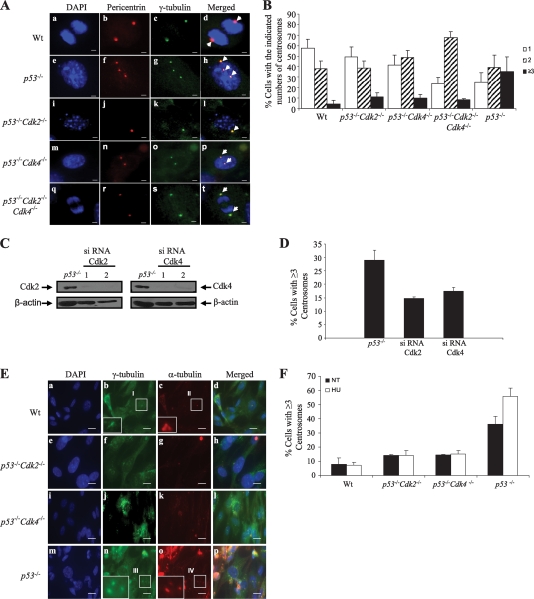
FIG. 4.
Ablation or siRNA-mediated silencing of Cdk2 and Cdk4 prevents centriole reduplication and centrosome amplification in p53−/− MEFs. (A) MEFs of the indicated genotypes were coimmunostained with antibodies recognizing pericentrin (red) (b, f, j, n, and r) and γ-tubulin (green) (c, g, k, o, and s). Nuclei were stained with DAPI (blue) (a, e, I, m, and q). (d, h, l, p, and t) Overlay images of the pericentrin and γ-tubulin immunostaining. Wt, wild type. (B) Proliferating E13.5 mouse embryonic fibroblasts of the indicated genotypes were fixed, processed, and coimmunostained with anti-pericentrin, anti-γ-tubulin, and the appropriate secondary antibodies. The graph presents averages ± standard deviations of the percentages of cells with one, two, and three or more centrosomes. Exactly 8 wild-type, 8 p53−/−, 5 p53−/− Cdk2−/−, 5 p53−/− Cdk4−/−, and 4 p53−/− Cdk2−/− Cdk4−/− embryos were analyzed. t test values for the percentage of cells in each population containing one centrosome (relative to that for the wild type) were 0.017174 for p53−/− MEFs, 0.137854 for p53−/− Cdk2−/− MEFs, 0.358121 for p53−/− Cdk4−/− MEFs, and 3.95E-05 for p53−/− Cdk2−/− Cdk4−/− MEFs. P values for the percentage of cells in each population containing two centrosomes relative to that for wild-type MEFs were 0.860687 for p53−/− MEFs, 0.9713 for p53−/− Cdk2−/− MEFs, 0.024679 for p53−/− Cdk4−/− MEFs, and 2.69E-05 for p53−/− Cdk2−/− Cdk4−/− MEFs. P values for the percentage of cells with ≥3 centrosomes relative to that for wild-type MEFs were 0.006967 for p53−/− MEFs, 0.232114 for p53−/− Cdk2−/− MEFs, 0.706722 for p53−/− Cdk4−/− MEFs, and 0.051209 for p53−/− Cdk2−/− Cdk4−/− MEFs. (C) Western blots of extracts from untransfected p53−/− MEFs or p53−/− MEFs transfected with Cdk2- or Cdk4-specific siRNAs were probed with the indicated primary antibodies. (D) Frequencies of centrosome amplification in control p53−/− MEFs and in p53−/− MEFs in which Cdk2 or Cdk4 was knocked down. Three independent MEFs were used. Centrosomes were detected as for panels A and B. The statistical significance of the averages (P ≤ 0.05) was established by an unequal-variance t test. P values for the percentage of cells with ≥3 centrosomes relative to that of control p53−/− MEFs were 0.002445 for MEFs transfected with a Cdk2-specific SiRNA and 0.006696 for MEFs transfected with a Cdk4-specific siRNA. (E) Proliferating MEFs of the indicated genotypes (3 per group) were either left untreated (NT) or treated with 2 mM HU for 48 h. To determine the presence of centrioles, the cells were subjected to cold treatment and brief extraction prior to fixation. This treatment destabilizes microtubules nucleated at centrosomes; hence, centrioles can be microscopically visualized by immunostaining for α-tubulin (a major component of centrioles) at a high magnification. Cells were coimmunostained with anti-γ-tubulin polyclonal (green) (b, f, j, and n) and anti-α-tubulin monoclonal (red) (c, g, k, and o) antibodies and were counterstained with DAPI (blue) (a, e, i, and m). (d, h, l, and p) Overlaid images of γ-tubulin and α-tubulin immunostaining. (Insets) Magnified images of the areas indicated. (F) Frequencies of centriole reduplication were established by counting cells with ≥3 separated centrioles in a population of at least 200 cells per group. P values for HU-treated compared to NT cells were 0.791492 for wild-type MEFs, 1 for p53−/− Cdk2−/− MEFs, 0.507158 for p53−/− Cdk4−/− MEFs, and 0.012161 for p53−/− MEFs.
Serum starvation and BrdU incorporation assay.
MEFs of the indicated genotypes plated in 60-mm-diameter dishes were first grown to confluence in 10% FBS-DMEM and then split into three groups. The cells in groups 1 and 2 were plated onto two-well tissue culture chamber slides, and those in the third group were plated onto 60-mm-diameter petri dishes. MEFs of different groups were first plated at high densities, then starved for 60 h by culturing in medium supplemented with 0.1% FBS, and finally released by the addition of 10% FBS for various times. The cells in group 1 were immediately fixed for centrosome staining at the time points indicated in Fig. 2. To measure the S-phase entry of the indicated genotypes, MEFs in group 2 were pulse-labeled with 20 μM bromodeoxyuridine (BrdU) (51-7581KZ; BD Pharmingen) and incubated for 30 min as described previously (64, 75). BrdU-positive cells were detected using primary antibodies against BrdU (NA61; Calbiochem) and an Alexa Fluor 555-conjugated secondary antibody (Molecular Probes). We counted 200 cells per group for the BrdU and centrosome assays. Lysates from cells in group 3 were obtained for Western blotting at the time points indicated in Fig. 2.
FIG. 2.
Ablation of Cdk2 or Cdk4 does not significantly alter the cell cycle. (A and B) Cells of the indicated genotypes were arrested in G0 and subsequently stimulated by addition of serum. Cells were pulse-labeled with BrdU 30 min prior to harvest and were harvested at the indicated time points after serum stimulation. Cells were stained with anti-BrdU antibodies and the appropriate secondary antibodies and were visualized using confocal microscopy and a 63× objective. Nuclei were counterstained with DAPI. This experiment was repeated twice; results of a representative experiment are presented. Frequencies represent BrdU-positive cells in a population of at least 200 cells per group. Wt, wild type. (C) Whole-cell extracts were prepared from MEFs collected at the indicated time points following serum addition and were analyzed by Western blotting using antibodies against cyclin A, cyclin D1, cyclin E, and β-actin as a control. Cyc, cyclin. (D) Western blotting was performed with MEFs cultured in DMEM containing 0.2% FBS for 48 h. The numbers 1 and 2 above the lanes represent the loading of the protein lysates of two independent MEFs of the indicated genotypes. Western blots were probed with antibodies against p21Waf1, p27Kip1, p57Kip2, and p16INK4A. β-Actin served as a loading control. (E) Western blots of proteins extracted from controls (wild-type MEFs) or of wild-type MEFs transfected with siRNAs specific to p53 were probed with p53, p57, p16, and β-actin (control).
Immunofluorescence.
Immunofluorescence was performed by following our published protocols (59). MEFs were plated at 4 × 104 per well into two-well tissue culture chamber slides and were grown for 2 to 3 days. Cells were fixed in cold 4% paraformaldehyde, washed in PBS, permeabilized in a 1% NP-40-PBS solution, and blocked in 5% bovine serum albumin (BSA) in PBS. Centrosomes were stained overnight at 4°C with monoclonal antibodies against pericentrin (611814; BD Biosciences) and/or γ-tubulin (ab11317; ABCAM). Chromosome breaks were detected using phosphorylated histone 2A variant X (γ-H2AX) (07-164; Upstate) with the appropriate Alexa Fluor-conjugated antibodies (Molecular Probes). Cells were also counterstained with 4′,6-diamidino-2-phenylindole (DAPI). For each experiment involving calculations of the frequencies of centrosomes, at least 200 cells from each chamber were counted per group.
Western blotting.
Western blotting was performed according to published protocols (59). Protein lysates were obtained by incubating cells in lysis buffer (50 mM HEPES [pH 7.9], 250 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.1% NP-40, 10% glycerol, 0.5 mM NaF, 0.1 mM NaVO4, 0.1 mM phenylmethylsulfonyl fluoride, 10 mM β-glycerophosphate, 0.1 mM dithiothreitol, 0.1 mg/ml aprotinin, 0.1 mg/ml leupeptin) for 30 min at 4°C. Samples were denatured at 95°C for 5 min in sodium dodecyl sulfate (SDS) sample buffer, resolved by SDS-polyacrylamide gel electrophoresis, and transferred to polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA). The blots were incubated in blocking buffer (5% [wt/vol] nonfat dry milk or BSA in Tris-buffered saline plus 0.1% Tween 20 [TBS-T]) for 1 h and were then probed overnight at 4°C with the primary antibodies. The blots were then rinsed in TBS-T and incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies at room temperature. The blots were then rinsed in TBS-T, and the antibody-antigen complex was visualized with the chemiluminescent HRP Antibody Detection reagent (Denville Scientific Inc., Metuchen, NJ). Western blotting for the detection of phosphorylated nucleophosmin was performed similarly, except that the serine/threonine phosphatase inhibitor calyculin A (Upstate, CA) was included in the culture medium 10 min prior to harvest at a concentration of 100 nM. The antibodies used in the various Western blotting experiments were as follows: anti-Cdk2 (sc-163; Santa Cruz), anti-Cdk4 (2906; Cell Signaling), anti-p53 (sc-6243; Santa Cruz), anti-p57Kip2 (sc-8298; Santa Cruz), anti-p16INK4A (sc-1207; Santa Cruz), anti-p21Waf1 (sc-397; Santa Cruz), anti-p27Kip1 (sc-528; Santa Cruz), anti-β-actin (4970; Cell Signaling), anti-cyclin A (ab38; Abcam), anti-cyclin D1 (2922; Cell Signaling), and anti-cyclin E (sc-481; Santa Cruz).
The micronucleus assay.
The micronucleus assay was performed as described previously (59). Briefly, 4 × 104 cells were plated into each well of a two-well chamber slide (177380; Nalge Nunc International). After 2 to 3 days in culture, cells were fixed in 4% paraformaldehyde and were stained with DAPI. Micronuclei appear as spherical structures with a morphology similar to that of the nucleus, except that their sizes range from 1/10 to 1/100 the size of a nucleus; 1,000 cells were counted for each genotype analyzed.
Transfections.
For transient transfection of wild-type and mutant NPM/B23, three independent p53−/− MEFs were cotransfected with a plasmid carrying either FLAG-tagged wild-type NPM/B23 or a FLAG-tagged substitution mutant (Thr199 → Ala) with a neomycin resistance gene (pcDNA3.1) by using Lipofectamine (Invitrogen, Carlsbad, CA). As a negative control, the empty vector was transfected. After transfection at 37°C overnight in a 5% CO2 incubator, cells were fed with fresh complete medium for 24 h. The cells were then treated with complete medium containing 2.5 mg/ml neomycin (Sigma, St. Louis, MO) for 7 days. G418-resistant cells were maintained in complete medium containing neomycin (1 mg/ml) for an additional 2 days and were replated for further culture in fresh complete medium for an additional 24 h.
Three independent Cdk4−/− MEFs were also transfected with a plasmid carrying an empty vector (pBABE-hygro) or encoding Cdk2 (pBABE-Hygro-Cdk2) by using Lipofectamine (Invitrogen) according to the manufacturer's protocol. After transfection (37°C overnight), cells were fed with fresh complete medium for 24 h and were then switched to selective medium (150 μg/ml hygromycin) for 4 days. Selected cells were directly plated onto two-chamber slides, fixed as previously described (59), and stained for centrosome analysis.
RNA interference.
Cdk2 and Cdk4 small interfering RNAs (siRNAs) (catalogue no. SC-29260 and SC-29262) were purchased from Santa Cruz Biotechnology, Inc. Three independent MEFs of each genotype were grown in a six-well plate until they became 60 to 80% confluent. The cells were then transfected with each siRNA according to the manufacturer's protocol, and 72 h after the transfection, the cells were used to prepare cell lysates and were plated onto a two-chamber slide for centrosome analysis.
We also knocked down p53 with two synthesized siRNA duplex sequences by transfecting three independent wild-type MEFs using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. The two siRNA duplex sequences targeting the p53 mRNA are ACCAF161020_1 (sense, 5′-rCrArCrArUrGrUrArCrUrUrGrUrArGrUrGrGrArUrGrGrUrGrGrUrA-3′; antisense, 5′-rCrCrArCrCrArUrCrCrArCrUrArCrArArGrUrArCrArUrGTG-3′) and ACCAF161020_2 (sense, 5′-rGrGrUrGrArArArUrArCrUrCrUrCrCrArUrCrArArGrUrGrGrUrUrU-3′; antisense, 5′-rArCrCrArCrUrUrGrArUrGr GrArGrArGrUrArUrUrUCrACC-3′).
All the siRNAs were prepared using a transcription-based method with the Silencer siRNA construction kit (Integrated DNA Technologies) according to the manufacturer's instructions. Three days after the addition of complete medium, cell lysates were prepared for appropriate assays.
Cyclin-dependent kinase 4 activity assay.
The cyclin-dependent kinase 4 assay was performed according to published methods (67). Briefly, the NPM peptide is phosphorylated because of Cdk4 activity, which is measured using luminometric estimation of ATP depletion. Immunoprecipitated (Ip) Cdk4, obtained from 150 μg of total-protein extract from wild-type and Cdk2−/− MEFs at different time points, was washed three times in cell lysis buffer and twice in kinase buffer and was then resuspended in 30 μl of kinase buffer, optimized to preserve the Cdk4 activity. A kinase reaction was performed by adding 20 μl of a mixture containing 0.5 μM ATP and 2 μg of NPM peptide (catalog no. ab39518; Abcam) as a substrate in kinase buffer. The reaction mixture was incubated for 30 min at 30°C, and then an equal volume of Kinase-Glo reagent was added. As reaction controls, the kinase reaction was performed with samples in the absence of the peptide substrate (no NPM), with no Ip-Cdk4, and with Ip-Cdk4 obtained from total-protein extracts from Cdk4−/− MEFs. Samples were then incubated for 20 min at room temperature, and the developed luminescence was recorded using the SpectraMax Gemini XS luminometer and expressed as relative light units.
Image acquisition and manipulation.
Slides were analyzed using a Zeiss Axioplan II microscope with a Plan-Apochromat 100× (numerical aperture, 1.4) oil immersion objective. Images were taken using a color digital camera (Axiocam HRC) and Zeiss Axiovision software. Confocal images were acquired with a Zeiss LSM 510 META point scanning laser confocal microscope mounted on a Zeiss Axioplan II upright microscope equipped with a Plan-Apochromat 63× (numerical aperture, 1.4) oil immersion objective. Images were captured by Zeiss Image Browser. All the samples were mounted in Fluoromount-G mounting medium (Southern Biotech) and were analyzed at room temperature.
RESULTS
Mouse embryonic fibroblasts proliferate despite the absence of Cdk2 and Cdk4.
To investigate how Cdk2 and Cdk4 individually or synergistically contribute to the regulation of normal centrosome duplication and how they mediate centrosome amplification, wild-type, p53−/−, Cdk2−/−, Cdk4−/−, Cdk2−/− Cdk4−/−, p53−/− Cdk2−/−, p53−/− Cdk4−/−, and p53−/− Cdk2−/− Cdk4−/− embryonic day 13.5 (E13.5) mouse embryonic fibroblasts (MEFs) were generated. Genotypes were determined by allele-specific PCR (Fig. 1A). Western blotting confirmed the presence or absence of p53, Cdk2, and Cdk4 (Fig. 1B). To establish whether Cdk2 and Cdk4 were upregulated as compensatory mechanisms for their loss, expression levels of Cdk2 in Cdk4−/− cells and of Cdk4 in Cdk2−/− cells were examined using wild-type cells as controls (Fig. 1C). There is no compensatory upregulation of Cdk2 or Cdk4 in Cdk4−/− or Cdk2−/− MEFs; their levels are the same as those in wild-type controls.
FIG. 1.
Genetic ablation of p53, Cdk2, or Cdk4 leads to absence of the respective protein expression. (A) PCR-based genotyping. Results of PCR analysis of genomic liver DNA from E.13.5 embryos generated by crossing p53+/− Cdk2+/− or p53+/− Cdk4+/− mice are shown. These gels included five double mutants (p53−/− Cdk2−/− [left panel] or p53−/− Cdk4−/− [right panel]), one Cdk2−/− mutant, one Cdk4−/− mutant, wild-type (Wt) embryos, and a control lacking DNA (H2O). M, molecular size marker; KO, knockout. (B) To confirm the genotyping data generated in panel A, Western blotting was performed using antibodies specific to p53, Cdk2, and Cdk4. β-Actin was used as a loading control (bottom). (C) Western blotting was conducted to determine the expression levels of Cdk4 in Cdk2−/− MEFs and of Cdk2 in Cdk4−/− MEFs. To ensure that equal amounts of proteins were loaded, β-actin was used to probe the same membrane.
Published observations indicate that early-passage Cdk2−/− and Cdk4−/− MEFs proliferate and that the kinetics of entry into S phase in Cdk2−/− or Cdk4−/− MEFs are moderately delayed relative to those of wild-type MEFs (5, 72). Additionally, Cdk2−/− Cdk4−/− MEFs senesce at earlier passages than the individual knockouts (5, 6, 52, 72). Ablation of p53 abrogates senescence associated with the single or combined loss of Cdk2 and Cdk4 at late passages (53). To rule out the possibility that any reductions in the frequency of centrosome amplification in p53−/− MEFs lacking Cdks are associated with major changes in the frequencies of proliferation, the G1/S transition in each genotype was investigated. The MEFs indicated in Fig. 2 were plated, serum starved, stimulated with serum, and harvested at the indicated time points for measurement of S-phase entry by BrdU incorporation (data available on request). Most MEFs peaked with 30 to 50% of cells in S phase at 12 h (Fig. 2A). In contrast, Cdk2−/− Cdk4−/− MEFs entered S phase with delayed kinetics, and fewer than 10% of the cells were actively proliferating at 16 h. Likewise, all MEFs lacking p53 and the Cdks reached their peak frequency of cells in S phase at 12 h (Fig. 2B).
Our goal was to identify deregulated cell cycle-regulatory molecules that may trigger the centrosome amplification observed in p53−/− MEFs. Various defects in the cell cycle-regulatory machinery result in deregulated Cdk activities and centrosome amplification. Those include overexpression of cyclins A, E, and D (27, 48-50, 59). To further investigate the molecular consequences of Cdk loss for regulatory molecules governing the G1/S transition, and whether deregulation of various cyclins accounted for centrosome amplification in p53-null cells, Western blotting was performed to analyze the expression of cyclin A, cyclin D1, and cyclin E at the indicated time points (Fig. 2C). Levels of cyclin E were robust throughout the cell cycle, but its accumulation was decreased in Cdk2−/− and Cdk2−/− Cdk4−/− MEFs from 4 to 12 h post-serum addition. Cyclin D1 levels reached maximal accumulation at 12 and 16 h. No major changes in cyclin D1 levels were observed, except for moderately diminished levels in Cdk4−/−, Cdk2−/− Cdk4−/−, p53−/− Cdk4−/−, and p53−/− Cdk2−/− Cdk4−/− MEFs relative to those in MEFs of other genetic groups at 12 and 16 h. Cyclin A expression was low in all the MEFs before 8 h post-serum addition; its expression levels peaked at 16 h post-serum addition, with similar expression in all MEFs. We conclude that except for minor changes in the expression of cyclins E and D1 when their respective catalytic partners, Cdk2 and Cdk4, are ablated, the temporal cyclin expression patterns are what we would expect of a normal cell cycle, consistent with the similar kinetics of entry into S phase for all genetic groups.
Reduced expression of certain CKIs can also result in elevated frequencies of centrosome amplification, and their overexpression can suppress centrosome amplification (42, 66, 68). For example, ectopic expression of p16INK4A, a Cdk4-specific inhibitor, prevents centriole duplication and centrosome amplification (42, 44). On the other hand, p27Kip1, recently reported to be a dual Cdk2 and Cdk4 inhibitor (54), prevents centrosome amplification triggered by gamma-irradiation (36, 66). In addition, ablation of p21Waf1, a Cdk2-specific inhibitor at physiological levels and a Cdk4 inhibitor at higher levels (29, 30, 35), results in centrosome amplification; its ectopic expression partly suppresses centrosome amplification in p53−/− MEFs (13, 39, 68). To determine whether CKIs may deregulate Cdk2 and Cdk4 in p53−/− MEFs, leading to centrosome amplification, we probed Western blots with antibodies against p21Waf1, p27Kip1, p57Kip2, and p16INK4A (Fig. 2D). To detect constitutive signaling triggered by the absence of p53, Western blotting was performed on serum-starved cells. Those analyses showed that in p53−/− MEFs, endogenous p21Waf1 was undetectable. In contrast, p27Kip1 levels in p53−/− MEFs were unchanged from those in wild-type and other MEFs. p57Kip2 and p16INK4A were overexpressed in p53−/− MEFs.
We also examined the expression level of p57Kip2 and p16INK4A by siRNA-mediated silencing of p53 in wild-type MEFs (Fig. 2E). p53-specific siRNA duplex sequences were synthesized and used to knock down the p53 gene in three independent wild-type MEFs. Western blot analysis revealed that depletion of p53 in wild-type cells did not lead to a major elevation of p57Kip2 levels but that steady-state p16INK4A levels were moderately increased. We conclude that the only major alteration in a cell cycle-regulatory molecule associated with centrosome amplification in p53−/− MEFs is the absence of p21Waf1, consistent with published results (68). We also conclude that the reported high frequencies of centrosome amplification in p53−/− MEFs (22) occurred despite robust levels of CKIs controlling Cdk2 and Cdk4 activities, including p16INK4A and p27Kip1, potent inhibitors of centrosome amplification and centriole duplication (36, 42, 44, 66).
Cells lacking Cdk2 and Cdk4 display abnormal centrosome cycles.
MEFs devoid of Cdk2 undergo minor defects in centriole duplication (14). This suggested to us that either the Cdks regulating the centrosome duplication cycle are redundant, as is the case with the Cdks regulating S-phase entry (2, 5, 6, 38, 52, 61, 72), or other Cdks or centrosomal kinases are solely responsible for orchestrating normal centrosome duplication. Since centrosome duplication licensing must be coordinated with entry into S phase (the latter initiated by cyclin D/Cdk4 and continued by cyclin E/Cdk2), and based on the brief expression overlap between cyclin D/Cdk4 and cyclin E/Cdk2 activities at the G1/S transition (37), we hypothesized that cyclin D1/Cdk4 is involved in that coordination. To assess the involvement of Cdk4 in normal centrosome duplication, and to explore whether Cdk2 and Cdk4 cooperate to affect centrosome duplication, we measured the frequencies of cells with 1, 2, or ≥3 centrosomes in early-passage (passage 2) MEFs devoid of Cdk2 and/or Cdk4 by using immunohistochemistry with antibodies against pericentrin and γ-tubulin, core components of the centrosome (Fig. 3A). Previous studies showed that normal ratios of centrosomes in wild-type MEFs are 60% cells with one centrosome to 40% cells with two centrosomes (68). Any deviation in the ratios of centrosomes within a population is indicative of defects in the various steps driving the centrosome duplication cycle: licensing, separation of centrioles, and duplication of centrioles (59, 68). A centrosome ratio favoring cells with one centrosome is indicative of defective licensing of the centrosome cycle, or centriole separation, while a centrosome ratio favoring cells with two centrosomes is indicative of premature centriole separation and duplication. Consistent with published results, Cdk2−/− MEFs did not display a statistically significant deviation in centrosome ratios from wild-type MEFs (48:46% versus 58:38%, respectively). In contrast, Cdk4−/− MEFs showed a significant divergence from wild-type MEFs in the ratio of cells with one centrosome to cells with two centrosomes (77:20% versus 58:38%). This accumulation of cells with one centrosome is not due to a longer G1 phase, since Cdk4−/− MEFs entered S phase with kinetics similar to those of wild-type MEFs (as presented in Fig. 2A). In addition, Cdk2−/− Cdk4−/− MEFs also displayed a severe deviation from wild-type MEFs in centrosome ratios (35:55% versus 58:38%).
FIG. 3.
Ablation of Cdk2 and Cdk4 or siRNA-mediated silencing of Cdk2 and Cdk4 leads to distinct centrosome cycle defects. (A) Proliferating E13.5 mouse embryonic fibroblasts of the indicated genotypes were fixed, processed, and coimmunostained with anti-pericentrin, anti-γ-tubulin, and the appropriate secondary antibodies. Averages ± standard deviations of percentages of cells with one, two, and three centrosomes are shown. Exactly 8 wild-type (Wt), 3 Cdk2−/−, 4 Cdk4−/−, and 3 Cdk2−/− Cdk4−/− embryos were analyzed. The statistical significance of the averages (P ≤ 0.05) was established by an unequal-variance t test. t test values for the percentages of cells in each population containing one centrosome relative to that for the wild type were 0.159885 for Cdk2−/− MEFs, 0.000518 for Cdk4−/− MEFs, and 0.000544 for Cdk2−/− Cdk4−/− MEFs. The P values for the percentages of cells in each population containing two centrosomes relative to that of wild-type MEFs were 0.122182 for Cdk2−/− MEFs, 0.000172 for Cdk4−/− MEFs, and 0.000528 for Cdk2−/− Cdk4−/− MEFs. The P values for the percentages of cells with ≥3 centrosomes relative to that of wild-type MEFs were 0.091487 for Cdk2−/− MEFs, 0.06122 for Cdk4−/− MEFs, and 0.000808 for Cdk2−/− Cdk4−/− MEFs. (B) MEFs of the indicated genotypes were grown in duplicate to confluence, followed by serum starvation for 60 h. Quiescent cells were stimulated with serum, and every 4 h for a period of 16 h, the numbers of cells with one and two centrosomes were scored. This experiment was repeated twice; results of a representative experiment are presented. The BrdU data are the same as those presented in Fig. 2A and B; they are shown here for purposes of clarity. (C) Western blots of proteins extracted from nontransfected wild-type cells, or from cells transfected with siRNAs against Cdk2 or Cdk4, and probed with antibodies against Cdk2 or Cdk4. The same membrane was probed with β-actin as a control. (D) Wild-type MEFs that either were left untransfected or were transfected with siRNAs against Cdk2 or Cdk4 were immunostained with anti-γ-tubulin antibodies, and frequencies were established by counting cells with one and two centrosomes in a population of at least 200 cells per group. Three independent MEF groups were used. The statistical significance of the averages (P ≤ 0.05) was established by an unequal-variance t test. t test values for the percentage of cells in each population containing one centrosome relative to the percentage with two centrosomes were 0.215535 for wild-type MEFs, 0.003271 for MEFs transfected with a siRNA against Cdk2, and 0.008772 for those transfected with a siRNA against Cdk4. (E) Three independent Cdk4−/− MEFs transfected with plasmids carrying either a control vector (pBABE-hygro) or pBABE-hygro-Cdk2 and plated after selection with hygromycin were immunostained using anti-γ-tubulin antibodies and the appropriate secondary antibodies. Frequencies were established by counting cells with one and two centrosomes in a population of at least 200 cells per group. t test values of the percentage of cells in each population containing one centrosome relative to the percentage with two centrosomes were 0.345271 for pBABE-hygro-transfected MEFs and 0.136406 for pBABE-hygro-Cdk2-transfected MEFs.
To assess the phase in the cell cycle at which the various centrosome defects occurred, we dissected the centrosome duplication kinetics in cells by comparative analysis of synchronized wild-type, Cdk2−/−, Cdk4−/−, and Cdk2−/− Cdk4−/− cells (Fig. 3B). MEFs of the indicated genotypes were grown in duplicate, followed by serum starvation for 60 h. Quiescent cells were stimulated with serum, and BrdU was included in the medium to monitor S-phase entry. Every 4 h for a period of 16 h, BrdU incorporation and the number of cells within the population with one or two centrosomes were scored. In accordance with the kinetics of entry into S phase, the percentage of wild-type MEFs with two centrosomes peaked at 12 h post-serum stimulation. Cdk2−/− MEFs reached maximal ratios of cells with two centrosomes earlier than wild-type MEFs, at 8 h poststimulation. In contrast to wild-type or Cdk2−/− MEFs, the percentages of Cdk4−/− MEFs with two centrosomes decreased throughout the cell cycle, as the cells steadily accumulated a centrosome content of one. Another intriguing defect was that in Cdk2−/− Cdk4−/− MEFs, in which ablation of Cdk2 overrode the defect in centrosome ratios in Cdk4−/− MEFs. Even though Cdk2−/− Cdk4−/− MEFs were unable to replicate their DNA efficiently, their centrosome duplication peaked at 8 h post-serum stimulation; thus, the centrosome and cell cycles were uncoupled in those cells.
We then explored whether transient downregulation of Cdk2 and Cdk4 with siRNAs in wild-type MEFs recapitulated the centrosome cycle defects in Cdk2−/− or Cdk4−/− MEFs (Fig. 3C and D). Wild-type MEFs were transfected with siRNA duplexes against Cdk2 or Cdk4. Seventy-two hours after transfection, cell lysates were obtained, and centrosome analyses were performed. Western blot analysis of the extracts from the transfected cells showed significant depletion of Cdk2 and Cdk4 (Fig. 3C). In contrast to the Cdk2−/− centrosome profile, which showed normal ratios of centrosomes, siRNA-mediated downregulation of Cdk2 led to the accumulation of cells with two centrosomes; however, as with Cdk4−/− MEFs, depletion of Cdk4 promoted more cells with one centrosome (Fig. 3D). Our results have identified one of the major triggers for normal centrosome duplication, which involves Cdk4, as well as the cooperation of Cdk2 and Cdk4 activities. Experiments that knocked out and knocked down Cdk2 or Cdk4 suggested that the functions of those Cdks are unique. To address whether ectopic expression of Cdk2 rescued the accumulation of cells with one centrosome observed for Cdk4−/− MEFs, we overexpressed Cdk2 in Cdk4−/− cells. As shown in Fig. 3E, Cdk2 significantly rescued the centrosome defects imposed by ablation of Cdk4, demonstrating that the centrosome defect imparted by ablated Cdk4 is reversed by overexpression of Cdk2; in this scenario, Cdk2 and Cdk4 are redundant.
Individual ablation of Cdk2 or Cdk4 abolishes centrosome amplification in p53−/− MEFs by preventing excessive centriole duplication.
The current models attempting to explain how the absence of p53 allows centrosome amplification propose that elevated Cdk2 activity is primarily responsible for centrosome amplification in those MEFs (21, 68). This was suggested by observations that p21Waf1−/− cells have elevated frequencies of centrosome amplification or that ectopic expression of p21Waf1 partly restored normal centrosome frequencies in p53−/− MEFs (13, 68). Since p21Waf1 influences cyclinD/Cdk4 positively by promoting its assembly but inhibits it at higher concentrations (3, 29, 30, 35), we speculated that Cdk4 may also mediate centrosome amplification in p53−/− MEFs. Therefore, we set out to explore the relative contributions of Cdk2 and/or Cdk4 to centrosome amplification in p53−/− MEFs. While wild-type cells did not display elevated frequencies of centrosome amplification, loss of p53 resulted in 40% of the cells displaying centrosome amplification (Fig. 4A and B). As predicted, ablation of Cdk2 in p53−/− MEFs prevented centrosome amplification (by approximately 70%). To our surprise, since no one has reported elevated Cdk4 activity in p53−/− MEFs, and since our Western blots presented in Fig. 2 did not reveal any changes in any cyclins or CKIs that might promote the deregulation of Cdk4, ablation of Cdk4 suppressed centrosome amplification in p53−/− MEFs to the same extent as ablation of Cdk2. To establish whether Cdk2 and Cdk4 cooperated to further decrease centrosome amplification, we calculated the frequencies of centrosome amplification in p53−/− Cdk2−/− Cdk4−/− MEFs. Indeed, p53−/− Cdk2−/− Cdk4−/− MEFs displayed frequencies of centrosome amplification similar to those of p53−/− Cdk2−/− or p53−/− Cdk4−/− MEFs. We next used siRNAs to silence the expression of Cdk2 or Cdk4 in p53−/− MEFs (Fig. 4C and D). Western blotting indicated that most of the Cdk2 or Cdk4 was depleted relative to the levels in the controls (Fig. 4C). Indeed, as with the combinatorial knockouts, siRNA-mediated inhibition of Cdk2 or Cdk4 suppressed centrosome amplification in p53−/− MEFs (Fig. 4D). We conclude that Cdk2 and Cdk4 are individually required to mediate centrosome amplification.
In normal centrosome duplication, cells enter G1 with a single centrosome composed of two centrioles: the mother centriole (older) and the daughter centriole (newer). To directly test whether Cdk2 and Cdk4 regulate centriole duplication, we performed a centriole reduplication assay. This assay involves challenging cells with hydroxyurea (HU) for 48 h, which inhibits DNA synthesis at late G1/early S phase. Since centrosomal checkpoints are functional in wild-type cells, centrosome reduplication is predicted to be absent in those cells (44). On the other hand, cells lacking certain checkpoint controls, such as those cells in which p53 is ablated, continue duplicating their centrioles within the centrosomes, resulting in multiple centrioles. For the centriole reduplication assay, proliferating MEFs of the indicated genotypes were either left untreated or treated with HU for 48 h. To determine the presence of centrioles, the cells were subjected to cold treatment and brief extraction prior to fixation. This treatment destabilizes microtubules nucleated at centrosomes; hence, centrioles can be microscopically visualized by immunostaining for α-tubulin, a major component of centrioles, at a high magnification. Coimmunostaining of cells subjected to cold treatment and brief extraction with anti-γ-tubulin (which detects the pericentriolar material [PCM]) and anti-α-tubulin revealed that wild-type cells stopped centrosome duplication after 48 h in culture in the presence of HU, while p53−/− cells continued centrosome duplication (Fig. 4E and F). On the other hand, ablation of either Cdk2 or Cdk4 in p53−/− cells completely halted centriole reduplication. The same experiment was repeated with 48-h cultures in the presence of mimosine or HU, with immunostaining done with γ-tubulin, and the results were the same: we detected the accumulation of centrosomes in p53−/− MEFs, and ablation of Cdk2 or Cdk4 prevented that accumulation (data not shown). These experiments demonstrated that ablation of Cdk2 and Cdk4 suppresses centrosome amplification by normalizing the centrosome reduplication defect triggered by ablation of p53. Importantly, this result identified one of the specific steps in the centrosome cycle affected by the absence of Cdk2 or Cdk4: centriole duplication.
Genetic ablation of Cdk2 or Cdk4 suppresses chromosome instability in p53−/− MEFs.
Ablation of p53 generates chromosome instability through centrosome amplification (22, 23, 43) and by allowing the generation of reactive oxygen species, which are predicted to result in double-strand DNA breaks (60). To establish how the absence of Cdk2 and Cdk4 modulates active chromosome instability in p53−/− MEFs, we used two assays: the micronucleus assay and γ-H2AX immunostaining. One of the initial cellular responses to the introduction of double-strand breaks is the phosphorylation of serine 139 of the carboxy-terminal tail of H2AX (56). The number of phosphorylated H2AX (γ-H2AX) molecules increases linearly with the severity of damage. Therefore, this assay represents a way to mark double-strand DNA breaks, a precursor to the chromosome breaks and recombinations leading to structural chromosomal abnormalities, the second major form of chromosome instability. Control experiments revealed that Adriamycin, a chemical that promotes DNA breaks, resulted in most cells in the population containing γ-H2AX foci (not shown). As shown in Fig. 5A and B, loss of p53 resulted in a significant elevation of the percentage of cells with γ-H2AX foci over that for wild-type controls. In contrast, ablation of Cdk2 or Cdk4 in p53−/− cells decreased the number of cells containing γ-H2AX relative to that observed for wild-type cells, consistent with published observations that loss of Cdk2 reduces DNA repair (62).
FIG. 5.
Ablation of Cdk2 and Cdk4 inhibits chromosome instability in cells lacking p53. (A and B) The frequencies of γ-H2AX foci (arrows) in cells with the indicated genotypes were calculated. Bars, 10 μm. The graph (B) shows the average percentage of γ-H2AX foci in each population of at least 200 cells. Error bars, standard deviations. Each group included 4 different MEFs. P values (relative to the wild-type control) were 0.015218 for p53−/− MEFs, 0.126173 for p53−/− Cdk2−/− MEFs, and 0.346771 for p53−/− Cdk4−/− MEFs. (C and D) Proliferating E13.5 mouse embryonic fibroblasts of the indicated genotypes were fixed, and nuclei were visualized with DAPI. Frequencies of micronucleus (insets and arrows) formation were calculated for at least 500 cells of each of the indicated genotypes. Each group included 4 different MEFs. P values (relative to the wild-type control) were 0.016122 for p53−/− MEFs, 0.137054 for p53−/− Cdk2−/− MEFs, and 0.370282 for p53−/− Cdk4−/− MEFs.
Our second assay to detect active genomic instability was the micronucleus assay. A micronucleus is a chromosome or chromosome fragment missegregated during mitosis as a consequence of spindle damage or as a consequence of lost centromeric sequences (acentric chromosomes are unable to bind mitotic fibers and are excluded from the segregating chromatids) (31, 46, 69). Following cytokinesis, micronuclei appear in the cytoplasm as DNA-containing spheres surrounded by a nuclear membrane (Fig. 5C, arrows). The extent of micronucleus formation reflects the frequency of cells in a population actively losing whole chromosomes (a type of chromosome instability dependent on centrosome amplification), as well as the frequency of fragmented chromosomes, which arise as a result of DNA breaks (57, 58). Our results indicated that ablation of Cdk2 or Cdk4 reduced micronucleus formation in p53−/− MEFs to wild-type levels (Fig. 5D). We conclude that the absence of Cdk2 or Cdk4 prevents chromosome instability, an abnormal phenotype strongly associated with tumor biogenesis and progression (1, 11, 16, 17, 20).
Cdk2 and Cdk4 signal centrosome amplification and chromosome instability through a common phosphorylation site in nucleophosmin, Thr199.
The nucleophosmin (NPM) protein, located at the centrosome, prevents premature centriole separation and duplication during early G1 similarly to the way in which nuclear Rb prevents premature entry from early G1 into S phase. Both proteins are phosphorylated in late G1 by the Cdks to relieve negative regulation (37, 51). Unphosphorylated NPM binds to unduplicated centrosomes (centrosomes with closely associated centrioles) to prevent premature centriole separation and duplication (51, 70). Upon phosphorylation by Cdk2 at the G1/S transition on T199, NPM disassociates from the centrosome, allowing centriole separation and duplication (51, 70). Our laboratory previously correlated unregulated cyclin E/Cdk2 activity with phosphorylation of NPM at T199 (59). We demonstrated that cyclin E/Cdk2 phosphorylation of NPMT199 in G0 rather than in late G1 rendered NPM functionally inactive, since it no longer had the ability to bind centrosomes at G0. This inability to bind centrosomes allowed a constitutive centrosome duplication cycle, where centrioles separated and duplicated uncontrollably, resulting in centrosome amplification. Since the loss of Cdk4 prevented centrosome amplification in p53−/− MEFs as efficiently as the ablation of Cdk2, we set out to establish whether they shared the same phosphorylation site in NPM to signal centrosome amplification and chromosome instability. Specifically, we tested whether suppression of centrosome amplification by ablation of Cdk2 and Cdk4 correlated with restoration of normal phosphorylation of NPMT199. MEFs were serum starved to mimic G0, and the phosphorylation status of NPMT199 was assessed by Western blotting (Fig. 6A). The following results provided important clues as to the mechanism by which ablation of Cdk2 or Cdk4 prevented centrosome amplification in p53−/− MEFs. First, while wild-type cells displayed a baseline level of NPM phosphorylation, ablation of Cdk2 resulted in a level of phosphorylation lower than that in wild-type cells. The relative level of phosphorylation of NPM in Cdk4−/− MEFs was identical to that in wild-type MEFs. Second, ablation of p53 resulted in constitutive phosphorylation of NPMT199; importantly, the absence of Cdk2 or Cdk4 reduced the hyperphosphorylation of NPMT199 in p53−/− MEFs to wild-type levels. These results suggested that ablation of Cdk2 or Cdk4 abrogated centrosome amplification in p53−/− MEFs by restoring the normal phosphorylation of NPMT199 and reestablishing normal centrosome duplication licensing. To test whether the Thr-199 phosphorylation of NPM in p53−/ MEFs was reduced by ablation of Cdk2 or Cdk4 under the conditions used for the centriole reduplication assay, MEFs were treated with HU for 48 h, followed by protein extraction. As shown in Fig. 6B, Western blotting indicated that in HU-arrested cells, ablation of Cdk2 and/or Cdk4 reduced the level of NPMT199 phosphorylation from that for p53−/− MEFs.
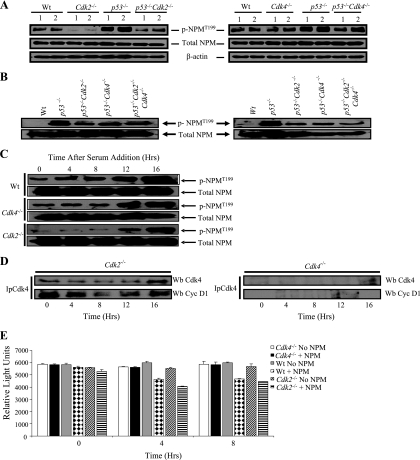
FIG. 6.
Cdk2 and Cdk4 affect the centrosome cycle and centrosome amplification through NPM. (A) E13.5 MEFs of the indicated genotypes were serum starved for 60 h. Cells were preincubated with calyculin A, a serine/threonine phosphatase inhibitor. (Top) Western blot analysis of protein fractions of G0-arrested MEFs probed with antibodies against phospho-NPMT199 (p-NPMT199). (Center and bottom) Blots were probed with antibodies against total nucleophosmin and β-actin to show equal loading. (B) MEFs of the indicated genotypes were treated with 2 mM HU for 48 h and were then preincubated with calyculin A, a serine/threonine phosphatase inhibitor, before protein extraction. Western blots of the protein extracts were probed with antibodies against NPMT199 or against total NPM (control). The MEFs for the left panel are independent of those for the right. (C) Western blot analyses of MEFs of the indicated genotypes that were serum arrested and released into the cell cycle for various times. Western blots of the protein extracts were probed with antibodies against NPMT199 or against total NPM (control). (D) Western blots of Cdk4 immunoprecipitated (Ip-Cdk4) from extracts of wild-type, Cdk2−/−, or Cdk4−/− MEFs were probed with antibodies against Cdk4 and cyclin D1. (E) Cdk4 kinase assays of protein lysates from wild-type, Cdk2−/−, and Cdk4−/− MEFs were carried out at various time points following serum addition. Results for the 0-, 4-, and 8-h time points are shown. The results are from three independent MEFs. The experiment was repeated at least twice, and results of a representative experiment are presented. The reaction mixtures either contained NPM peptide (+ NPM) or contained no NPM peptide (no NPM). Luminescence was recorded by the SpectraMax Gemini XS luminometer using the SoftMax program. P values of kinase assays comparing NPM to no NPM at each indicated time point were 0.107203829 for lysates from wild-type MEFs at 0 h, 0.037437678 for those from Cdk2−/− MEFs at 0 h, 0.000111335 for those from wild-type MEFs at 4 h, 0.002861355 for those from Cdk2−/− MEFs at 4 h, 0.000761355 for those from wild-type MEFs at 8 h, and 0.000449084 for those from Cdk2−/− MEFs at 8 h. The P values from the kinase assays performed with Cdk4−/− MEFs were greater than 0.05 at any given time point.
NPM/B23 is a direct substrate of Cdk2 (51). To establish the status of NPMT199 phosphorylation in wild-type, Cdk2−/−, and Cdk4−/− MEFs throughout the cell cycle, we performed Western blotting at various time points following release from serum starvation (Fig. 6C). Phosphorylation of NPMT199 was moderately lower in Cdk4−/− MEFs than in wild-type MEFs at 4 and 8 h. In contrast, phosphorylation of NPMT199 in Cdk2−/− MEFs between 0 and 12 h was severely diminished. Because Western blotting suggested that NPM was indeed a target of Cdk4, we set out to demonstrate that Cdk4 can directly phosphorylate NPM. To that end, wild-type, Cdk2−/−, and Cdk4−/− MEFs were serum starved, stimulated by the addition of serum, and harvested at different time points, followed by protein extraction. The presence of Cdk4 (34 kDa) and its cofactor cyclin D1 (36 kDa) in Ip-Cdk4 was demonstrated by analysis of Western blots probed with anti-cyclin D1 and anti-Cdk4 antibodies (Fig. 6D). As expected, immunoprecipitations using antibodies against Cdk4 and cyclin D1 with Cdk4−/− MEFs did not pull down any Cdk4 or cyclin D1, demonstrating the specificity of the antibodies. After showing that the antibodies specifically pulled down Cdk4, we performed luminescent kinase assays with the Kinase Glo kit (Promega), as previously described (67), immunoprecipitating cyclin D/Cdk4 complexes with antibodies against Cdk4 and incubating immunoprecipitates with purified NPM (Fig. 6E). In this assay, reduction of luminescence by the addition of kinase buffer and purified NPM peptide to the Cdk4 immunoprecipitates indicates kinase activity. To rule out the possibility that any contaminating Cdk2 activity would result in phosphorylation of NPM, we performed immunoprecipitation with synchronized Cdk2−/− MEFs. As an additional control, we performed kinase assays with Cdk4−/− MEFs. For extracts of wild-type or Cdk2−/− MEFs from 4 and 8 h post-serum stimulation, considerable kinase activity was detected in samples containing NPM peptide compared to samples without the substrate (no NPM) (Fig. 6E). This phosphorylation was slightly reduced, but still statistically significant, in wild-type MEFs at 12 and 16 h after serum addition (data available on request). We then validated our experiment by including Ip-Cdk4 from extracts of Cdk4−/− MEFs at the indicated time points; the extract from the Cdk4−/− MEFs where no Cdk4 was precipitated showed no kinase activity compared to samples without the substrate (no NPM). These results demonstrated that NPM is a Cdk4 target and that the maximal phosphorylation corresponded to the point of the cell cycle where centrosome cycle licensing occurs: mid/late G1. However, unlike the phosphorylation of NPM by Cdk2, which occurs both in the nucleus and in the cytoplasm (59), due to a centrosome localization signal in cyclin E (41), phosphorylation of NPM by Cdk4 is predicted to be nuclear, because localization of Cdk4 and of cyclin D1 was strictly nuclear, and we did not find any Cdk4 or cyclin D1 within centrosomes (data available on request).
To gather more-direct evidence that the phosphorylation of NPMT199 by Cdk2 and Cdk4 is critical to centrosome amplification, plasmids encoding FLAG epitope-tagged wild-type NPM and the mutant NPM/B23 lacking the T199 phosphorylation site (NPMT199A) were transfected into E13.5 MEFs derived from p53 mutant embryos (Fig. 7). As control, the vector alone was transfected. Following neomycin selection, cells were examined for the level of expression of exogenous NPM/B23 by Western blot analysis using an anti-FLAG antibody. This analysis revealed that NPM- and NPMT199A-transfected cells expressed similar protein levels (Fig. 7A). In addition, we set out to establish whether wild-type NPM or NPMT199A modulated the frequencies of centrosome amplification in p53−/− MEFs (Fig. 7B). This analysis revealed that while p53−/− MEFs carrying the vector control and those expressing wild-type NPM had the same frequencies of centrosome amplification, the mutant lacking the G1 Cdk phosphorylation site displayed greatly reduced frequencies of centrosome amplification. The extent of inhibition was almost identical to that in p53−/− Cdk2−/− or p53−/− Cdk4−/− MEFs (as presented in Fig. 4B). In summary, we have demonstrated that Cdk2 and Cdk4 hyperphosphorylate NPM and that this hyperphosphorylation is critical to centrosome amplification.
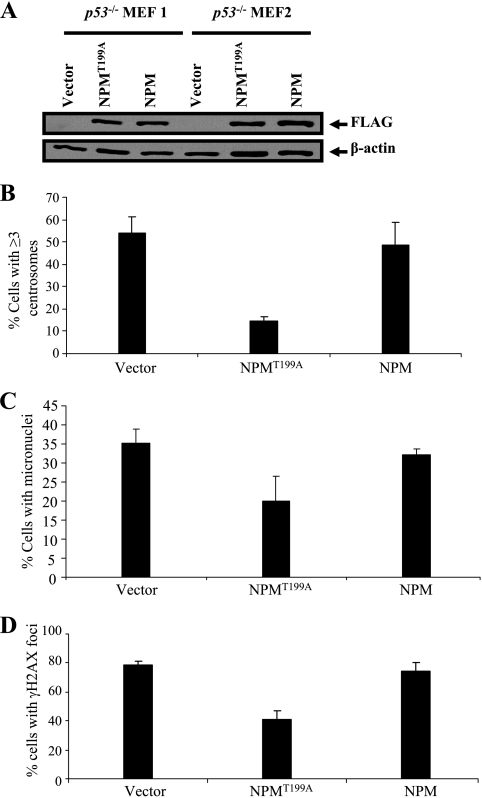
FIG. 7.
NPMT199A suppresses centrosome amplification and chromosome instability. (A) Passage 2 p53−/− MEFs were transiently transfected with plasmids encoding FLAG epitope-tagged NPM and the NPM/B23 mutant (NPMT99A). As a control, an empty vector was transfected. After neomycin selection, cell lysates were obtained and then probed with anti-FLAG antibodies. (B) The transfectants described in the legend to panel A were fixed, immunostained with anti-γ-tubulin polyclonal antibodies, and detected with Alexa Fluor 488-conjugated secondary antibodies. Cells were counterstained with DAPI. The number of cells with ≥3 centrosomes in a population of at least 200 cells was statistically analyzed by fluorescence microscopy. Each group included three transfected MEFs. P values (relative to the result for the transfected vector control) were 0.006963 for NPMT199A and 0.560677 for wild-type NPM. (C) Proliferating E13.5 mouse embryonic fibroblasts of the indicated genotypes were fixed, and nuclei were visualized with DAPI. The frequencies of micronucleus formation in a population of 500 cells were calculated for the indicated genotypes. P values (relative to the result for the transfected vector control) were 0.011338 for NPMT199A and 0.353737 for wild-type NPM. (D) Frequencies of γ-H2AX foci in cells with the indicated genotypes were calculated. P values (relative to the result for the transfected vector control) were 0.002591 for NPMT199A and 0.476327 for wild-type NPM.
In addition, we investigated whether the suppression of centrosome amplification by NPMT199A restored genomic stability. Cells expressing the vector and wild-type NPM had the same frequencies of micronucleus formation, while those expressing NPMT199A had reduced frequencies (Fig. 7C). Likewise, NPMT199A inhibited the formation of γ-H2AX double-stranded foci (Fig. 7D). We conclude from these experiments that ablation of either Cdk2 or Cdk4 prevents the generation of chromosome breaks and chromosome losses in p53−/− MEFs. We propose a new paradigm for how centrosome amplification and chromosome instability arise in p53−/− MEFs: the presence of both Cdk2 and Cdk4 is absolutely required in order to hyperphosphorylate NPMT199 to generate centrosome amplification and chromosome instability (Fig. 8).
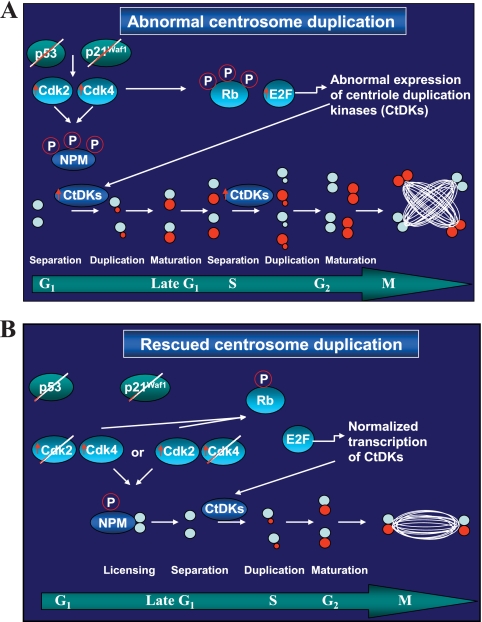
FIG. 8.
Model explaining how the ablation of Cdks prevents centrosome amplification. (A) Ablation of p53 results in undetectable levels of p21Waf1, leading to hyperactive Cdk2 and Cdk4. Hyperactive Cdks cross talk to the centrosome in two modes. First, hyperactive Cdks hyperphosphorylate Rb in the nucleus, leading to uncontrolled E2F-dependent transcription of molecules that influence various steps in the centrosome duplication cycle: those involved in centriole splitting, as well as centriole duplication kinases (CtDKs). Second, hyperactive Cdks constitutively phosphorylate NPMT199, resulting in excessive licensing of centrosome duplication. Uncontrolled expression of CtDKs and the inability of NPM to suppress normal centrosome duplication lead to faster centrosome duplication cycles within a single cell cycle, resulting in the formation of multiple centrosomes. (B) When Cdk2 or Cdk4 is deleted from p53−/− MEFs, Rb is underphosphorylated, and the E2F-dependent transcription of CtDKs is restored. In addition, underphosphorylated NPM restores normal centrosome licensing and prevents excessive centriole duplication. This restricts the centrosome duplication cycle to one per cell cycle, thus resulting in normal centrosome numbers.
DISCUSSION
In this study, we investigated the relative contributions of the G1 Cdks—Cdk2 and Cdk4—to centrosome amplification in p53−/− MEFs and to the regulation of the centrosome cycle. How do Cdk2 and Cdk4 affect the centrosome cycle? Ablation of Cdk2 leads to moderate defects in centrosome duplication, suggesting redundancy in the control of this process (14); our results with asynchronously growing Cdk2−/− MEFs confirmed those results. However, experiments measuring the centrosome cycle at various time points throughout the cell cycle in Cdk2−/− and Cdk4−/− MEFs, as well as transient downregulation of Cdk2 and Cdk4 using siRNAs, uncovered distinct centrosome cycle defects, suggesting that the functions of these Cdks are nonredundant. For example, while Cdk2 deficiency promoted the early separation and duplication of centrosomes, the absence of Cdk4 promoted the accumulation of cells with one centrosome that failed to separate and duplicate. The accumulation of cells with one centrosome in Cdk4−/− MEFs was not compensated for by passage, since the accumulation of cells with one centrosome was also observed in MEFs silenced with a siRNA directed against Cdk4. The accumulation of cells with one centrosome in Cdk4−/− MEFs was not due to a block in G1; these MEFs entered S phase with kinetics similar to those of wild-type and Cdk2−/− MEFs. On the other hand, inhibition of Cdk2 with siRNAs led to many cells in the population harboring two centrosomes, suggesting that that defect was compensated for in Cdk2−/− MEFs by Cdk4, other Cdks, or centrosomal kinases. Our observations showed that concomitant ablation of Cdk2 and Cdk4 caused a severe accumulation of cells with two centrosomes; experiments following the cell and centrosome cycles in Cdk2−/− Cdk4−/− MEFs confirmed that centriole separation and duplication were premature. The fact that the centrosome cycle defect in Cdk2−/− Cdk4−/− MEFs was much more severe than that in other genetic groups suggests that Cdk2 and Cdk4 cooperate to regulate the centrosome cycle. The accumulation of cells with two centrosomes in asynchronously growing Cdk2−/− Cdk4−/− MEFs was not a complete block, since 40% of cells still contained one centrosome. Upon serum arrest, the ratio of the number of Cdk2−/− Cdk4−/− MEFs with one centrosome to that with two centrosomes was similar to that for wild-type MEFs, and upon serum addition, centrosomes separated and duplicated prematurely, indicating an active centrosome cycle in Cdk2−/− Cdk4−/− MEFs. In those MEFs, the cell and centrosome cycles were uncoupled: centriole separation and duplication preceded entry into S phase. Thus, as in the cell cycle, in the absence of Cdk2 and Cdk4, other Cdks or centrosome duplication kinases also play a role in driving the centrosome duplication cycle. That redundancy is also supported by the baseline phosphorylation of NPMT199 in p53−/− Cdk2−/− Cdk4−/− MEFs cultured with HU, demonstrating that NPMT199 is phosphorylated by another kinase in the absence of Cdk2 and Cdk4. Those results indicated that the same redundancy operating within the kinases controlling S phase also exists in the regulation of the centrosome cycle.
Because NPM phosphorylation at Thr199 by Cdk2 is essential to signal entry into the centrosome cycle (51, 70), we assessed whether the centrosome defects observed in Cdk2−/− and Cdk4−/− MEFs were due in part to changes in NPMT199 phosphorylation. As reported previously, NPMT199 is a canonical target of Cdk2; our Western blot analyses revealed that at early time points (0 to 8 h) following the addition of serum to Cdk2−/− MEFs, the phosphorylation of NPMT199 was greatly diminished from that for wild-type MEFs. The phosphorylation of NPMT199 increased in Cdk2−/− MEFs at 12 h following serum addition. The fact that cyclin D1 levels peaked at 12 h of serum addition in Cdk2−/− MEFs indicated that perhaps cyclin D1/Cdk4 was responsible for that phosphorylation. On the other hand, levels of NPMT199 were moderately reduced in Cdk4−/− MEFs at early time points after serum addition (0 to 8 h). Our Cdk4 kinase assays using NPM as a substrate confirmed that NPM is indeed a Cdk4 target and that its phosphorylation is cell cycle regulated. In serum-arrested cells we did not detect phosphorylation of NPM; that phosphorylation commenced at 4 and 8 h following serum addition and was detected up to 16 h after serum addition. However, the fact that the reduction in NPMT199 phosphorylation was not nearly as great in Cdk4−/− MEFs as in Cdk2−/− MEFs suggests that the Cdk2 present in Cdk4−/− MEFs actively phosphorylates NPMT199 and that the phosphorylation of NPMT199 is more efficiently carried out by Cdk2 than by Cdk4.
We speculate that cells have evolved compensatory mechanisms such that when they are devoid of Cdk2, Cdk4 can indeed license the centrosome cycle. However, at this point we cannot rule out the possibility that Cdk2 phosphorylates NPMT199 to prime Cdk4 phosphorylation of NPM in other sites that would control centrosome licensing at late G1. Indeed, there is evidence in the literature that phosphorylation of NPM at Thr234 and Thr237 by Cdk1 stabilizes NPM in the centrosome during M phase (9). A second possible reason why Cdk4−/− MEFs display cells with one centrosome is that Cdk4 may directly phosphorylate and regulate the activity of a centriole separase, or it may regulate the Rb/E2F-dependent transcription of a centriole separase. Centriole separases are poorly understood; one centrosome separase normally active at M phase that displays centriole separase activity when ectopically expressed during interphase is Nek2A (18). A third possibility is that Cdk4 modulates other licenser factors besides NPM. The only other known centrosome-regulatory protein that may display licensing activity is CP110. Like NPM, CP110 is a phosphorylation target of Cdk2, and inhibition of its activity causes centrosome amplification (10). However, unlike NPM, which disassociates from centrosomes upon Cdk phosphorylation, CP110 is present in the centrosome to cap the synthesis of a new centriole (34). Thus, we think it unlikely that Cdk4 utilizes CP110 to license the centrosome cycle. Nevertheless, the activity inhibited by the ablation of Cdk4 that leads to the accumulation of cells with one centrosome is reversible, since ablation of Cdk2 (in Cdk2−/− Cdk4−/− MEFs), ectopic expression of Cdk2, or ablation of p53 does not allow the accumulation of cells with one centrosome.
What causes the accumulation of two centrosomes in cells in which Cdk2 is knocked down, and in cells in which Cdk2 and Cdk4 are ablated? Our experiments following the cell and centrosome cycles in Cdk2−/− MEFs and in Cdk2−/− Cdk4−/− MEFs revealed premature separation and duplication of centrosomes. The premature accumulation of cells with two centrosomes was present in early cell cycle stages of Cdk2−/− MEFs, but the premature accumulation was restored at later passages cycle. Perhaps Cdk2 alone and Cdk2 and Cdk4 together impose negative regulation of centriole separases or of other centrosome kinases. For example, Cdk2 targets other than NPM may prevent the binding of centriole separases to the centriole pair. When Cdk2 alone or both Cdk2 and Cdk4 are ablated, those centriole separases and/or centriole duplication kinases may associate with the centrosomes prematurely to signal early centriole separation and duplication. Other targets of Cdk2 that regulate various steps within the centrosome duplication cycle include CP110 and mMps-1 (10, 19). Plk4 has not been reported to be directly regulated by Cdk2 but requires Cdk2 activity for its maximum activity (26). Our future experiments will identify the centriole separases, centrosome cycle licensers, or centriole duplication kinases deregulated in Cdk2−/−, Cdk4−/−, and Cdk2−/− Cdk4−/− MEFs at the transcriptional or posttranslational level.
Our first major finding was that Cdk2 and Cdk4 regulate distinct steps in the centrosome cycle. The second major finding is that either Cdk2 or Cdk4 signals centrosome amplification in p53-null MEFs. Ablation of p53 was proposed to lead to centrosome amplification, in part by preventing p21Waf1 from controlling Cdk2 (13, 39, 68). Using a genetic approach, we demonstrated that, indeed, Cdk2 is a major mediator of centrosome amplification in p53−/− MEFs. A unique finding was that ablation of Cdk4 was equally effective at suppressing centrosome amplification. This was unexpected, since no one has reported elevated Cdk4 activity in p53−/− MEFs. Western blot analysis assessing the relative levels of cyclins or CKIs that promote or inhibit Cdk2 or Cdk4 activities failed to identify any deregulated cell cycle-regulatory molecule that would promote Cdk4 activity. For example, levels of cyclin D1 were similar in all groups. On the other hand, p16 was upregulated in p53−/− MEFs, which would be consistent with decreased Cdk4 activity.
How does Cdk4 mediate centrosome amplification in p53−/− MEFs? At low concentrations, p21Waf1 is a specific inhibitor of Cdk2 and promotes the assembly of cyclin D/Cdk4 (35), while at higher concentrations it equally inhibits Cdk2, Cdk3, Cdk4, and Cdk6 (30, 35). In fact, there is precedent for the involvement of p21Waf1 in inhibiting Cdk2 and Cdk4 activities triggered by the Ras oncogene, since mice lacking p21Waf1 and overexpressing MMTV-H-RasG12V display higher Cdk2 and Cdk4 activities than those expressing H-RasG12V alone (4). Perhaps a similar scenario exists within the centrosome, as p21Waf1 may inhibit both Cdk2 and Cdk4 activities to regulate centrosome duplication; thus, ablation of p53 and the concomitant absence of endogenous p21Waf1 may hyperactivate Cdk2 and Cdk4. Another possibility is that ablation of Cdk2 or Cdk4 changes the CKI/Cdk ratios within the cell, allowing more p21Waf1 to bind to and inhibit the Cdk that is still present in the cell; this compensatory mechanism has been observed in Cdk4−/− MEFs, in which more p27Kip1 is available to bind and inhibit Cdk2 activity (72). However, observations that p53−/− cells are devoid of detectable endogenous p21Waf1 render unlikely the possibility that more p21Waf1 is available to inhibit Cdk2 activity in p53−/− Cdk4−/− MEFs to prevent centrosome amplification. Other Cdks that may impose that kind of compensatory regulation are p16 and p57, which are overexpressed in p53−/− MEFs. Nevertheless, since p16 and p57 did not accumulate upon transient knockdown of p53, and because the silencing of Cdk2 or Cdk4 was able to suppress centrosome amplification in the same cells, that level of indirect compensatory inhibition of Cdk2 or Cdk4 by the CKIs is unlikely. Still, because our experiments have not ruled out that type of compensatory mechanism, we will develop composite p53, Cdk, and CKI knockdowns and knockouts to directly address the individual roles of those CKIs in preventing the hyperactivity of the remaining Cdk.
The following evidence supports a direct role for Cdk2 and Cdk4 in mediating centrosome amplification in p53-null cells. First, concomitant loss of Cdk2 and Cdk4 in p53−/− MEFs did not reduce the frequencies of centrosome amplification more than the ablation of each individual kinase, rendering unlikely the possibility that the compensatory mechanism imposed by CKIs on Cdk2 or Cdk4 kinase activities is responsible for the suppression of centrosome amplification when either kinase is ablated. Second, concomitant ablation of Cdk2 and Cdk4 in p53-deficient MEFs cultured in HU reduced NPMT199 hyperphosphorylation to the same extent as ablation of Cdk2 or Cdk4 alone. In addition, rendering the NPMT199 site nonphosphorylatable by mutating the Cdk2/Cdk4 phosphorylation site (NPMT199A) suppressed centrosome amplification to the same extent as ablation of Cdk2 or Cdk4, demonstrating that ablation of Cdks prevents maximum centrosome amplification. Therefore, we conclude that both Cdk2 and Cdk4 are individually required to signal centrosome amplification in cells in which p53 has been deleted.
How do Cdk2 and Cdk4 deregulate centrosome duplication, resulting in centrosome amplification? The model depicted in Fig. 8, based on published observations (44, 51, 59, 70) and the observations reported here, shows two ways in which the G1 Cdks could affect centrosome amplification: first, by triggering unregulated phosphorylation of Rb and the release of E2Fs, and second, by the direct phosphorylation of centrosomal targets. Although we did not perform a wide screen for all the E2F targets that may result in centrosome amplification in p53−/− MEFs, those E2F targets that we analyzed by Western blotting were unchanged (cyclins A, E, and D). Thus, in the present study, we tested the second scenario.
In this study, we resolved how Cdk2 and Cdk4 directly impinge on centrosome amplification through NPM. Our experiments indicated that ablation of Cdk2 or Cdk4 decreased NPMT199 hyperphosphorylation in p53−/− MEFs arrested in G0 (serum arrested) or in late G1 (under an HU block). Those experiments suggested that restoration of the normal phosphorylation of NPMT199 normalized the licensing of the centrosome cycle, as well as the excessive centriole reduplication, in p53−/− MEFs to prevent centrosome amplification. Our experiments showing that the introduction of a mutant NPM lacking Cdk2 and Cdk4 phosphorylation sites (NPMT199A) abrogated centrosome amplification in p53−/− MEFs to the same extent as the ablation of Cdk2 or Cdk4 demonstrated that, indeed, the Cdk2, Cdk4→NPMT199 pathway is central to the prevention of centrosome amplification. Based on our observations that Cdk2 and Cdk4 cross talk to the centrosome through the same phosphorylation site in NPM, Thr199, we propose a novel paradigm: that full-fledged phosphorylation of NPM by both Cdk2 and Cdk4 in p53−/− MEFs is critical to the induction of centrosome amplification. How do Cdk2 and Cdk4 prevent centriole reduplication? Our experiments showing that ablation of Cdk2 or Cdk4 prevented centriole reduplication clearly identified an important step within the centrosome cycle that impinges on centrosome amplification. Western blot analyses showing that the hyperphosphorylation of NPMT199 is abrogated in p53−/− Cdk2−/−, p53−/− Cdk4−/−, and p53−/− Cdk2−/− Cdk4−/− MEFs indicated that the restoration of normal centrosome cycle licensing is key to the prevention of centriole reduplication. Another putative mechanism is that the Cdks control the expression or activity of centriole duplication kinases, including Plk4, Plk2, and mMps-1. So far, Western blotting and real-time PCR have not detected increased levels of Plk4 in p53−/− MEFs; we have yet to explore whether loss of p53 influences Plk4's kinase activity. Thus, future experiments will address how the activities of centriole duplication kinases are modulated by Cdk2 and Cdk4.
A third major finding was that ablation of Cdk2 or Cdk4 in p53−/− MEFs inhibits two precursors to the two major types of chromosome instability: the formation of micronuclei (precursors to aneuploidy) and double-strand DNA breaks (precursors to chromosome rearrangements). We conclude that both G1 Cdks are critical to centrosome amplification and chromosome instability in p53−/− MEFs. Our data have important implications for cancer therapy, since the 50% of human cancers that harbor p53 mutations can be treated with small-molecule inhibitors of Cdk2 or Cdk4, currently under development or undergoing clinical trials (63). By using inhibitors specific to either Cdk, we can remove the ability of p53 ablation to generate chromosome instability, an abnormal phenotype strongly associated with a poor prognosis and resistance to chemotherapy (8, 15), thus stopping cancer progression in its tracks. One could accomplish this inhibition without the proliferative toxicity predicted to occur when Cdk2 and Cdk4 are concomitantly inhibited and without the mitotic and developmental defects triggered by Cdk1 inhibition (6, 61, 63).
Acknowledgments
We thank Carla G. Saavedra for scientific editing and Joi Carmichael for assistance with mouse husbandry. Thanks to Peter J. Stambrook and Paul W. Doetsch for critical discussions of the manuscript. Special thanks to Kenji Fukasawa for NPM constructs, and to Gustavo W. Leone for Cdk2 expression vectors. We acknowledge Adam Marcus from the Winship Cancer Institute's imaging core facility for assistance.
H. I. Saavedra was supported by NCI award K01CA104079, a Georgia Cancer Coalition Distinguished Scholar Award, and the Department of Radiation Oncology, Emory University School of Medicine. H. Kiyokawa was supported by NIH-R01-CA100204 and NIH-R01CA112282. M. K. Harrison was supported by the Genetics and Molecular Biology Program's NIH Predoctoral Training Grant (GM008490-16).
Footnotes
Published ahead of print on 23 November 2009.
REFERENCES
- 1.Albertson, D. G., C. Collins, F. McCormick, and J. W. Gray. 2003. Chromosome aberrations in solid tumors. Nat. Genet. 34:369-376. [DOI] [PubMed] [Google Scholar]
- 2.Aleem, E., H. Kiyokawa, and P. Kaldis. 2005. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat. Cell Biol. 7:831-836. [DOI] [PubMed] [Google Scholar]
- 3.Ball, K. L., S. Lain, R. Fahraeus, C. Smythe, and D. P. Lane. 1997. Cell-cycle arrest and inhibition of Cdk4 activity by small peptides based on the carboxy-terminal domain of p21WAF1. Curr. Biol. 7:71-80. [DOI] [PubMed] [Google Scholar]
- 4.Bearss, D. J., R. J. Lee, D. A. Troyer, R. G. Pestell, and J. J. Windle. 2002. Differential effects of p21(WAF1/CIP1) deficiency on MMTV-ras and MMTV-myc mammary tumor properties. Cancer Res. 62:2077-2084. [PubMed] [Google Scholar]
- 5.Berthet, C., E. Aleem, V. Coppola, L. Tessarollo, and P. Kaldis. 2003. Cdk2 knockout mice are viable. Curr. Biol. 13:1775-1785. [DOI] [PubMed] [Google Scholar]
- 6.Berthet, C., K. D. Klarmann, M. B. Hilton, H. C. Suh, J. R. Keller, H. Kiyokawa, and P. Kaldis. 2006. Combined loss of Cdk2 and Cdk4 results in embryonic lethality and Rb hypophosphorylation. Dev. Cell 10:563-573. [DOI] [PubMed] [Google Scholar]
- 7.Reference deleted.
- 8.Castedo, M., A. Coquelle, I. Vitale, S. Vivet, S. Mouhamad, S. Viaud, L. Zitvogel, and G. Kroemer. 2006. Selective resistance of tetraploid cancer cells against DNA damage-induced apoptosis. Ann. N. Y. Acad. Sci. 1090:35-49. [DOI] [PubMed] [Google Scholar]
- 9.Cha, H., C. Hancock, S. Dangi, D. Maiguel, F. Carrier, and P. Shapiro. 2004. Phosphorylation regulates nucleophosmin targeting to the centrosome during mitosis as detected by cross-reactive phosphorylation-specific MKK1/MKK2 antibodies. Biochem. J. 378:857-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Z., V. B. Indjeian, M. McManus, L. Wang, and B. D. Dynlacht. 2002. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev. Cell 3:339-350. [DOI] [PubMed] [Google Scholar]
- 11.Coleman, W. B., and G. J. Tsongalis. 1999. The role of genomic instability in human carcinogenesis. Anticancer Res. 19:4645-4664. [PubMed] [Google Scholar]
- 12.Doxsey, S. 2002. Duplicating dangerously: linking centrosome duplication and aneuploidy. Mol. Cell 10:439-440. [DOI] [PubMed] [Google Scholar]
- 13.Duensing, A., L. Ghanem, R. A. Steinman, Y. Liu, and S. Duensing. 2006. p21(Waf1/Cip1) deficiency stimulates centriole overduplication. Cell Cycle 5:2899-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duensing, A., Y. Liu, M. Tseng, M. Malumbres, M. Barbacid, and S. Duensing. 2006. Cyclin-dependent kinase 2 is dispensable for normal centrosome duplication but required for oncogene-induced centrosome overduplication. Oncogene 25:2943-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duesberg, P., R. Li, R. Sachs, A. Fabarius, M. B. Upender, and R. Hehlmann. 2007. Cancer drug resistance: the central role of the karyotype. Drug Resist. Updat. 10:51-58. [DOI] [PubMed] [Google Scholar]
- 16.Duesberg, P., and D. Rasnick. 2000. Aneuploidy, the somatic mutation that makes cancer a species of its own. Cell Motil. Cytoskeleton 47:81-107. [DOI] [PubMed] [Google Scholar]
- 17.Duesberg, P., D. Rasnick, R. Li, L. Winters, C. Rausch, and R. Hehlmann. 1999. How aneuploidy may cause cancer and genetic instability. Anticancer Res. 19:4887-4906. [PubMed] [Google Scholar]
- 18.Faragher, A. J., and A. M. Fry. 2003. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol. Biol. Cell 14:2876-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisk, H. A., and M. Winey. 2001. The mouse Mps1p-like kinase regulates centrosome duplication. Cell 106:95-104. [DOI] [PubMed] [Google Scholar]
- 20.Fukasawa, K. 2005. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 230:6-19. [DOI] [PubMed] [Google Scholar]
- 21.Fukasawa, K. 2008. p53, cyclin-dependent kinase and abnormal amplification of centrosomes. Biochim. Biophys. Acta 1786:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukasawa, K., T. Choi, R. Kuriyama, S. Rulong, and G. F. Vande Woude. 1996. Abnormal centrosome amplification in the absence of p53. Science 271:1744-1747. [DOI] [PubMed] [Google Scholar]
- 23.Fukasawa, K., F. Wiener, G. F. Vande Woude, and S. Mai. 1997. Genomic instability and apoptosis are frequent in p53 deficient young mice. Oncogene 15:1295-1302. [DOI] [PubMed] [Google Scholar]
- 24.Geng, Y., W. Whoriskey, M. Y. Park, R. T. Bronson, R. H. Medema, T. Li, R. A. Weinberg, and P. Sicinski. 1999. Rescue of cyclin D1 deficiency by knockin cyclin E. Cell 97:767-777. [DOI] [PubMed] [Google Scholar]
- 25.Geng, Y., Q. Yu, E. Sicinska, M. Das, J. E. Schneider, S. Bhattacharya, W. M. Rideout, R. T. Bronson, H. Gardner, and P. Sicinski. 2003. Cyclin E ablation in the mouse. Cell 114:431-443. [DOI] [PubMed] [Google Scholar]
- 26.Habedanck, R., Y. D. Stierhof, C. J. Wilkinson, and E. A. Nigg. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7:1140-1146. [DOI] [PubMed] [Google Scholar]
- 27.Hanashiro, K., M. Kanai, Y. Geng, P. Sicinski, and K. Fukasawa. 2008. Roles of cyclins A and E in induction of centrosome amplification in p53-compromised cells. Oncogene 27:5288-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harbour, J. W., R. X. Luo, A. Dei Santi, A. A. Postigo, and D. C. Dean. 1999. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 98:859-869. [DOI] [PubMed] [Google Scholar]
- 29.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 30.Harper, J. W., S. J. Elledge, K. Keyomarsi, B. Dynlacht, L. H. Tsai, P. Zhang, S. Dobrowolski, C. Bai, L. Connell-Crowley, E. Swindell, et al. 1995. Inhibition of cyclin-dependent kinases by p21. Mol. Biol. Cell 6:387-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hennig, U. G., N. L. Rudd, and D. I. Hoar. 1988. Kinetochore immunofluorescence in micronuclei: a rapid method for the in situ detection of aneuploidy and chromosome breakage in human fibroblasts. Mutat. Res. 203:405-414. [DOI] [PubMed] [Google Scholar]
- 32.Hinchcliffe, E. H., C. Li, E. A. Thompson, J. L. Maller, and G. Sluder. 1999. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science 283:851-854. [DOI] [PubMed] [Google Scholar]
- 33.Hinchcliffe, E. H., and G. Sluder. 2002. Two for two: Cdk2 and its role in centrosome doubling. Oncogene 21:6154-6160. [DOI] [PubMed] [Google Scholar]
- 34.Kleylein-Sohn, J., J. Westendorf, M. Le Clech, R. Habedanck, Y. D. Stierhof, and E. A. Nigg. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell 13:190-202. [DOI] [PubMed] [Google Scholar]
- 35.LaBaer, J., M. D. Garrett, L. F. Stevenson, J. M. Slingerland, C. Sandhu, H. S. Chou, A. Fattaey, and E. Harlow. 1997. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 11:847-862. [DOI] [PubMed] [Google Scholar]
- 36.Lacey, K. R., P. K. Jackson, and T. Stearns. 1999. Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. U. S. A. 96:2817-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lundberg, A. S., and R. A. Weinberg. 1998. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol. Cell. Biol. 18:753-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malumbres, M., R. Sotillo, D. Santamaria, J. Galan, A. Cerezo, S. Ortega, P. Dubus, and M. Barbacid. 2004. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell 118:493-504. [DOI] [PubMed] [Google Scholar]
- 39.Mantel, C., S. E. Braun, S. Reid, O. Henegariu, L. Liu, G. Hangoc, and H. E. Broxmeyer. 1999. p21(cip-1/waf-1) deficiency causes deformed nuclear architecture, centriole overduplication, polyploidy, and relaxed microtubule damage checkpoints in human hematopoietic cells. Blood 93:1390-1398. [PubMed] [Google Scholar]
- 40.Matsumoto, Y., K. Hayashi, and E. Nishida. 1999. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr. Biol. 9:429-432. [DOI] [PubMed] [Google Scholar]
- 41.Matsumoto, Y., and J. L. Maller. 2004. A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science 306:885-888. [DOI] [PubMed] [Google Scholar]
- 42.McDermott, K. M., J. Zhang, C. R. Holst, B. K. Kozakiewicz, V. Singla, and T. D. Tlsty. 2006. p16(INK4a) prevents centrosome dysfunction and genomic instability in primary cells. PLoS Biol. 4:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meraldi, P., R. Honda, and E. A. Nigg. 2002. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 21:483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meraldi, P., J. Lukas, A. M. Fry, J. Bartek, and E. A. Nigg. 1999. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1:88-93. [DOI] [PubMed] [Google Scholar]
- 45.Mikule, K., B. Delaval, P. Kaldis, A. Jurcyzk, P. Hergert, and S. Doxsey. 2007. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat. Cell Biol. 9:160-170. [DOI] [PubMed] [Google Scholar]
- 46.Miller, B. M., H. F. Zitzelsberger, H. U. Weier, and I. D. Adler. 1991. Classification of micronuclei in murine erythrocytes: immunofluorescent staining using CREST antibodies compared to in situ hybridization with biotinylated gamma satellite DNA. Mutagenesis 6:297-302. [DOI] [PubMed] [Google Scholar]
- 47.Montagna, C., E. R. Andrechek, H. Padilla-Nash, W. J. Muller, and T. Ried. 2002. Centrosome abnormalities, recurring deletions of chromosome 4, and genomic amplification of HER2/neu define mouse mammary gland adenocarcinomas induced by mutant HER2/neu. Oncogene 21:890-898. [DOI] [PubMed] [Google Scholar]
- 48.Mussman, J. G., H. F. Horn, P. E. Carroll, M. Okuda, P. Tarapore, L. A. Donehower, and K. Fukasawa. 2000. Synergistic induction of centrosome hyperamplification by loss of p53 and cyclin E overexpression. Oncogene 19:1635-1646. [DOI] [PubMed] [Google Scholar]
- 49.Nakayama, K., H. Nagahama, Y. A. Minamishima, M. Matsumoto, I. Nakamichi, K. Kitagawa, M. Shirane, R. Tsunematsu, T. Tsukiyama, N. Ishida, M. Kitagawa, K. Nakayama, and S. Hatakeyama. 2000. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 19:2069-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelsen, C. J., R. Kuriyama, B. Hirsch, V. C. Negron, W. L. Lingle, M. M. Goggin, M. W. Stanley, and J. H. Albrecht. 2005. Short term cyclin D1 overexpression induces centrosome amplification, mitotic spindle abnormalities, and aneuploidy. J. Biol. Chem. 280:768-776. [DOI] [PubMed] [Google Scholar]
- 51.Okuda, M., H. F. Horn, P. Tarapore, Y. Tokuyama, A. G. Smulian, P. K. Chan, E. S. Knudsen, I. A. Hofmann, J. D. Snyder, K. E. Bove, and K. Fukasawa. 2000. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103:127-140. [DOI] [PubMed] [Google Scholar]
- 52.Ortega, S., I. Prieto, J. Odajima, A. Martin, P. Dubus, R. Sotillo, J. L. Barbero, M. Malumbres, and M. Barbacid. 2003. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat. Genet. 35:25-31. [DOI] [PubMed] [Google Scholar]
- 53.Padmakumar, V. C., E. Aleem, C. Berthet, M. B. Hilton, and P. Kaldis. 2009. Cdk2 and Cdk4 activities are dispensable for tumorigenesis caused by the loss of p53. Mol. Cell. Biol. 29:2582-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ray, A., M. K. James, S. Larochelle, R. P. Fisher, and S. W. Blain. 2009. p27Kip1 inhibits cyclin D-cyclin-dependent kinase 4 by two independent modes. Mol. Cell. Biol. 29:986-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reddy, H. K., R. V. Mettus, S. G. Rane, X. Grana, J. Litvin, and E. P. Reddy. 2005. Cyclin-dependent kinase 4 expression is essential for neu-induced breast tumorigenesis. Cancer Res. 65:10174-10178. [DOI] [PubMed] [Google Scholar]
- 56.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 57.Saavedra, H. I., K. Fukasawa, C. W. Conn, and P. J. Stambrook. 1999. MAPK mediates RAS-induced chromosome instability. J. Biol. Chem. 274:38083-38090. [DOI] [PubMed] [Google Scholar]
- 58.Saavedra, H. I., J. A. Knauf, J. M. Shirokawa, J. Wang, B. Ouyang, R. Elisei, P. J. Stambrook, and J. A. Fagin. 2000. The RAS oncogene induces genomic instability in thyroid PCCL3 cells via the MAPK pathway. Oncogene 19:3948-3954. [DOI] [PubMed] [Google Scholar]
- 59.Saavedra, H. I., B. Maiti, C. Timmers, R. Altura, Y. Tokuyama, K. Fukasawa, and G. Leone. 2003. Inactivation of E2F3 results in centrosome amplification. Cancer Cell 3:333-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sablina, A. A., A. V. Budanov, G. V. Ilyinskaya, L. S. Agapova, J. E. Kravchenko, and P. M. Chumakov. 2005. The antioxidant function of the p53 tumor suppressor. Nat. Med. 11:1306-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santamaría, D., C. Barriere, A. Cerqueira, S. Hunt, C. Tardy, K. Newton, J. F. Caceres, P. Dubus, M. Malumbres, and M. Barbacid. 2007. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448:811-815. [DOI] [PubMed] [Google Scholar]
- 62.Satyanarayana, A., M. B. Hilton, and P. Kaldis. 2008. p21 inhibits Cdk1 in the absence of Cdk2 to maintain the G1/S phase DNA damage checkpoint. Mol. Biol. Cell 19:65-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shapiro, G. I. 2006. Cyclin-dependent kinase pathways as targets for cancer treatment. J. Clin. Oncol. 24:1770-1783. [DOI] [PubMed] [Google Scholar]
- 64.Sharma, N., C. Timmers, P. Trikha, H. I. Saavedra, A. Obery, and G. Leone. 2006. Control of the p53-p21CIP1 axis by E2f1, E2f2 and E2f3 is essential for G1/S progression and cellular transformation. J. Biol. Chem. 281:36124-36131. [DOI] [PubMed] [Google Scholar]
- 65.Sherr, C. J., and J. M. Roberts. 2004. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 18:2699-2711. [DOI] [PubMed] [Google Scholar]
- 66.Sugihara, E., M. Kanai, S. Saito, T. Nitta, H. Toyoshima, K. Nakayama, K. I. Nakayama, K. Fukasawa, M. Schwab, H. Saya, and M. Miwa. 2006. Suppression of centrosome amplification after DNA damage depends on p27 accumulation. Cancer Res. 66:4020-4029. [DOI] [PubMed] [Google Scholar]
- 67.Tagliati, F., A. Bottoni, A. Bosetti, M. C. Zatelli, and E. C. degli Uberti. 2005. Utilization of luminescent technology to develop a kinase assay: Cdk4 as a model system. J. Pharm. Biomed. Anal. 39:811-814. [DOI] [PubMed] [Google Scholar]
- 68.Tarapore, P., H. F. Horn, Y. Tokuyama, and K. Fukasawa. 2001. Direct regulation of the centrosome duplication cycle by the p53-p21Waf1/Cip1 pathway. Oncogene 20:3173-3184. [DOI] [PubMed] [Google Scholar]
- 69.Thomson, E. J., and P. E. Perry. 1988. The identification of micronucleated chromosomes: a possible assay for aneuploidy. Mutagenesis 3:415-418. [DOI] [PubMed] [Google Scholar]
- 70.Tokuyama, Y., H. F. Horn, K. Kawamura, P. Tarapore, and K. Fukasawa. 2001. Specific phosphorylation of nucleophosmin on Thr(199) by cyclin-dependent kinase 2-cyclin E and its role in centrosome duplication. J. Biol. Chem. 276:21529-21537. [DOI] [PubMed] [Google Scholar]
- 71.Toledo, F., and G. M. Wahl. 2006. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer 6:909-923. [DOI] [PubMed] [Google Scholar]
- 72.Tsutsui, T., B. Hesabi, D. S. Moons, P. P. Pandolfi, K. S. Hansel, A. Koff, and H. Kiyokawa. 1999. Targeted disruption of CDK4 delays cell cycle entry with enhanced p27(Kip1) activity. Mol. Cell. Biol. 19:7011-7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ussar, S., and T. Voss. 2004. MEK1 and MEK2, different regulators of the G1/S transition. J. Biol. Chem. 279:43861-43869. [DOI] [PubMed] [Google Scholar]
- 74.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 75.Wu, L., C. Timmers, B. Maiti, H. I. Saavedra, L. Sang, G. T. Chong, F. Nuckolls, P. Giangrande, F. A. Wright, S. J. Field, M. E. Greenberg, S. Orkin, J. R. Nevins, M. L. Robinson, and G. Leone. 2001. The E2F1, E2F2, and E2F3 transcription factors are essential for cellular proliferation. Nature 414:457-462. [DOI] [PubMed] [Google Scholar]
- 76.Xiong, Y., G. J. Hannon, H. Zhang, D. Casso, R. Kobayashi, and D. Beach. 1993. p21 is a universal inhibitor of cyclin kinases. Nature 366:701-704. [DOI] [PubMed] [Google Scholar]
- 77.Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature 411:1017-1021. [DOI] [PubMed] [Google Scholar]