Abstract
All eukaryotic cells have to maintain cholesterol concentrations within defined margins in order to function normally. Perturbing cholesterol homeostasis can result in a wide range of cellular and systemic defects, including cardiovascular diseases, as well as Niemann-Pick and Tangier diseases. Here, we show that DHR96 is indispensable for mediating the transcriptional response to dietary cholesterol and that it acts as a key regulator of the Niemann-Pick type C gene family, as well as of other genes involved in cholesterol uptake, metabolism, and transport. DHR96 mutants are viable and phenotypically normal on a standard medium but fail to survive on diets that are low in cholesterol. DHR96 mutants have aberrant cholesterol levels, demonstrating a defect in maintaining cholesterol homeostasis. Remarkably, we found that a high-cholesterol diet phenocopied the genomic profile of the DHR96 mutation, indicating that DHR96 resides at the top of a genetic hierarchy controlling cholesterol homeostasis in insects. We propose a model whereby DHR96 is activated when cellular cholesterol concentrations drop below a critical threshold in order to protect cells from severe cholesterol deprivation.
Cholesterol is best known for its adverse effects on human health, since an excess of the substance is a significant risk factor for developing atherosclerosis, heart disease, and stroke. On the other hand, cholesterol is critically important for higher organisms, mostly because it performs a range of vital cellular functions. In particular, cholesterol functions as an indispensable precursor for steroid hormones, and it contributes to the fluidity and integrity of the lipid bilayer. In addition, cholesterol has important roles in cell signaling, since it is required for the covalent modification of hedgehog proteins and constitutes a critical component of lipid rafts. To ensure that all of these processes function properly, cellular cholesterol concentrations have to be maintained within specific limits. Exceeding healthy levels of free (i.e., unesterified) cholesterol may result in cytotoxicity (40), whereas insufficient levels may result in dramatic malformations and behavioral problems, as observed in the Smith-Lemli-Opitz syndrome, a developmental disorder caused by a mutation that affects cholesterol biosynthesis (12, 50).
While the mechanisms that control cholesterol biosynthesis have been characterized in great detail, we are only beginning to understand the processes that regulate cholesterol uptake, removal, and turnover. In vertebrates, cholesterol homeostasis is maintained by controlling dietary uptake, by adjusting cellular influx and efflux, and through the regulation of key steps in the cholesterol biosynthetic pathway. Two vertebrate transcription factor families, sterol-regulatory element binding proteins (SREBPs) and liver X receptors (LXRs), are involved in regulating cholesterol homeostasis. A series of studies have identified SREBP1a, -1c, and -2 as critical regulators of fatty acid and cholesterol biosynthetic pathways (6, 46, 49). These proteins are proteolytically activated by a decline in cellular cholesterol levels and are subsequently imported into the nucleus, where they act as transcription factors. In contrast, the mechanism by which the nuclear receptors LXRα (NR1H3) and LXRβ (NR1H2) regulate gene expression relies on direct binding to cholesterol metabolites that function as agonistic ligands (30, 32). Nuclear receptors form a large superfamily of proteins that function simultaneously as transcription factors and receptors for small lipophilic compounds. Members of this protein family directly regulate gene expression in response to fat-soluble ligands, such as steroid hormones, certain vitamins, fatty acids, oxysterols, and bile acids. Ligands that bind with high affinity to LXR include 24(S),25-epoxy-cholesterol, an oxysterol, which is sufficient to modulate the expression of LXR target genes in vivo (5). LXRα controls genes required for cholesterol efflux, indicating that the receptor acts to protect cells against high cholesterol levels (3). Mice lacking functional LXRα display an abnormal response to dietary cholesterol and accumulate hepatic cholesterol when reared on a cholesterol-rich diet (31).
We used the fruit fly, Drosophila melanogaster, to study the regulation of cholesterol homeostasis in a simple genetic model organism. Similar to vertebrates, Drosophila requires cholesterol as a precursor for steroid hormones, such as the molting hormone ecdysone, and as a structural component in plasma membranes. Drosophila, like all other insects, cannot synthesize cholesterol and has to retrieve a suitable sterol from the diet (9, 17, 18). The sterol content of a particular diet therefore constitutes an important developmental variable for the survival of Drosophila and allows the study of mutants that fail to survive on low-cholesterol media.
The Drosophila genome harbors a single ortholog of the SREBP gene family; however, some notable functional differences exist between the fly and vertebrate proteins. The Drosophila SREBP pathway is activated by phosphatidylethanolamine instead of cholesterol and regulates fatty acid synthesis pathways (10, 22, 37), suggesting that a different protein controls cholesterol homeostasis in flies. So far, however, no nuclear receptor or other transcriptional regulator has been linked to cholesterol homeostasis in Drosophila. Vertebrate LXRs have two close homologs in the fly, the Ecdysone Receptor (EcR) and Drosophila Hormone Receptor 96 (DHR96) genes (23). Specifically, the ancestral LXR gene, predating the split of the vertebrate and insect lineages, gave rise to seven human genes (LXRα/β, FXRα/β, CAR, VDR, and SXR) and two Drosophila genes (EcR and DHR96) (see Fig. S1 at http://www.biology.ualberta.ca/king-jones_lab/Bujold_et_al_2010_Supplemental_data_MCB.pdf). Of the two fly homologs, LXR has greater similarity to EcR, which binds with high affinity to 20-hydroxyecdysone (hereafter referred to as 20E), the metabolically active form of ecdysone. It appears unlikely, however, that EcR functions as a cholesterol sensor, as it has widespread roles in orchestrating 20E-driven developmental transitions, including molting and metamorphosis. In contrast to EcR, DHR96 has no apparent developmental function and plays a role in regulating detoxification responses (21), indicating that this nuclear receptor exerts control over metabolic processes. We showed previously that DHR96 is strongly expressed in tissues important for nutrient traffic, storage, and excretion, suggesting it may act as a nutrient sensor (21). Furthermore, we recently demonstrated that DHR96 has the ability to bind cholesterol in vivo, suggesting that the receptor could act as a sterol sensor in a manner comparable to that of LXRα (18a).
Certain target genes of LXRα have confirmed roles in maintaining cholesterol homeostasis. One such target gene is ABCA1, which encodes an ABC transporter that plays a critical role in reverse cholesterol transport, a process by which excess cellular cholesterol is transported by HDL from peripheral tissues to the liver (8). Patients with a mutation in both copies of the ABCA1 gene suffer from Tangier disease, a disorder that is characterized by severely reduced HDL levels and accumulation of cholesterol in peripheral tissues (28, 29).
Some evidence suggests that LXR target genes may include the Niemann-Pick disease Type C 1 and 2 genes (NPC1 and -2), since they are upregulated by synthetic LXR agonists (33). The NPC1 and NPC2 proteins participate in the transport of unesterified cholesterol from the late endosomal/lysosomal compartment to the cytosol. Mutations in either NPC1 or NPC2 result in a usually fatal lysosomal storage disorder with characteristic accumulation of cholesterol and sphingolipids (7, 24, 26). A gut-specific member of the NPC1 protein family, NPC1L1, is critical for intestinal absorption of dietary cholesterol and plant sterols, but it is unclear whether liver X receptors transcriptionally regulate the gene (1, 45). The Drosophila genome includes a set of highly conserved Niemann-Pick disease genes, including two NPC1 homologs. NPC1a is widely expressed and was shown to play a critical role in transporting sterols to the ring gland, thus allowing ecdysone production to occur (13). In contrast, NPC1b expression is restricted to the midgut, and a recent study demonstrated that the protein plays an important role in the intestinal uptake of cholesterol, like its vertebrate ortholog, NPC1L1 (44). In addition to the NPC1 homologs, eight genes with significant sequence similarity to NPC2 have been recently described in Drosophila, among which Npc2a and Npc2b function in a redundant manner to coordinate sterol homeostasis (19). No studies have yet addressed whether transcriptional regulation of Drosophila NPC genes might contribute to maintaining cholesterol homeostasis.
Here, we show that the Drosophila nuclear receptor DHR96 is required to ensure survival on a low-cholesterol diet and that it is necessary to maintain appropriate cholesterol levels. Importantly, we show that genes with roles in maintaining cholesterol homeostasis are strongly dependent on DHR96 function, including homologs of vertebrate NPC1L1, NPC2, and ABCA1. In this paper, we use a combination of genetics, nutritional studies, and genomics as a systemic approach for studying the molecular mechanisms by which cholesterol homeostasis is achieved.
MATERIALS AND METHODS
Population studies.
For population studies, we transferred 50 rinsed or dechorionated Drosophila embryos to each vial, and each condition was tested in triplicate. We scored the appearance of pupae every 24 h and maintained the vials until the pupae developed into adults. To prepare lipid-depleted or untreated “4-24” medium (Carolina Biological Supply Company) (hereafter referred to as C424), flakes were ground using a household blade grinder. Small batches were combined and thoroughly mixed to ensure equal distribution of methylparaben powder in untreated C424. To extract lipids, 200 g of ground C424 powder was transferred to a 4-liter Erlenmeyer flask and treated six times for 12 h each time with 1 liter of chloroform. The lipid-depleted C424 was then air dried until no traces of chloroform were detectable. Finally, methylparaben was added to a final concentration of 1% of the wet weight. To add sterols, each vial received a total of 200 μl of ethanol containing the appropriate amount of sterol on the surface of 1 g of C424 powder. After the ethanol was allowed to evaporate, each vial was mixed vigorously with 5 ml water until the medium was set.
Larval collections and RNA extraction.
For the C424-versus-standard-medium microarray, 100 eggs were transferred to petri dishes containing C424 medium or standard medium. The larvae were raised at room temperature for 4 days, collected as mid-second instars (25 per sample), flash frozen, and stored at −80°C. Larval staging was verified through quantitative real-time PCR (qPCR) using the DHR4 and β-FTZ-F1 genes, which are genes induced during the second half of the larval stage, as timing controls. Roche UPL probes and TaqMan Universal PCR Master Mix (ABI) were used according to the manufacturer's instructions. RNA was extracted from frozen larvae using a slightly modified Trizol protocol (Invitrogen) in which the RNA was chloroform extracted an additional time and ethanol precipitated using LiCl to remove organic contaminants, salts, and genomic DNA.
The cholesterol gradient qPCR studies were carried out in a similar manner, with the following exceptions: C424 medium was first lipid extracted with chloroform (see below), air dried, and then supplemented with either 0 μg, 50 μg, or 200 μg cholesterol per gram dry weight.
To prepare microarray samples from larvae that were exposed to high cholesterol, 200 eggs were transferred to bottles containing a standard medium supplemented with either 0% or 1% cholesterol, and eight mid-L3 larvae per sample were collected. For timing larvae to the mid-third-instar stage, we added 0.05% bromophenol blue to the medium and selected for larvae with dark-blue guts, as described by others (2, 25). RNA extraction followed the modified Trizol protocol, but instead of LiCl, we used sodium acetate (NaOAC) for precipitation. Total-RNA samples were purified using Qiagen RNeasy columns before cDNA synthesis.
cDNA preparation and preamplification.
cDNA was prepared from 1 μg of total RNA using either a Stratagene Affinity Script qPCR cDNA Synthesis Kit or an ABI High Capacity Reverse Transcription Kit. For microfluidics-based qPCR on the Fluidigm Biomark, we carried out preamplification of cDNA. For this, we used the equivalent of 5 ng of total RNA to amplify cDNA samples with the TaqMan Pre-Amp 2× Master Mix (Applied Biosystems) by following the procedures recommended by Fluidigm.
Microarray analysis.
To generate labeled cRNA from 200 ng of starting RNA, we used an Affymetrix GeneChip 3′ IVT Express kit according to the manufacturer's instructions. For each experimental condition, we prepared samples in triplicate. Affymetrix gene expression arrays (Drosophila Genome 2.0) were hybridized and scanned at the University of Alberta microarray facility. Raw data analysis was conducted by using LIMMA (47) and gcRMA (48) downloaded from the bioconductor (15) web page, while data mining and gene ontology (GO) statistics were based on Affymetrix annotation files that were analyzed with Microsoft Access and Microsoft Excel.
Microfluid-based high-throughput qPCR.
High-throughput qPCR (2,304 reactions per run) was performed on preamplified cDNA samples and analyzed on 48.48 dynamic arrays (Fluidigm). Sample mixtures were prepared according to the manufacturer's instructions using Fluidigm DA Sample Reagent, TaqMan Universal PCR Master Mix, and preamplified cDNA samples. Assay mixtures were prepared following the manufacturer's instructions using Fluidigm DA Assay Reagent in combination with Roche UPL probes and oliguncleotides (IDT). The primer design (melting temperature [Tm] = 60 ± 1°C) and probe selection were based on the Roche online assay design center. The microfluidic qPCRs were run according to thermal-cycling parameters recommended by Applied Biosystems for the TaqMan Universal PCR Master Mix. For each experimental condition, we tested four samples in triplicate. We included five housekeeping control genes per run: rp49 (CG7939), α-tubulin 84B (CG1913), metallothionein A (CG9470), RNA polymerase II 140-kDa subunit (CG3180), and tropomyosin 1 (CG4898). ΔΔCT values were calculated individually for each control gene, and differential gene expression was determined as the geometric mean of all the ΔΔCT values (43).
Lipid measurements.
Total-cholesterol concentrations were determined with the Amplex Red cholesterol assay kit (Invitrogen) following the instructions of the manufacturer. Cholesteryl ester concentrations were measured using gas chromatography (GC). Lipid extractions were carried out as described previously (35). For either of these assays, 50 L2 larvae and 8 L3 larvae were collected per sample, flash frozen, and homogenized with a motorized pestle with 500 μl of phosphate-buffered saline (PBS). For gas chromatography, samples were mixed with tridecanoin as an internal standard and analyzed on an Agilent 6890 Gas Chromatography System using a Zebron ZB-5 (Phenomenex) column. Data were visualized with Agilent Chemstation software. All measurements were normalized to the sample protein concentration as determined by the bicinchoninic acid (BCA) protein assay reagent (Pierce).
RESULTS
DHR96 mutants are highly sensitive to cholesterol deprivation.
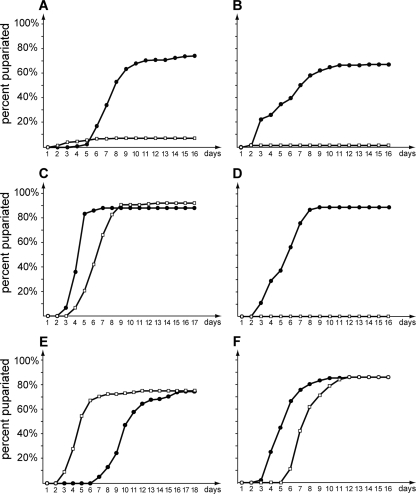
The close structural and phylogenetic relationships of the vertebrate liver X receptors to their fly counterpart, DHR96, suggest that the latter may play a role in controlling cellular and/or systemic cholesterol levels in Drosophila. Therefore, a mutation in DHR96 may affect the ability to properly regulate cholesterol homeostasis, raising the question of how DHR96 mutants might respond to low or high levels of dietary cholesterol. On a standard cornmeal diet (hereafter referred to as standard medium/diet), DHR96 mutants appear normal and display no obvious phenotypes, suggesting that DHR96 is not essential when the animals are maintained under optimal laboratory conditions. However, when DHR96 mutants are reared on a potato tuber-derived medium that is naturally low in cholesterol (C424), ∼90% of the population arrest development at the L2 stage, while 75 to 90% of the controls reach the adult stage (Fig. 1A). The inability of DHR96 mutants to survive on C424 medium could indicate a failure to properly retrieve a vital nutrient from the diet. To examine this possibility, we supplemented C424 medium with low concentrations of yeast, since yeast is a sufficient food source for Drosophila. Supplementation with 1% yeast completely restored the viability of DHR96 mutants (Fig. 1C). Remarkably, supplementation with yeast extract failed to rescue DHR96 mutants, suggesting that a vital nutrient is lost during the extraction procedure (Fig. 1D). Yeast extract is water soluble and consequently contains substantially fewer lipids than live yeast, suggesting that DHR96 mutants fail to survive on C424 medium because it is low in, or lacks, specific classes of lipids. The only known essential lipid class required in a Drosophila diet is sterols that can be metabolized to cholesterol by the fly (36). We therefore tested whether dietary supplementation of C424 medium with cholesterol would result in a rescue of DHR96 mutants. Indeed, reconstituting C424 with 0.1% or 0.01% cholesterol was sufficient to rescue DHR96 mutants (Fig. 1E and F), with survival rates similar to those observed with yeast supplementation (Fig. 1C). In contrast, a nonsterol source like oleic acid failed to restore viability in DHR96 mutants (Fig. 1B), suggesting that the lethality associated with the DHR96 mutation arises from a defect in sterol homeostasis and does not reflect a global defect in lipid metabolism. We also showed that DHR96 mutants could not be rescued with 20E or desmosterol or combinations thereof; however, ergosterol, the main yeast sterol, and stigmasterol, a widespread plant sterol, could rescue DHR96 mutants (A. Gopalakrishnan, N. Premji, M. Bujold, and K. King-Jones, unpublished data).
FIG. 1.
DHR96 mutants are hypersensitive to cholesterol deprivation. Larvae were reared on a low-cholesterol medium (C424) with no added nutrients (A) or C424 medium that was supplemented with 0.1% oleic acid (B), 1% yeast (C), 2.5% yeast extract (D), 0.01% cholesterol (E), or 0.1% cholesterol (F). Survival was quantified by determining the number of emerging pupae every 24 h. Pupae were followed to adulthood to ensure successful completion of development. Solid circles, wild type; open squares, DHR96 mutants.
We concluded that the presence of cholesterol or other suitable sterols is a prerequisite for the survival of DHR96 mutants, suggesting that these animals suffer from widespread defects that include, but are not limited to, impaired steroid hormone production.
DHR96 mutants have aberrant cholesterol levels.
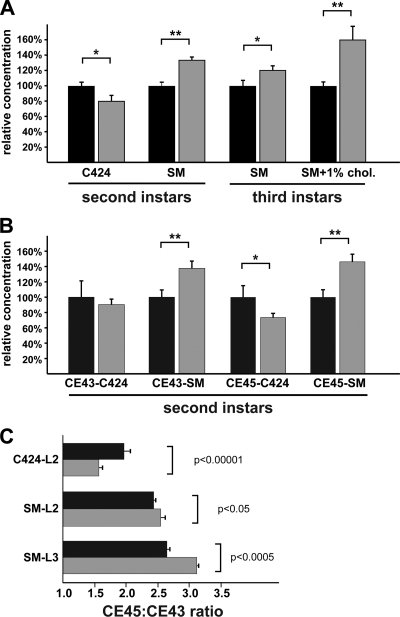
The fact that DHR96 mutants can be rescued by cholesterol supplementation may indicate a defect in either absorption or distribution of cholesterol. To address this possibility, we measured total cholesterol levels in lipid extracts isolated from L2 and L3 larvae using the Amplex Red kit (Molecular Probes). First, we compared the wild type and the DHR96 mutants reared on C424 medium to examine cholesterol levels in these animals. We found that the cholesterol concentration in DHR96 mutants was ∼20% lower than in the wild type on this medium, indicating a significant but not dramatic decrease due to the loss of DHR96 function (Fig. 2). In contrast, when larvae were reared on standard medium, we observed 20 to 35% higher cholesterol levels in DHR96 mutants than in the wild type, and this effect was observed in L2 and L3 larvae (Fig. 2A). This suggests that DHR96 mutants accumulate cholesterol on diets with sufficiently high concentrations of the compound. To test whether this cholesterol accumulation phenotype could be aggravated when animals were reared on a high-cholesterol diet, we repeated the experiment with L3 larvae retrieved from a standard medium that was supplemented with 1% cholesterol. Under these conditions, DHR96 mutants accumulated cholesterol at ∼45% above wild-type levels, indicating that the increased load of dietary cholesterol translated into a corresponding elevation of total cholesterol in mutant animals. In conclusion, DHR96 mutants do not appear to suffer from an inability to absorb cholesterol. However, it is clear that cholesterol uptake is not properly regulated in DHR96 mutants, as evidenced by the accumulation of cholesterol when the animals were reared on media with relatively high cholesterol concentrations.
FIG. 2.
DHR96 mutants display aberrant cholesterol levels.(A) Total cholesterol levels in L2 and L3 larvae reared on C424, standard medium (SM), or standard medium supplemented with 1% cholesterol (SM + 1% chol). (B) Relative abundances of cholesteryl esters measured in L2 larvae collected from C424 medium and L3 larvae reared on standard medium. CE43 and CE45, cholesteryl esters with 43 or 45 carbon atoms. (C) Relative ratios of CE43 to CE45 in the wild type and DHR96 mutants measured on different diets. L2 larvae were reared on C424 and standard medium, while L3 larvae were collected from standard medium only. Black bars, wild type; gray bars, DHR96 mutants. *, P < 0.05; **, P < 0.01. The error bars indicate standard errors.
To validate these findings, we measured cholesteryl esters in mutant and wild type animals using GC. This approach allowed us to quantify the relative abundances of two prevalent cholesteryl ester classes: cholesteryl esters with 43 carbon atoms (CE43) are predominantly composed of cholesteryl palmitate, while the CE45 subclass may contain cholesteryl oleate as well as cholesteryl stearate. Any perturbation in the concentrations of these cholesterol metabolites may indicate aberrant expression of enzymes that generate or cleave cholesteryl esters and would suggest that this process is regulated by DHR96. Similar to the results for total cholesterol levels, we found lower abundances of both CE43 and CE45 cholesteryl esters in DHR96 mutants that were reared on C424 medium but significantly higher concentrations of these metabolites when mutants were maintained on a standard medium (Fig. 2B). Cholesteryl esters with 45 carbon atoms are more abundant than their CE43 counterparts, and this ratio varies in the wild type by a maximum of 35%, depending on the food source used (the ratios are between 2.0 and 2.55) (Fig. 2C). In contrast, the CE45/CE43 ratios fluctuate to a greater degree in DHR96 mutants. When mutant larvae are reared on C424 medium, the relative abundance of CE45 drops by ∼50% compared to the wild type, while extracts isolated from mutants that were raised on a standard medium have ∼25% higher ratios than controls. Overall CE45/CE43 ratios vary by ∼100% in DHR96 mutants, almost 3-fold higher than in the wild type.
Summing up, we found that total cholesterol and cholesteryl ester levels dropped moderately but significantly when DHR96 mutants were reared on a low-cholesterol medium and that the opposite was true when mutants were raised on standard or high-cholesterol medium. This suggests a role for DHR96 in the regulation of cholesterol homeostasis and in the metabolism of cholesteryl esters.
DHR96 is required for expression of sterol metabolism genes.
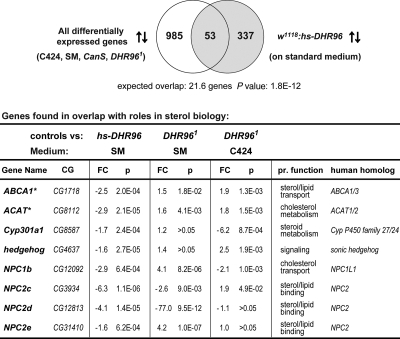
To identify genes that are differentially regulated by DHR96 on C424 medium compared to a standard diet, we compared the genomic responses of mutant and wild-type L2 larvae on these different medium types using Affymetrix 2.0 microarrays. We filtered data sets for genes that showed significant changes in expression between the wild type and DHR96 mutants and/or responded to dietary differences, which yielded a total of 1,038 genes. To extract a more meaningful subset, we compared this set to 390 genes that are significantly affected by ectopic DHR96 expression (21). This comparison yielded a highly significant overlap of 53 genes (P value, 1.8E−12) (Fig. 3), which we expect to be highly enriched for genes doubly dependent on DHR96 and differences in the diet, suggesting that these gene responses are mediated by DHR96. Remarkably, this set of 53 genes harbored 8 genes with known links to sterol metabolism and transport, suggesting that the genes respond to different levels of dietary cholesterol via DHR96 (Fig. 3, table). Among these 8 genes, 4 are homologs of genes associated with human Niemann-Pick disease type C (NPC1b and NPC2c, -d, and -e). Furthermore, we identified hedgehog, which encodes a signaling molecule that is covalently modified by cholesterol, as well as CYP301A1, a cytochrome P450 gene with a predicted function in mitochondrial steroid metabolism because of its close relationship to three genes of the “Halloween” group that act in the ecdysteroid biosynthesis pathway (16). Finally, we found CG8112 and CG1718 among this group. Upon conducting a bidirectional BLAST search, these genes turned out to be most closely related to human ACAT1 and -2 and human ABCA1 and -3, respectively.
FIG. 3.
Comparison of microarray data sets identified genes with roles in sterol biology. Two microarray data sets were compared. The first set harbored 1,038 differentially expressed genes obtained from an analysis of either wild-type or mutant L2 larvae that were reared on C424 and standard medium (SM). The second set contained a list of 390 genes that were affected in response to ectopic expression of DHR96 (hs-DHR96 transgenic line [21]). The resulting overlap (53 genes) contained ∼2.5-fold more genes than one would expect, on average, when random sets of the same size are compared. This enrichment is significant, as indicated by the P value derived from a χ2 calculation. The arrows indicate up- and downregulated genes. The table lists genes with known or predicted functions with links to sterol biology. An asterisk indicates that the gene name is not listed in Flybase. CG, computed gene; FC, fold change; P, P value, based on Student's t test calculated by the LIMMA software package; pr. function, predicted function. Controls for the gain-of-function array were w1118 larvae, whereas controls for the C424/SM array were CanS larvae.
Acyl coenzyme A (CoA)/cholesterol acyltransferase 1 and 2 (ACAT1 and -2), are enzymes that catalyze the reaction by which free cholesterol is esterified with long-chain fatty acids, effectively lowering the cellular concentration of free cholesterol and allowing the sequestration of cholesteryl esters into lipid droplets for future use. To our knowledge, no transcriptional regulators of the ACAT genes have been identified in any model system, and the finding that DHR96 is required for the regulation of Drosophila ACAT (CG8112) may suggest that LXRs might have a similar role in vertebrates.
As indicated above, ABCA1 is an ABC transporter with important roles in the reverse cholesterol transport pathway. ABCA3, on the other hand, encodes a lipid transporter required for surfactant function at birth (38). Presumably, CG1718 is the single fly ortholog of both human genes, and we refer to it as ABCA1 from here on, since our data suggest that it has a role in cholesterol homeostasis.
In summary, the presence of 8 genes in a set of 53 genes with links to sterol biology is highly significant. For instance, there are a total of 10 NPC genes in the fly genome (8 NPC2 and 2 NPC1 homologs), and the probability of finding 4 out of 10 by chance in a set of 53 genes is very low (P value, 6.5E−124).
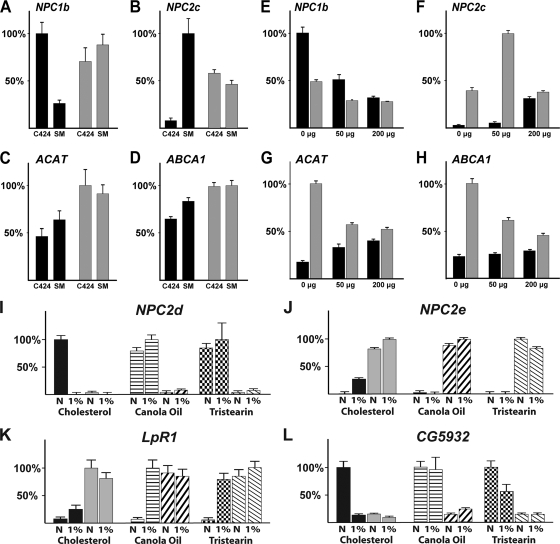
To validate these microarray results, we analyzed four genes (ACAT, ABCA1, NPC2c, and NPC1b) (Fig. 4A to D; also see Fig. S2 at http://www.biology.ualberta.ca/king-jones_lab/Bujold_et_al_2010_Supplemental_data_MCB.pdf) using qPCR. In the wild type, we found that NPC1b was expressed at an ∼4-fold-higher level on C424 medium than on standard medium, suggesting that NPC1b levels are elevated on a low-cholesterol diet in order to improve intestinal absorption of sterols. In contrast, NPC1b mRNA levels were very similar in DHR96 mutants regardless of the medium, indicating that the gene has lost the ability to appropriately respond to dietary cholesterol levels. The net effect is that NPC1b expression is roughly 3-fold higher in DHR96 mutants than in controls when standard medium is used (Fig. 4A). Given its role in cholesterol uptake, the high levels of NPC1b in DHR96 mutants might account for the higher cholesterol levels we observed in mutant animals collected from a standard or high-cholesterol diet (Fig. 2A and B).
FIG. 4.
Dietary cholesterol regulates cholesterol metabolism genes in a concentration- and DHR96-dependent manner. (A to D) Staged L2 larvae were collected from either untreated C424 medium or a standard medium (SM). (E to H) Staged L2 larvae were collected from lipid-depleted C424 medium that was supplemented with either 0 μg, 50 μg, or 200 μg cholesterol per gram (dry weight). For each gene, the highest expression level was normalized to 100%. Black bars, wild type (CanS); gray bars, DHR96 mutants. (I to L) Staged mid-L3 larvae were collected from media that contained 1% cholesterol, 1% canola oil, or 1% tristearin. LpR1, LDL-receptor-related protein. The error bars indicate standard errors. N, standard medium with no added lipid.
In many respects, the expression profiles of NPC2c, ACAT, and ABCA1 appear to be opposite to that of NPC1b. For instance, these genes are different from NPC1b in that they are expressed at a higher level in wild-type controls reared on a standard diet than in those reared on C424 medium, suggesting they might be induced by dietary sterols. While this difference is substantial for NPC2c (∼12.5-fold) (Fig. 4B; also see Fig. S2 at http://www.biology.ualberta.ca/king-jones_lab/Bujold_et_al_2010_Supplemental_data_MCB.pdf), we observed only moderately higher levels for ACAT and ABCA1 (40% and 30% increases, respectively) (Fig. 4C and D). In neither case did we see a corresponding increase in the DHR96 mutants, indicating that DHR96 is necessary for this response. In addition, expression levels of NPC2, ACAT, and ABCA1 in DHR96 mutants reared on C424 medium were consistently higher than those of control animals. This also contrasts with NPC1b, where this relationship is reversed. Despite the difference between NPC1b and the other three genes tested here, we found that the transcriptional responses of all four genes to cholesterol were either muted or absent in DHR96 mutants.
Taken together, our results suggest that the differences in sterol compositions and concentrations between C424 and standard medium are responsible for the transcriptional responses in genes associated with different aspects of sterol homeostasis. Importantly, these transcriptional responses are dependent on DHR96, suggesting that DHR96 coordinates the regulation of genes involved in sterol homeostasis and metabolism.
Cholesterol regulates gene expression in Drosophila.
One would expect that the differences between C424 medium and a standard diet are complex and that consequently one would observe transcriptional changes of genes in many metabolic pathways. The fact that DHR96 mutants reared on C424 food can be rescued by cholesterol supplementation and the finding that genes linked to sterol metabolism pathways require DHR96 for proper regulation suggest that a critical distinction between these two media is the sterol concentration and composition. To test directly which genes respond to dietary cholesterol, we examined qPCR-based gene expression profiles of wild-type and mutant L2 larvae reared on lipid-depleted C424 medium that was supplemented with either 0 μg, 50 μg, or 200 μg of cholesterol per vial, which corresponds to 0%, 0.00083%, and 0.0033% cholesterol (wet weight), respectively. For this experiment, we selected 34 genes, mainly from our existing microarray data but also including lipid and cholesterol metabolism genes described in gene ontology files and publications (see Table S1 at http://www.biology.ualberta.ca/king-jones_lab/Bujold_et_al_2010_Supplemental_data_MCB.pdf). Using this approach, we found that NPC1b, NPC2c, ACAT, and ABCA1 responded specifically to changes in dietary cholesterol concentrations. In particular, NPC1b expression was repressed by dietary cholesterol, while NPC2c, ACAT, and ABCA1 levels increased with higher cholesterol concentrations (Fig. 4E to H). These results precisely track the changes we observed between C424 and standard medium (Fig. 4A to D), strongly suggesting that these differences are indeed due to cholesterol. On lipid-depleted medium with no added cholesterol, NPC1b levels in DHR96 mutants were 50% lower than in controls (Fig. 4E). Similar to our findings for C424 medium (compare C424 expression levels in Fig. 4A to D), we observed that NPC2c, ACAT, and ABCA1 expression levels were strongly elevated in DHR96 mutants when the animals were maintained on very low cholesterol concentrations, as seen in lipid-depleted C424 medium supplemented with 0 μg and 50 μg cholesterol (Fig. 4F to H).
In addition to NPC1b, NPC2c, ACAT, and ABCA1, we found the following genes to be repressed by cholesterol: Lip3, which encodes a predicted cholesterol ester hydrolase; CG5932, which encodes a gastric triacylglyceride lipase; CG31148, which encodes a glucosylceramidase with roles in sphingolipid metabolism (see below); and FANCL, which encodes a predicted ubiquitin E3 ligase (see Fig. S4 at http://www.biology.ualberta.ca/king-jones_lab/Bujold_et_al_2010_Supplemental_data_MCB.pdf). In contrast, Cyp12d1, a cytochrome P450 gene closely related to CYP301a1 (41) (Fig. 3), appears to be induced by cholesterol, while CG10514, a gene encoding a DUF227 domain (domain of unknown function 277), was not significantly affected by changes in the diet but displayed low levels of expression in DHR96 mutants, regardless of the condition (see Fig. S4 at the URL given above).
Taken together, we found that NPC1b is downregulated by dietary cholesterol, while NPC2c, ACAT, and ABCA1 are upregulated by cholesterol. Similarly, NPC1b is clearly downregulated in DHR96 mutants, while NPC2c, ACAT, and ABCA1 are substantially upregulated in mutant animals.
Exposure to high cholesterol triggers distinct transcriptional responses.
We expected that gene responses to cholesterol might be concentration dependent, due to the diverse gene functions associated with cholesterol homeostasis. Specifically, a gene that responds within a defined concentration range might be unresponsive to doses that lie outside this range, and vice versa. To complement our expression analysis that was based on lipid-depleted C424 supplemented with 0, 50, or 200 μg cholesterol, we used microarray analysis to examine the transcriptional effects of a high-cholesterol diet in wild-type animals and DHR96 mutants reared on a standard medium, to which we added either 0% or 1% cholesterol (wet weight). In total, we found 73 genes in the wild type that were upregulated by high cholesterol and 55 genes that were downregulated, based on a greater-than-1.5-fold change and a P value of less than 0.01.
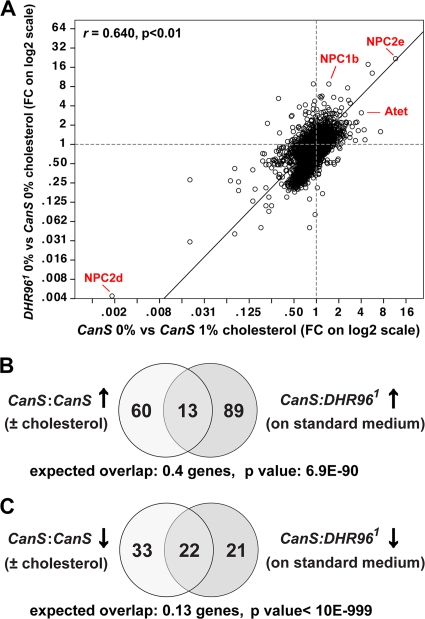
The two genes most strongly affected by high cholesterol in the wild type, whether based on fold change or significance, are NPC2d and NPC2e, suggesting that these Niemann-Pick gene homologs also have roles in sterol transport in Drosophila (Fig. 5A also see Tables S2 and S3 at http://www.biology.ualberta.ca/king-jones_lab/Bujold_et_al_2010_Supplemental_data_MCB.pdf). Interestingly, NPC2e is strongly induced by cholesterol, while NPC2d expression is essentially shut down, which may suggest that the two genes have opposing functions. To validate these findings, we tested (i) whether we could recapitulate the results using qPCR and (ii) whether these responses were specific to cholesterol. For the latter, we reared larvae on media containing either 1% canola oil or 1% tristearin instead of cholesterol. Canola oil is mainly composed of monounsaturated fatty acids; oleic acid, linoleic acid, and some trace amounts of plant sterols (27). Tristearin, on the other hand, is a naturally occurring triglyceride that contains only stearic acid as a fatty acid and has no toxicity in Drosophila (11). We found that both NPC2d and NPC2e are highly specific to cholesterol (Fig. 4I and J). NPC2d was not repressed by canola oil or by tristearin, while NPC2e failed to be induced by these substances, suggesting that both genes play specific roles in cholesterol homeostasis.
FIG. 5.
A high-cholesterol diet phe nocopies the transcriptional response caused by the DHR96 mutation. (A) Global statistical analysis of two microarray data sets based on a Pearson correlation. Fold changes (FC) of all genes, affected either by the DHR96 mutation alone (y axis) or by feeding a 1% cholesterol diet to wild-type animals (x axis), were plotted against each other using the SPSS software package. Linear fold changes are shown on log2-based axes. The correlation between the two data sets is indicated by the r value, and the resultant significance is indicated by the P value (two-tailed Student's t test). Some genes mentioned in the text are indicated in red. (B) Comparison of microarray data sets representing 73 genes upregulated in response to high cholesterol (left circle) or 102 upregulated due to a mutation in the DHR96 gene (right circle). The P value indicates the significance of the overlap, based on a χ2 test. (C) Analysis similar to that in panel B; however, here the data sets comprise 55 genes downregulated in response to high cholesterol (left circle) and 43 genes downregulated in DHR96 mutants that were maintained on standard medium. Average expected overlaps and P values based on χ2 calculations are indicated.
LpR1, which encodes a homolog of the LDL receptor protein family, is upregulated ∼3-fold by cholesterol but displays much stronger induction by canola oil and tristearin, demonstrating that this gene responds to a wide range of lipids (Fig. 4K). CG5932 encodes a midgut-specific lipase, but despite an apparent role in fat breakdown rather than cholesterol homeostasis, this gene is clearly specifically regulated by cholesterol. In particular, CG5932 shows very strong repression in the presence of cholesterol but is completely unaffected by canola oil; however, the gene shows some repression by tristearin (Fig. 4L).
Other significantly induced genes include Atet, which codes for an ABC transporter, and egghead, which encodes a beta-1,4-mannosyltransferase required for glycosphingolipid biosynthesis (see Table S2 at http://www.biology.ualberta.ca/king-jones_lab/Bujold_et_al_2010_Supplemental_data_MCB.pdf). Some genes are effectively repressed by cholesterol, such as FANCL, which encodes a ubiquitin E3 ligase, and CG10300, which encodes a protein that harbors a Sec14p-like lipid-binding domain (see Table S2 at the URL given above).
We used gene ontology statistics to further characterize the results obtained from the high-cholesterol microarrays. In the wild type, relatively few groups were induced by cholesterol. Most notably, we found 4 homeobox-containing genes (twin of eyeless, dorsotonalis, NK7.1, and CG10037), 3 genes encoding a DUF227 domain (“domain of unknown function 227”), and two sulfotransferase genes (Table 1, rows 6, 13, and 3, respectively). Based on the group terms “DNA-binding/transcription factor” and “nucleus,” we identified 15 and 19 transcripts, respectively, among the 73 cholesterol-induced genes (Table 1, rows 22 and 24), suggesting that the exposure to high cholesterol triggered the induction of a complex transcription factor network. Groups that stand out among the 55 genes repressed by cholesterol are five cytochrome P450 enzymes, three glutathione transferases, and two UDP-glucoronosyl/UDP-glucosyl transferases, all protein families that are linked to detoxification responses, suggesting that high levels of dietary cholesterol do not trigger a detoxification response. Enriched GO terms are “metabolic process” with 16 genes (row 27) and “catalytic activity” with 12 genes (row 23), perhaps suggesting that high concentrations of specific nutrients trigger the downregulation of enzymes that are only required when small quantities of the nutrient are available.
TABLE 1.
Enrichment of gene families, gene ontology terms, and related array results in data sets derived from “0% versus 1% cholesterol” microarray experimentsa
| Row | Gene list | Category | Set size (n) | WT vs WT + 1% cholesterol up (73) |
WT vs WT + 1% cholesterol down (55) |
DHR96 vs WT (L3, 0%) up (102) |
DHR96 vs WT (L3, 0%) down (43) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overlap | Expect | P value | Overlap | Expect | P value | Overlap | Expect | P value | Overlap | Expect | P value | ||||
| Protein domains | |||||||||||||||
| 1 | Acyl-CoA-binding protein | IPR000582 | 7 | 0 | 0.0 | >0.01 | 2 | 0.0 | 1.4E−43 | 0 | 0.0 | >0.01 | 0 | 0.0 | >0.01 |
| 2 | Insect cuticle protein | IPR000618 | 104 | 0 | 0.4 | >0.01 | 3 | 0.3 | 9.4E−07 | 0 | 0.6 | >0.01 | 0 | 0.2 | >0.01 |
| 3 | Sulfotransferase | IPR000863 | 10 | 2 | 0.0 | 2.1E−23 | 1 | 0.0 | 1.3E−08 | 1 | 0.1 | 4.7E−05 | 1 | 0.0 | 1.0E−10 |
| 4 | Cytochrome P450 | IPR001128 | 92 | 0 | 0.4 | >0.01 | 5 | 0.3 | 5.9E−20 | 0 | 0.5 | >0.01 | 4 | 0.2 | 1.2E−16 |
| 5 | Cell. retinaldehyde binding | IPR001251 | 24 | 0 | 0.1 | >0.01 | 1 | 0.1 | 4.4E−04 | 2 | 0.1 | 2.1E−07 | 2 | 0.1 | 9.6E−17 |
| 6 | Homeobox | IPR001356 | 114 | 4 | 0.4 | 8.0E−08 | 0 | 0.3 | >0.01 | 4 | 0.6 | 1.6E−05 | 0 | 0.3 | >0.01 |
| 7 | SCD/R | IPR002198 | 56 | 0 | 0.2 | >0.01 | 4 | 0.2 | 2.2E−21 | 0 | 0.3 | >0.01 | 1 | 0.1 | >0.01 |
| 8 | UDP | IPR002213 | 35 | 0 | 0.1 | >0.01 | 2 | 0.1 | 2.9E−09 | 0 | 0.2 | >0.01 | 2 | 0.1 | 1.1E−11 |
| 9 | NAM/LA amidase | IPR002502 | 11 | 0 | 0.0 | >0.01 | 0 | 0.0 | >0.01 | 2 | 0.1 | 1.7E−15 | 0 | 0.0 | >0.01 |
| 10 | Chitin-binding peritrophin A | IPR002557 | 84 | 0 | 0.3 | >0.01 | 0 | 0.2 | >0.01 | 3 | 0.5 | 1.5E−04 | 0 | 0.2 | >0.01 |
| 11 | NPC2-like domain | IPR003172 | 8 | 1 | 0.0 | 3.7E−08 | 1 | 0.0 | 1.7E−10 | 1 | 0.0 | 4.2E−06 | 1 | 0.0 | 3.8E−13 |
| 12 | Glutathione transferase | IPR004046 | 42 | 0 | 0.2 | >0.01 | 3 | 0.1 | 2.0E−16 | 0 | 0.2 | >0.01 | 1 | 0.1 | 3.5E−03 |
| 13 | DUF227 | IPR004119 | 48 | 3 | 0.2 | 6.5E−11 | 3 | 0.1 | 2.1E−14 | 1 | 0.3 | >0.01 | 4 | 0.1 | 6.3E−32 |
| 14 | DUF725 | IPR007931 | 10 | 0 | 0.0 | >0.01 | 0 | 0.0 | >0.01 | 4 | 0.1 | 1.2E−64 | 0 | 0.0 | >0.01 |
| Array results | |||||||||||||||
| 15 | WT vs WT + 1% cholesterol | Up | 73 | 73 | 73 | NA | 0 | 0 | NA | 13 | 0.4 | 6.9E−90 | 0 | 0.2 | >0.01 |
| 16 | WT vs WT + 1% cholesterol | Down | 55 | 0 | 0 | NA | 55 | 55 | NA | 1 | 0.3 | >0.01 | 22 | 0.1 | <1E−999 |
| 17 | w1118 vs hs-DHR96 | Up | 153 | 0 | 0.6 | >0.01 | 1 | 0.4 | >0.01 | 0 | 0.8 | >0.01 | 1 | 0.4 | >0.01 |
| 18 | w1118 vs hs-DHR96 | Down | 241 | 7 | 0.9 | 2.7E−10 | 4 | 0.7 | 7.8E−05 | 9 | 1.3 | 1.2E−11 | 6 | 0.6 | 1.5E−13 |
| 19 | WT vs DHR96 (L2, 0%) | Up | 67 | 8 | 0.3 | 2.7E−52 | 3 | 0.2 | 2.2E−10 | 21 | 0.4 | 1.3E−258 | 0 | 0.2 | >0.01 |
| 20 | WT vs DHR96 (L2, 0%) | Down | 81 | 1 | 0.3 | >0.01 | 7 | 0.2 | 4.1E−44 | 2 | 0.4 | >0.01 | 9 | 0.2 | 1.2E−93 |
| GO description | |||||||||||||||
| 21 | Protein import into nucleus | 0000059/60 | 65 | 4 | 0.3 | 7.4E−14 | 0 | 0.2 | >0.01 | 1 | 0.4 | >0.01 | 0 | 0.1 | >0.01 |
| 22 | DNA-binding/TXN factor | 0003677/3700 | 1,023 | 15 | 4.0 | 1.2E−08 | 1 | 3.0 | >0.01 | 8 | 5.6 | >0.01 | 1 | 2.3 | >0.01 |
| 23 | Catalytic activity | 0003824 | 1,004 | 2 | 3.9 | >0.01 | 12 | 2.9 | 5.5E−08 | 6 | 5.5 | >0.01 | 11 | 2.3 | 3.6E−09 |
| 24 | Nucleus | 0005634 | 1,811 | 19 | 7.0 | 2.0E−06 | 2 | 5.3 | >0.01 | 11 | 9.8 | >0.01 | 2 | 4.1 | >0.01 |
| 25 | Protein phosphorylation | 0006468 | 315 | 6 | 1.2 | 1.3E−05 | 0 | 0.9 | >0.01 | 2 | 1.7 | >0.01 | 0 | 0.7 | >0.01 |
| 26 | Sphingolipid metabolism | Multiple terms | 33 | 1 | 0.1 | >0.01 | 0 | 0.1 | >0.01 | 5 | 0.2 | 3.2E−30 | 0 | 0.1 | >0.01 |
| 27 | Metabolic process | Multiple terms | 1471 | 3 | 5.7 | >0.01 | 16 | 4.3 | 4.3E−09 | 13 | 8.0 | >0.01 | 13 | 3.4 | 4.5E−08 |
Pairwise comparisons between gene sets derived from the high-cholesterol microarray (columns) and gene lists representing InterPro domains (rows 1 to 14), additional microarray results (categories are relative to wild type [WT]) (rows 15 to 20), and gene ontology terms (categories are GO terms) (rows 21 to 27). The number of genes in a set either is given in parentheses (columns) or can be found in a separate column (n), with a value for each row. Each pairwise comparison results in an observed overlap, an expected overlap (expect). and a P value (based on a χ2 test) that indicates the significance between the observed and expected overlaps. UDP, UDP-glucoronosyl/UDP-glucosyl transferase; NAM/LA amidase, N-acetylmuramoyl-l-alanine amidase; SCD/R, short-chain dehydrogenase/reductase; TXN, transcription; NA, not applicable; 0%, no cholesterol added to standard medium; 1%, cholesterol added to standard medium (wet weight).
We also compared the high-cholesterol array results to other microarray data, specifically DHR96 gain- and loss-of-function arrays (rows 17 to 20). Apart from genes that are upregulated due to ectopic expression of DHR96, we saw strong correlations between these different microarrays, indicating that DHR96 partially regulates the same genes that are affected by treatment with cholesterol. For the GO analysis of DHR96 mutants, we analyzed data from animals reared on a standard diet only. In the set upregulated by the DHR96 mutation, we found 4 homeobox-containing genes. Two of these transcription factor genes, dorsotonalis and NK7.1, are identical to those found to be upregulated by cholesterol, suggesting there is a common mechanism by which these transcription factors are regulated. The most significant GO term is “sphingolipid metabolism,” a group of 5 affected genes (row 26), which strongly suggests that DHR96 plays a role in sphingolipid processes, as well. Like sterols, sphingolipids play important roles in plasma membranes and cell signaling, and both molecule classes are enriched in lipid rafts. Furthermore, sphingolipids accumulate along with cholesterol in patients with Niemann-Pick type C disease (20), suggesting that homeostatic control of both lipid classes may be regulated by shared pathways. Transcripts affected by the DHR96 mutation with predicted roles in sphingolipid metabolism include egghead, CG16708 (a predicted d-erythrosphingosine kinase), CG15534 and CG15533 (both encode sphingomyelin phosphodiesterases), and ifc (a stearoyl-CoA 9-desaturase).
Exposure to high cholesterol phenocopies the DHR96 mutation.
If one compares the transcriptional consequences of a high-cholesterol diet to the effects of the DHR96 mutation alone, one notices that 53 of the 55 genes downregulated by cholesterol display corresponding lower levels of expression in DHR96 mutants than in controls (see Table S2 at http://www.biology.ualberta.ca/king-jones_lab/Bujold_et_al_2010_Supplemental_data_MCB.pdf) and that 72 of 73 genes that are upregulated in response to 1% cholesterol have higher levels in DHR96 mutants (see Table S2 at the URL given above), even though mutant animals were maintained on a standard diet without added cholesterol. To test whether any of these corresponding changes are significant in DHR96 mutants, we compared only significant gene sets. We found 102 genes that are significantly upregulated in the DHR96 mutant (Table 1), and a comparison with the set of 73 cholesterol-induced genes showed 13 overlapping genes, roughly 33 times higher than expected (P = 6.9E−90) (Fig. 5B and Table 1, row 15). Even more notable is the overlap of 22 genes when one compares the 43 genes that are downregulated in DHR96 mutants with the 55 genes repressed by high cholesterol (P < 10E−999) (Fig. 5C and Table 1, row 16). These data suggest that the administration of cholesterol through the diet phenocopies many of the transcriptional changes that are caused by disrupting DHR96 function. This observation is further corroborated by the fact that the actual fold changes are equally similar under both conditions, for instance, the top genes induced and repressed by high cholesterol, NPC2e and NPC2d, respectively, are also the most strongly affected genes in DHR96 mutants (see Tables S2 and S3 at http://www.biology.ualberta.ca/king-jones_lab/Bujold_et_al_2010_Supplemental_data_MCB.pdf).
To examine whether this correlation can be observed on a genome-wide level, we carried out a Pearson correlation for all fold changes observed under high cholesterol in the wild type or by the DHR96 mutation alone (Fig. 5A). We found that the correlation holds even in this global analysis, with a Pearson coefficient (r) of 0.640 and a P value of <0.01 based on a two-tailed Student t test. In agreement with this statistical analysis, we found that most genes fall into the lower left and upper right quadrants, as one would expect if the majority of genes are coordinately regulated.
To rule out a bias in the microarray design, we carried out a repeat experiment with the following differences: independent samples were collected and analyzed by a second experimenter, and gene expression was examined by microfluidics-based qPCR (Fluidigm) rather than microarray analysis. Each sample was analyzed for the expression of 34 genes (not counting controls), which were selected based on microrarray results, predicted functions, and published studies. To determine gene expression differentials with high reliability, we calculated relative fold changes based on the geometric mean of 5 internal control genes per sample (43). Of the 34 genes analyzed, 26 displayed greater than 1.2-fold changes in wild-type animals reared on standard medium with or without 1% cholesterol, while the remaining 8 genes showed no significant differences. In DHR96 mutants, all 26 genes showed gene expression changes in the same direction and of magnitude similar to those observed in the wild type in response to cholesterol (see Fig. S3 at http://www.biology.ualberta.ca/king-jones_lab/Bujold_et_al_2010_Supplemental_data_MCB.pdf). The strong correlation between the magnitudes of the gene expression changes confirmed our finding that feeding a high-cholesterol diet phenocopies the DHR96 mutation.
Finally, if we compare completely independent microarray studies, namely, data sets from mutant L2 larvae to wild-type L3 larvae reared on standard medium and 1% cholesterol, respectively, we still observe highly significant correlations (Table 1, rows 19/20), indicating that the transcriptional consequences of cholesterol are similar to those of the DHR96 mutation.
Taken together, we provide several lines of evidence demonstrating that the global transcriptional changes triggered by a high-cholesterol diet phenocopy the effects on gene expression caused by the DHR96 mutation alone. This not only corroborates a role for DHR96 in coordinating the global response to cholesterol, it also suggests that cholesterol in fact suppresses DHR96 function, since the removal of DHR96 seems to be equivalent to elevating cholesterol levels.
Cholesterol downregulates DHR96 transcription.
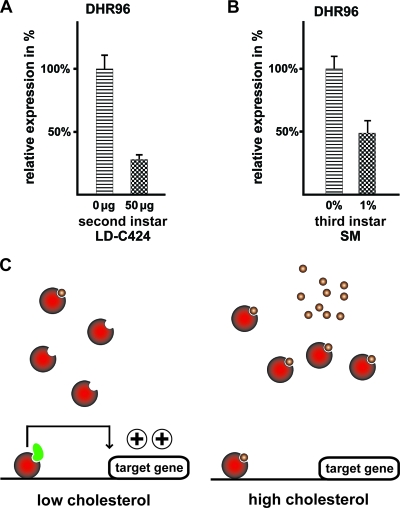
A simple explanation for the fact that dietary cholesterol can phenocopy the DHR96 mutation could be that cholesterol inactivates and/or downregulates DHR96. Given the fact that DHR96 can bind to cholesterol (18a), this may suggest that DHR96 is directly inactivated by a cholesterol metabolite. Alternatively, cholesterol may transcriptionally downregulate DHR96, possibly through an autoregulatory mechanism. We therefore analyzed DHR96 expression in wild-type larvae reared on different medium types supplemented or not with cholesterol. We found that DHR96 was repressed roughly 4-fold by cholesterol in L2 larvae reared on lipid-depleted medium supplemented with as little as 50 μg per vial (Fig. 6A). Similarly, wild-type L3 larvae reared on standard medium had two-times-higher levels of DHR96 mRNA than their counterparts reared on a 1% cholesterol diet (Fig. 6B). These data suggest that DHR96 transcription is downregulated by cholesterol in a concentration-dependent manner. Since DHR96 itself mediates responses to dietary cholesterol, it is possible that the DHR96 gene is controlled by an autoregulatory feedback loop.
FIG. 6.
Cholesterol represses DHR96. (A) qPCR analysis based on wild-type L2 larvae raised on lipid-depleted (LD) C424 medium with or without 50 μg cholesterol. (B) qPCR analysis based on wild-type L3 larvae reared on a standard medium (SM) with or without 1% cholesterol. (C) Model for DHR96 function. For simplicity, DHR96 is depicted as a monomeric protein. (Left) Under conditions of low dietary cholesterol levels, DHR96 protein remains active due to low ligand concentrations. This allows coactivators to bind (green) and transcription to occur. (Right) High levels of dietary cholesterol result in high concentrations of a presumptive DHR96 ligand (small circles). These ligands deactivate DHR96 by preventing coregulators from binding.
DISCUSSION
The lethal phenotype of DHR96 mutants.
In the present work, we demonstrated that DHR96 is a critical regulator of cholesterol homeostasis and that it mediates transcriptional changes in response to dietary cholesterol. A central question is why DHR96 mutants fail to survive on a low-cholesterol diet. The most likely answer is that DHR96 mutants are unable to recognize that they are ingesting a low-cholesterol diet and therefore fail to implement the necessary transcriptional programs that are required to adapt to conditions of severe cholesterol paucity. For instance, genes that function to increase cellular cholesterol levels, such as NPC1b and Lip3, are transcriptionally upregulated in wild-type larvae when cholesterol concentrations decline. In DHR96 mutants, however, the upregulation of NPC1b and Lip3 never reaches wild-type expression levels (Fig. 4A and E; see also Fig. S4 at http://www.biology.ualberta.ca/king-jones_lab/Bujold_et_al_2010_Supplemental_data_MCB.pdf). NPC1b is specifically expressed in the gut and is required for cholesterol absorption, while the predicted function of the Lip3 protein is the hydrolyzing of cholesteryl esters, resulting in the liberation of free cholesterol. This suggests that at least two genes required for increasing cellular cholesterol levels are submaximally expressed in DHR96 mutants.
It appears likely that insufficient cholesterol absorption in DHR96 mutants merely contributes to the failure to survive on low cholesterol, because we found only a 20% reduction of total cholesterol and cholesteryl ester levels in mutant animals compared to controls (Fig. 2A and B). With this in mind, it is interesting that genes with roles in reducing cellular cholesterol concentrations are substantially overexpressed in DHR96 mutants, suggesting that cellular cholesterol efflux is increased in mutant animals compared to the wild type. The ACAT and ABCA1 genes fall into this category, since ACAT reduces the pool of free cellular cholesterol through an esterification reaction that adds a fatty acid to the molecule, while ABCA1 encodes an ATP transporter that is involved in the active removal of cholesterol by pumping it across the cell membrane. Wild-type ACAT and ABCA1 are downregulated in response to declining cholesterol concentrations, which is in agreement with the idea that under conditions of dangerously low cholesterol levels, genes that increase cholesterol efflux must be turned off. On lipid-depleted medium, ACAT and ABCA1 mRNA levels are substantially higher in DHR96 mutants than in controls (Fig. 4G and H), suggesting that mutant cells actively reduce cellular cholesterol concentrations under conditions of low dietary cholesterol. At the same time, cholesterol uptake is reduced as well, thus aggravating the situation.
A model for DHR96 function.
The fact that treatment with 1% cholesterol largely phenocopies the genome-wide effects of the DHR96 mutation strongly suggests that DHR96 functions at the top of a gene network controlling the systemic response to varying levels of dietary sterols. In a recent study, we showed that DHR96 can bind cholesterol in vivo, suggesting that this nuclear receptor is a cellular sensor for varying sterol levels (18a). This would suggest that cholesterol or a very similar metabolite acts as a direct ligand for DHR96; however, a key question is whether such a ligand would act as an agonist or an antagonist. The “constitutive androstane receptor” (CAR), which is one of the three vertebrate nuclear receptor orthologs of DHR96 (see Fig. S1 at http://www.biology.ualberta.ca/king-jones_lab/Bujold_et_al_2010_Supplemental_data_MCB.pdf), displays the unusual feature that it acts as a constitutively active transcription factor in the absence of a ligand. Androstane metabolites, however, act as inverse agonists and deactivate murine CAR upon ligand binding (14, 42). Prior to this finding, all nuclear receptors were believed to be activated by ligand binding; however, it remains unclear how widespread this mode of nuclear receptor inactivation is.
Our observations are best explained by an inverse agonist mechanism similar to what has been described for CAR (Fig. 6C). Given the fact that DHR96 is only required for survival when the animal is feeding on a low-cholesterol diet, it follows that cholesterol metabolites that might act as ligands for DHR96 are scarce under these conditions. This suggests that DHR96 is active in the absence of a ligand. Conversely, the DHR96 gene is not required for survival when cholesterol concentrations are sufficiently high, suggesting that the DHR96 protein is inactive when potential ligands are abundant, again favoring the view of an inverse agonist mechanism. Perhaps the strongest argument for a mechanism through an inverse agonist derives from the fact that it provides the simplest explanation for the fact that a high-cholesterol diet is able to phenocopy the DHR96 mutation. In accordance with the finding that DHR96 binds to cholesterol (18a), this model predicts that a high-cholesterol diet would result in a widespread deactivation of DHR96 receptor molecules, essentially turning off DHR96 activity. Consequently, one would expect that the molecular consequences of deactivated DHR96 protein (via a high-cholesterol treatment) or removing functional protein altogether (via a null mutation) are very similar indeed. The observation that DHR96 mRNA levels decline in response to increasing cholesterol levels (Fig. 6A and B) is also compatible with the idea of inverse agonism. Since DHR96 mRNA is possibly regulated by its own protein, one would predict that increasing cholesterol levels reduce DHR96 activity, which in turn results in reduced transcription of the DHR96 gene itself.
The cholesterol gene network and DHR96.
Nutrigenomics is a powerful strategy for identifying genes that act in nutrient-dependent pathways (4, 34), and our studies represent a first step toward the identification of genes with hitherto unknown roles in cholesterol homeostasis. We show here that the expression of four Niemann-Pick genes—NPC1b and NPC2c, -d, and -e—is strongly dependent on the concentration of dietary cholesterol. Other identified genes with predicted roles in sterol biology are ACAT, ABCA1, Lip3, and Cyp12d1 (see Fig. S4 at http://www.biology.ualberta.ca/king-jones_lab/Bujold_et_al_2010_Supplemental_data_MCB.pdf). We also found cholesterol-responsive genes with no known links to cholesterol homeostasis. For instance, CG5932, encoding a gastric lipase; FANCL, encoding a ubiquitin E3 ligase; and CG31148, encoding a glucosylceramidase, were all downregulated in response to increasing cholesterol concentrations (see Fig. S4 at the URL given above). A recent study showed that DHR96 mutants are resistant to diet-induced obesity, which is at least in part due to the role of DHR96 in regulating CG5932, confirming that this nuclear receptor also has important roles in controlling lipid metabolism (39). In addition, a study of mice demonstrated that the “Idol” ubiquitin E3 ligase is transcriptionally induced by LXR and triggers proteolytic degradation of the LDL receptor via ubiquitination, thereby downregulating cellular cholesterol uptake (51). Future work may provide insight into whether FANCL has comparable roles in regulating cholesterol homeostasis in Drosophila.
The present study provides new avenues to model cholesterol homeostasis in Drosophila. By studying the functions of nuclear receptor DHR96 and the genomic responses to dietary cholesterol, we are beginning to unravel the regulatory networks that control cholesterol absorption, transport, and metabolism. Our work also provides a starting point for identifying novel genes with roles in cholesterol homeostasis, and this study provides the framework to conduct comparative genomics to analyze conserved lipid signaling pathways in other organisms.
Acknowledgments
This work was supported by CIHR, NSERC, and AHFMR operating grants, as well as AIF and CFI infrastructure grants.
We thank A. Waskiewicz, D. Pilgrim, and M. Kostiuk for critical comments on the manuscript. We thank R. Lehner for providing training and support for the GC analysis and S. Eaton for helpful discussions regarding sterol biology and fly media. We also thank M. Feldmann for access to the microplate reader.
Footnotes
Published ahead of print on 23 November 2009.
REFERENCES
- 1.Altmann, S. W., H. R. Davis, Jr., L. J. Zhu, X. Yao, L. M. Hoos, G. Tetzloff, S. P. Iyer, M. Maguire, A. Golovko, M. Zeng, L. Wang, N. Murgolo, and M. P. Graziano. 2004. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science 303:1201-1204. [DOI] [PubMed] [Google Scholar]
- 2.Andres, A. J., and C. S. Thummel. 1994. Methods for quantitative analysis of transcription in larvae and prepupae, p. 565-573. In L. S. B. Goldstein and E. A. Fyrberg (ed.), Drosophila melanogaster: practical uses in cell and molecular biology. Academic Press, New York, NY. [DOI] [PubMed]
- 3.Baldan, A., D. D. Bojanic, and P. A. Edwards. 2009. The ABCs of sterol transport. J. Lipid Res. 50(Suppl.):S80-S85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, M., A. Hamm, and M. J. Pankratz. 2004. Linking nutrition to genomics. Biol. Chem. 385:593-596. [DOI] [PubMed] [Google Scholar]
- 5.Bjorkhem, I. 2009. Are side-chain oxidized oxysterols regulators also in vivo? J. Lipid Res. 50(Suppl.):S213-S218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, M. S., and J. L. Goldstein. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331-340. [DOI] [PubMed] [Google Scholar]
- 7.Carstea, E. D., J. A. Morris, K. G. Coleman, S. K. Loftus, D. Zhang, C. Cummings, J. Gu, M. A. Rosenfeld, W. J. Pavan, D. B. Krizman, J. Nagle, M. H. Polymeropoulos, S. L. Sturley, Y. A. Ioannou, M. E. Higgins, M. Comly, A. Cooney, A. Brown, C. R. Kaneski, E. J. Blanchette-Mackie, N. K. Dwyer, E. B. Neufeld, T. Y. Chang, L. Liscum, J. F. Strauss III, K. Ohno, M. Zeigler, R. Carmi, J. Sokol, D. Markie, R. R. O'Neill, O. P. van Diggelen, M. Elleder, M. C. Patterson, R. O. Brady, M. T. Vanier, P. G. Pentchev, and D. A. Tagle. 1997. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 277:228-231. [DOI] [PubMed] [Google Scholar]
- 8.Chawla, A., W. A. Boisvert, C. H. Lee, B. A. Laffitte, Y. Barak, S. B. Joseph, D. Liao, L. Nagy, P. A. Edwards, L. K. Curtiss, R. M. Evans, and P. Tontonoz. 2001. A PPAR gamma-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol. Cell 7:161-171. [DOI] [PubMed] [Google Scholar]
- 9.Clark, A. J., and K. Bloch. 1959. Absence of sterol synthesis in insects. J. Biol. Chem. 234:2578-2582. [PubMed] [Google Scholar]
- 10.Dobrosotskaya, I. Y., A. C. Seegmiller, M. S. Brown, J. L. Goldstein, and R. B. Rawson. 2002. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science 296:879-883. [DOI] [PubMed] [Google Scholar]
- 11.Etges, W. J., C. L. Veenstra, and L. L. Jackson. 2006. Premating isolation is determined by larval rearing substrates in cactophilic Drosophila mojavensis. VII. Effects of larval dietary fatty acids on adult epicuticular hydrocarbons. J. Chem. Ecol. 32:2629-2646. [DOI] [PubMed] [Google Scholar]
- 12.Fitzky, B. U., H. Glossmann, G. Utermann, and F. F. Moebius. 1999. Molecular genetics of the Smith-Lemli-Opitz syndrome and postsqualene sterol metabolism. Curr. Opin. Lipidol. 10:123-131. [DOI] [PubMed] [Google Scholar]
- 13.Fluegel, M. L., T. J. Parker, and L. J. Pallanck. 2006. Mutations of a Drosophila NPC1 gene confer sterol and ecdysone metabolic defects. Genetics 172:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forman, B. M., I. Tzameli, H. S. Choi, J. Chen, D. Simha, W. Seol, R. M. Evans, and D. D. Moore. 1998. Androstane metabolites bind to and deactivate the nuclear receptor CAR-β. Nature 395:612-615. [DOI] [PubMed] [Google Scholar]
- 15.Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert, L. 2004. Halloween genes encode P450 enzymes that mediate steroid hormone biosynthesis in Drosophila melanogaster. Mol. Cell Endocrinol. 215:1-10. [DOI] [PubMed] [Google Scholar]
- 17.Hobson, R. P. 1935. On a fat-soluble growth factor required by blow-fly larvae: distribution and properties. Biochem. J. 29:1292-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hobson, R. P. 1935. On a fat-soluble growth factor required by blow-fly larvae: identity of the growth factor with cholesterol. Biochem. J. 29:2023-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Horner, M. A., K. Pardee, S. Liu, K. King-Jones, G. Lajoie, A. Edwards, H. M. Krause, and C. S. Thummel. 2009. The Drosophila DHR96 nuclear receptor binds cholesterol and regulates cholesterol homeostasis. Genes Dev. 23:2711-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, X., J. T. Warren, J. Buchanan, L. I. Gilbert, and M. P. Scott. 2007. Drosophila Niemann-Pick type C-2 genes control sterol homeostasis and steroid biosynthesis: a model of human neurodegenerative disease. Development 134:3733-3742. [DOI] [PubMed] [Google Scholar]
- 20.Ioannou, Y. A. 2005. Guilty until proven innocent: the case of NPC1 and cholesterol. Trends Biochem. Sci. 30:498-505. [DOI] [PubMed] [Google Scholar]
- 21.King-Jones, K., M. A. Horner, G. Lam, and C. S. Thummel. 2006. The DHR96 nuclear receptor regulates xenobiotic responses in Drosophila. Cell Metab. 4:37-48. [DOI] [PubMed] [Google Scholar]
- 22.Kunte, A. S., K. A. Matthews, and R. B. Rawson. 2006. Fatty acid auxotrophy in Drosophila larvae lacking SREBP. Cell Metab. 3:439-448. [DOI] [PubMed] [Google Scholar]
- 23.Laudet, V., and F. Bonneton. 2005. Evolution of nuclear hormone receptors in insects, p. 287-318. In L. I. Gilbert, K. Iatrou, and S. S. Gill (ed.), Molecular insect science, vol. 3. Elsevier, Oxford, United Kingdom. [Google Scholar]
- 24.Loftus, S. K., J. A. Morris, E. D. Carstea, J. Z. Gu, C. Cummings, A. Brown, J. Ellison, K. Ohno, M. A. Rosenfeld, D. A. Tagle, P. G. Pentchev, and W. J. Pavan. 1997. Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science 277:232-235. [DOI] [PubMed] [Google Scholar]
- 25.Maroni, G., and C. Stamey. 1983. Use of blue food to select synchronous late third instar larvae. Drosoph. Inf. Serv. 59:142-143. [Google Scholar]
- 26.Naureckiene, S., D. E. Sleat, H. Lackland, A. Fensom, M. T. Vanier, R. Wattiaux, M. Jadot, and P. Lobel. 2000. Identification of HE1 as the second gene of Niemann-Pick C disease. Science 290:2298-2301. [DOI] [PubMed] [Google Scholar]
- 27.O'Brian, R. 2004. Fats and oils: formulating and processing for applications, 2nd ed., p. 31-34. CRC Press, Boca Raton, FL.
- 28.Oram, J. F. 2002. Molecular basis of cholesterol homeostasis: lessons from Tangier disease and ABCA1. Trends Mol. Med. 8:168-173. [DOI] [PubMed] [Google Scholar]
- 29.Oram, J. F., and R. M. Lawn. 2001. ABCA1. The gatekeeper for eliminating excess tissue cholesterol. J. Lipid Res. 42:1173-1179. [PubMed] [Google Scholar]
- 30.Peet, D. J., B. A. Janowski, and D. J. Mangelsdorf. 1998. The LXRs: a new class of oxysterol receptors. Curr. Opin. Genet. Dev. 8:571-575. [DOI] [PubMed] [Google Scholar]
- 31.Peet, D. J., S. D. Turley, W. Ma, B. A. Janowski, J. M. Lobaccaro, R. E. Hammer, and D. J. Mangelsdorf. 1998. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell 93:693-704. [DOI] [PubMed] [Google Scholar]
- 32.Repa, J. J., and D. J. Mangelsdorf. 2002. The liver X receptor gene team: potential new players in atherosclerosis. Nat. Med. 8:1243-1248. [DOI] [PubMed] [Google Scholar]
- 33.Rigamonti, E., L. Helin, S. Lestavel, A. L. Mutka, M. Lepore, C. Fontaine, M. A. Bouhlel, S. Bultel, J. C. Fruchart, E. Ikonen, V. Clavey, B. Staels, and G. Chinetti-Gbaguidi. 2005. Liver X receptor activation controls intracellular cholesterol trafficking and esterification in human macrophages. Circ. Res. 97:682-689. [DOI] [PubMed] [Google Scholar]
- 34.Ruden, D. M., M. De Luca, M. D. Garfinkel, K. L. Bynum, and X. Lu. 2005. Drosophila nutrigenomics can provide clues to human gene-nutrient interactions. Annu. Rev. Nutr. 25:499-522. [DOI] [PubMed] [Google Scholar]
- 35.Sahoo, D., T. C. Trischuk, T. Chan, V. A. Drover, S. Ho, G. Chimini, L. B. Agellon, R. Agnihotri, G. A. Francis, and R. Lehner. 2004. ABCA1-dependent lipid efflux to apolipoprotein A-I mediates HDL particle formation and decreases VLDL secretion from murine hepatocytes. J. Lipid Res. 45:1122-1131. [DOI] [PubMed] [Google Scholar]
- 36.Sang, J. 1978. The nutritional requirements of Drosophila, p. 159-190. In M. Ashburner and T. R. F. Wright (ed.), The genetics and biology of Drosophila, vol. 2a. Academic Press, New York, NY.
- 37.Seegmiller, A. C., I. Dobrosotskaya, J. L. Goldstein, Y. K. Ho, M. S. Brown, and R. B. Rawson. 2002. The SREBP pathway in Drosophila: regulation by palmitate, not sterols. Dev. Cell 2:229-238. [DOI] [PubMed] [Google Scholar]
- 38.Shulenin, S., L. M. Nogee, T. Annilo, S. E. Wert, J. A. Whitsett, and M. Dean. 2004. ABCA3 gene mutations in newborns with fatal surfactant deficiency. N. Engl. J. Med. 350:1296-1303. [DOI] [PubMed] [Google Scholar]
- 39.Sieber, M. H., and C. S. Thummel. 2009. The DHR96 nuclear receptor controls triacylglycerol homeostasis in Drosophila. Cell Metab. 10:481-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tabas, I. 2002. Consequences of cellular cholesterol accumulation: basic concepts and physiological implications. J. Clin. Invest. 110:905-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tijet, N., C. Helvig, and R. Feyereisen. 2001. The cytochrome P450 gene superfamily in Drosophila melanogaster: annotation, intron-exon organization and phylogeny. Gene 262:189-198. [DOI] [PubMed] [Google Scholar]
- 42.Tzameli, I., and D. D. Moore. 2001. Role reversal: new insights from new ligands for the xenobiotic receptor CAR. Trends Endocrinol Metab. 12:7-10. [DOI] [PubMed] [Google Scholar]
- 43.Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe, and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voght, S. P., M. L. Fluegel, L. A. Andrews, and L. J. Pallanck. 2007. Drosophila NPC1b promotes an early step in sterol absorption from the midgut epithelium. Cell Metab. 5:195-205. [DOI] [PubMed] [Google Scholar]
- 45.Wang, D. Q. 2007. Regulation of intestinal cholesterol absorption. Annu. Rev. Physiol. 69:221-248. [DOI] [PubMed] [Google Scholar]
- 46.Wang, X., R. Sato, M. S. Brown, X. Hua, and J. L. Goldstein. 1994. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell 77:53-62. [DOI] [PubMed] [Google Scholar]
- 47.Wettenhall, J. M., and G. K. Smyth. 2004. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics 20:3705-3706. [DOI] [PubMed] [Google Scholar]
- 48.Wu, Z., R. Irizarry, R. Gentlemen, F. Martinez-Murillo, and F. Spencer. 2004. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99:909-917. [Google Scholar]
- 49.Yokoyama, C., X. Wang, M. R. Briggs, A. Admon, J. Wu, X. Hua, J. L. Goldstein, and M. S. Brown. 1993. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell 75:187-197. [PubMed] [Google Scholar]
- 50.Yu, H., and S. B. Patel. 2005. Recent insights into the Smith-Lemli-Opitz syndrome. Clin. Genet. 68:383-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zelcer, N., C. Hong, R. Boyadjian, and P. Tontonoz. 2009. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science 325:100-104. [DOI] [PMC free article] [PubMed] [Google Scholar]