Abstract
The Polycomb group proteins (PcGs) play a vital role throughout development by maintaining precise gene expression patterns. In Drosophila melanogaster, PcG-mediated gene silencing is achieved through DNA elements called Polycomb response elements (PREs); however, the mechanism for establishing silencing and the requirements and composition of a working PRE are not fully understood. We have used the computer program jPREdictor to uncover PREs located within the invected (inv) locus. The functionalities of these predicted PREs were tested in two different assays: one analyzing their abilities to maintain expression of a β-galactosidase reporter gene and the other evaluating their abilities to establish pairing-sensitive silencing of the mini-white reporter in the vector pCaSpeR. We have identified two previously uncharacterized PREs at the inv gene and demonstrate that they produce similar results in the two assays. Our results indicate that clusters of protein binding sites do not accurately predict PREs and provide new insight into the DNA sequence requirements for the binding of the PcG protein Pho. Finally, our data show that PREs and regulatory DNA from different genes can function together to establish PcG-mediated silencing, highlighting the versatility of PREs despite discrepancies in the number and location of DNA binding sites.
Polycomb group (PcG) genes encode a large group of conserved proteins that act on chromatin to repress transcription. Originally identified in Drosophila melanogaster as silencers of homeotic genes, PcG proteins act as transcriptional repressors for many diverse targets. PcG proteins act in multiple protein complexes to modify chromatin with marks typically associated with gene repression, including trimethylation of histone H3 at lysine 27 and ubiquitination of H2A at lysine 119 (for a review, see reference 32). Histone deacetylases may also be involved. The exact protein complexes and mechanisms of PcG protein function are areas of intense investigation, with new complexes still being discovered (for a review, see references 32 and 38). Genetic evidence suggests that not all PcG proteins work on all targets (41), so it is probable that, in addition to having shared targets, different PcG protein complexes may regulate different genes. While all PcG proteins are specifically associated with chromatin, the only known PcG proteins with sequence-specific DNA binding activity are Pleiohomeotic (Pho) and the related protein Pho-like (Phol) (3, 5). Pho is present in a complex with dSfmbt (Pho-RC) and is thought to play a key role in recruitment of other PcG protein complexes (24).
In Drosophila, PcG proteins are recruited to specific cis-regulatory DNA elements called Polycomb response elements (PREs). These elements are typically found upstream of the transcript region and vary greatly in size and sequence (for a review, see reference 31). Extensive analysis of PREs at a variety of genes, including the biothorax complex (BX-C) genes, engrailed (en), polyhomeotic (PH), and others, has revealed that PREs consist of multiple short motifs (34). DNA binding proteins implicated in PRE function include the GAGA factor (GAF) (42) and Pipsqueak (Psq) (27), which bind the same sequence, and Zeste (20, 36), Pho (5, 28), Sp1/Krüppel-like factor (KLF) family members (4), DSP1 (9), and Grainyhead (Grh) (2).
As mentioned previously, the Pho subunit of Pho-RC is a DNA binding protein and has been shown to be a central component of PREs and PcG protein function. In vitro binding studies of the Pho protein and its mammalian homolog, YY1, have resulted in the generation of a core consensus sequence for Pho binding (GCCAT) (14, 21, 29). Recent chromatin immunoprecipitation (ChIP)-on-chip experiments have led to the development of longer, more stringent versions of the Pho consensus sequence (26, 33). Once the Pho-RC complex is bound to PREs, it is believed to assist in actively recruiting the other PcG complexes to DNA, placing Pho in a role of great importance for the establishment of PcG-mediated silencing. While it is easy to speculate that Pho is a central component of PcG function, it is interesting that the removal of Pho consensus sequences from PREs does not always result in the complete loss of PcG-mediated silencing (4, 5); furthermore, while Pho mutants exhibit phenotypes associated with Polycomb (PC), they are able to survive to the pharate adult stage (16). The identification of Phol, a factor with a high degree of homology to Pho and some degree of functional redundancy, helps explain why the pho mutant phenotype is not more severe (3).
Since deletion of any one individual DNA binding site often does not have a dramatic effect on PRE activity, it has been speculated that the multitude of binding sites work together to cooperatively recruit the factors necessary to establish gene silencing. Furthermore, it has been difficult to determine which combination of binding sites is required for PRE activity. To try and gain a better understanding of what key elements and binding sites may be required for PRE function, Ringrose et al. (35) analyzed experimentally defined PREs throughout the genome in an attempt to find similarities that might allow for detection of additional PREs. Their findings support the idea that the DNA binding sites work cooperatively, as PREs are likely to contain multiple binding sites close to one another, with multiple Pho binding sites being the strongest predictor of a PRE.
One particular target of PcG regulation, en, has been found to have two PREs and has been the subject of many studies regarding PcG function and PRE activity (1, 5, 22). en is part of a gene complex which includes another gene, invected (inv). The two genes have been shown previously to code for proteins with similar sizes and sequences, each containing a homeodomain (6, 18). Results from additional experiments indicate that the two genes are coregulated (17, 18). These data led us to the question of whether inv is regulated by its own set of PREs or whether en PREs regulate both genes. To date, no PREs have been identified at inv, so we sought to predict new PREs at inv by utilizing the established prediction methods. Here, we report the finding of two new PREs at inv. The PREs were identified using a combination of the jPREdictor program (13) and published ChIP-on-chip data (7). While the jPREdictor program helped us identify one of the PREs, it failed to identify the second PRE, presumably because this particular PRE lacks canonical Pho consensus sequences. Despite the lack of consensus Pho binding sites, we show that Pho binds to various regions of this PRE fragment in vitro, indicating that Pho is able to bind a variety of DNA sequences.
MATERIALS AND METHODS
PRE prediction.
A 30-kb DNA sequence (from FlyBase D. melanogaster release 5.9) surrounding the inv transcription start site was entered into the jPREdictor program (http://bibiserv.techfak.uni-bielefeld.de/jpredictor/). The frequencies of individual and paired DNA binding motifs (as described in reference 35) were determined, and the numerical data output from the jPREdictor program was plotted in Microsoft Excel. The coordinates of the identified fragments, including the fragment-specific primer sequences (with restriction sites and FLP recombination target [FRT] sequences not included), are listed in Table 1.
TABLE 1.
inv DNA fragments identified in this studya
| inv fragment | Fragment end | Genome coordinate | DNA sequence |
|---|---|---|---|
| 1 | 5′ | 7356766 | AAGAGAGAGAGGCAGCAA |
| 3′ | 7358046 | CCCAACTCTGTTCCACTT | |
| 2 | 5′ | 7353060 | ATCAATTAAGGCGGCTCC |
| 3′ | 7355512 | GAAATGCCAGCGATGAAA | |
| 3 | 5′ | 7358942 | CTACTGCCTTAAACTGGG |
| 3′ | 7360944 | CGTCCACATCTTCATCAC | |
| 4 | 5′ | 7362343 | CAGTAGAAACCCAGCAAG |
| 3′ | 7363955 | GAGATCTTCGGGTGAGAA | |
| 5 | 5′ | 7363514 | GCTCAAGCTGACTATTCC |
| 3′ | 7365058 | TGCCAGGATCTACTACAG | |
| 6 | 5′ | 7364681 | AGGAGCATCCGGTAAATC |
| 3′ | 7366548 | GCCACCACAACAATAAAC | |
| 7 | 5′ | 7366301 | CGCTGATGGATGTCAAAC |
| 3′ | 7368213 | AATTGGACCTGCTCCTTC |
The 5′ and 3′ ends of the DNA sequence for each test inv fragment are shown, and the coordinates for these sequences on chromosome 2R in the D. melanogaster genome (release version 5.9; FlyBase) are indicated.
Plasmid construction.
All constructs for the pairing-sensitive silencing (PSS) assay were derived from the pCaSpeR4 parent construct by using KpnI/XbaI sites for cloning of the inv fragments into pCaSpeR4. The SD10 construct was used to generate constructs for the embryonic PRE activity assay. SD10 was generated by cutting construct P[en2] (10) with SphI to remove a 2-kb fragment of DNA including PSE1 and PSE2 fragments. The inv fragments were generated by PCR using primers that contained SphI sites and FRT sites. Fragment insertion and orientation were verified by DNA sequencing.
Transgenic lines.
All constructs were injected into w1118 embryos by Genetic Services (Sudbury, MA), and transformants were identified by the presence of eye color. PRE deletion (ΔPRE) lines were generated by crossing transgenic lines to a line with heat-sensitive FLP and heat shocking embryos for 1 h at 37°C 2 days post-egg laying. Removal of the inv inserts was verified by PCR.
Embryo stainings.
Preparation of embryos for immunoperoxidase staining was done as described by DiNardo et al. (11). Anti-β-galactosidase (anti-β-Gal) primary antibody (1:15,000) was used with anti-rabbit secondary antibody (1:200), and antibodies were detected using an ABC elite kit (Vector Labs).
Gel mobility shift assay.
Full-length Pho was synthesized in vitro from the T7link/PHO vector (14) by using the TNT coupled transcription/translation system (Promega). Gel shifts were performed as described by Americo et al. (1), except that in vitro-synthesized full-length Pho was used in place of nuclear extract.
RESULTS
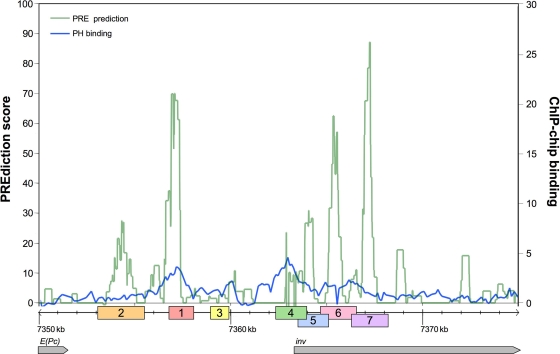
To better understand the coregulation of the en and inv genes, we sought to identify potential PREs upstream of and within the inv transcript. Previous studies by Ringrose et al. (35) concluded that combinations of specific PRE binding sites, including those for Pho, GAF, Zeste, and EN1, can predict that a DNA fragment is a PRE. This prediction analysis was subsequently presented in a format accessible online as the jPREdictor program (13). The DNA sequence around the inv gene (kb −12 to +13) was entered into the jPREdictor program, and prediction scores, graphically displayed in Fig. 1, were generated using the standard PC combined-motif analysis included in the program. The PRE prediction scores ranged from 0 to 90, with an average background score of 6.2.
FIG. 1.
Predicted PREs at inv. A DNA sequence from the 3′ end of the Enhancer of PC gene [E(Pc)] through the first 12 kb of the inv transcript was processed through the jPREdictor program, with scoring for multiple DNA binding motifs within a 250-bp window (green line). Published ChIP-on-chip data for PH, represented as the amount of change (n-fold) on a log2 scale, were aligned with the prediction data and plotted on the same graph (blue line) (7). DNA regions corresponding to potential PREs are represented by boxes along the x axis and numbered 1 through 7. y axis scales are adjusted for each data set to maximize the visual analysis of the data. The major transcription start site for inv as described by Coleman et al. (6) is shown.
Recent evidence from multiple groups has shown that the jPREdictor program, while useful in narrowing down DNA fragments that may have PRE functions, is not very accurate in identifying PREs, as it misidentifies many and misses some (33, 37). In addition, while our scores were not extremely low, they were not as high as scores for some well-defined PREs, such as PRE1 and PRE2 at en, which scored 235 and 134, respectively (data not shown). To help us better differentiate between real and background peaks in our prediction, we took advantage of published genome-wide ChIP-on-chip data (7) to give us a better understanding of the protein-DNA interactions around inv. Protein binding data for the PC and Polyhomeotic (PH) PcG proteins (7) were aligned with the prediction data and plotted on the graph in Fig. 1. As shown previously, the PC binding data reflect a general coating of PC along the length of the DNA, with no isolated peaks (data not shown). In contrast, the PH binding profile shows two main peaks—one approximately 4 kb upstream of the inv transcription start site and the second at the beginning of the inv transcript. The intergenic PH peak coincided with one of the strongest peaks identified by the jPREdictor program, suggesting that this particular prediction has a good chance of representing a biologically functioning PRE. Interestingly, the second PH binding peak, located around the transcription start site, did not align with any significant PRE prediction peaks. The remaining PRE prediction peaks did not coincide with any unique aspects of the PH or PC binding profiles. From these various data sets, we identified six DNA fragments (numbered 1, 2, and 4 through 7) that might be inv PREs and one control fragment (numbered 3) (Fig. 1).
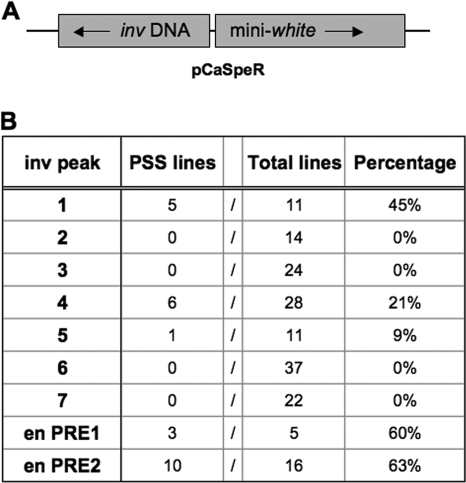
Predicted PRE fragments exhibit PSS.
The phenomenon of PSS was fortuitously discovered during studies of a 2.6-kb fragment of en DNA, subsequently found to contain two PREs (22, 23). Each of these PREs is able to repress the expression of the mini-white gene that is present in the constructs as a reporter gene. This repression is called PSS because it is much stronger in flies homozygous for a PRE-containing mini-white transgene than in flies heterozygous for the transgene. In practical terms, this often means that heterozygotes have orange eyes and that homozygotes have white eyes. Many other PRE fragments have since been shown to exhibit this ability to silence the mini-white gene, making PSS a good assay to screen for PREs among our inv DNA fragments. DNA fragments were inserted upstream of the mini-white gene in the orientation opposite of their orientation toward their own promoter (Fig. 2A). Since PRE fragments have been shown previously to be orientation independent (1, 25), these fragments should be able to function in either direction. Transgenic flies were obtained, and the number of fly lines exhibiting PSS compared to the total number of transgenic lines was determined for each inv fragment (Fig. 2B). Data on PSS by the well-characterized en PREs, PRE1 and PRE2, are included in Fig. 2B for comparison.
FIG. 2.
PSS by inv fragments. (A) Schematic of the pCaSpeR4 construct region involved in the PSS assay. inv test DNA fragments are present (3′-5′ orientation) upstream of the mini-white gene (5′-3′ orientation). (B) Results from the PSS assay. The number of fly lines exhibiting a decrease in eye color in homozygotes (PSS lines) versus heterozygotes is shown compared to the total number of transgenic fly lines analyzed (total lines). The percentage represents the percentage of total fly lines that exhibited PSS. PSS data for en PRE1 and PRE2 fragments (1, 22) are shown at the bottom of the table for reference.
The inv DNA fragments gave a range of results, from strong PSS activity (fragment 1) to moderate PSS activity (fragment 4) to little (fragment 5) or no PSS activity. Fragments 1 and 4 had the highest levels of PSS activity. Figure 1 shows that they are both located at the peak PH binding sites identified by ChIP, indicating that PcG proteins are able to bind to these DNA sequences in vivo and would likely contribute to their ability to silence mini-white in our assay. Fragment 5 overlaps with fragment 4 and appears to contain a low level of activity. Interestingly, while fragment 1 corresponds to a strong PRE prediction, indicative of having multiple PRE consensus binding sites present, fragment 4 has a much lower prediction score of 26.7, a comparatively insignificant score. This finding will be further discussed later in this study. It is also interesting that some of the fragments that have the highest scores for PRE prediction, such as fragments 6 and 7, have no PSS activity.
Maintenance of en-like expression patterns by predicted fragments.
The PSS assay gives a good indication of the abilities of the predicted fragments to invoke downregulation of a reporter gene, in this case, the mini-white gene. While the PRE-containing mini-white transgene is present in every cell in the embryo, the mini-white gene is poised for activation only in the eye and can be assayed only with this tissue. In embryos, PcG proteins act on genes that are active in some cells and repressed in others and the PcG proteins must differentiate between genes that should be kept off and those that should be left on. Thus, while the PSS assay gives a good indication of whether or not the inv fragments are capable of silencing a reporter gene, it does not tell us whether these DNA fragments are capable of recruiting PcG proteins in a discriminatory manner to maintain specific gene-silencing patterns.
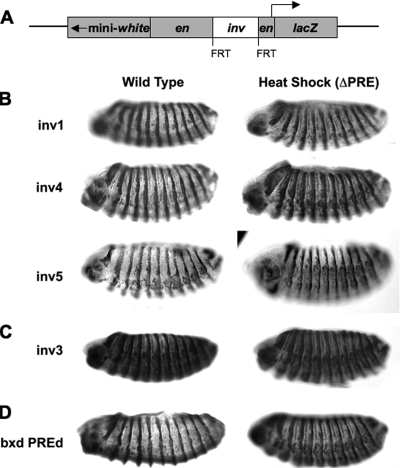
To specifically assay for PRE activity, we tested the abilities of the inv DNA fragments to maintain a specific pattern of expression of a reporter gene. We began with a vector that contains the en promoter and 8 kb of en regulatory DNA cloned upstream of the β-Gal reporter gene (P[en2] [10]) and that maintains expression in en pattern-like stripes throughout embryonic development. This construct contains regulatory DNA for en stripes and a 2-kb DNA fragment that contains en PRE1 and PRE2. We removed the 2-kb fragment that contains these PREs to generate our vector, SD10. Without the en PREs, β-Gal is expressed in stripes early in development but severe misexpression occurs during later developmental stages (10). Thus, our test inv fragments will restore the en stripe pattern late in development only if they function similarly to the en PREs.
Each inv DNA fragment was inserted into the SD10 construct between the upstream en regulatory DNA and the en promoter, yielding SD10-inv constructs. With the exception of fragment 4, all fragments were inserted in the same orientation as the en DNA and the β-Gal gene. Since fragment 4 contains the inv promoter, it was inserted in the reverse orientation to circumvent potential problems with the β-Gal reporter gene. In addition, each inv fragment was flanked by FRT sites to allow for specific excision of the fragments. These constructs were injected into fly embryos, and expression patterns of β-Gal in fixed embryos of transgenic fly lines were analyzed. While multiple lines were analyzed for each construct, representative results are shown in Fig. 3B and C.
FIG. 3.
Maintenance of en promoter-driven β-Gal expression patterns. (A) Schematic of the SD10 construct region involved in the PSS assay. The gray shaded boxes represent the portions of the parent SD10 construct. The lacZ reporter gene was placed under the control of the en promoter. An additional en regulatory sequence including enhancers was present further upstream. The mini-white gene was included for identification of transgenic fly lines and was placed in the reverse orientation to prevent interference with the transcription of lacZ. A 2-kb fragment containing the en PREs required for maintaining stripes was removed and replaced with the test PRE fragments (inv). FRT sites present on each end of the inv fragments allow for removal of the insert by FLP recombinase. (B, C, and D) An overnight collection of embryos was probed with anti-β-Gal primary antibody to observe patterns of lacZ expression. Wild-type embryos positive for PRE activity (B and D) and those with no PRE activity (C) are shown. After removal of the PRE fragments by heat shock, embryos were collected and stained in the same manner as the wild-type embryos (heat shock [ΔPRE]). Lateral views of stage 14 embryos are shown anterior left, dorsal up.
The results obtained from this assay correlated well with those obtained from the PSS assay. Fragments 2, 3, 6, and 7 had no activity in this assay (an example of these results is presented in Fig. 3C). In general, the embryos carrying these fragments showed large amounts of misexpression in between stripes, indicating a lack of PcG-mediated repression of the en promoter in our transgene. However, some SD10-inv constructs exhibited significant variability in the degree of misexpression produced among the independent transgenic fly lines. For example, while most lines generated from the SD10 construct carrying fragment 7 (SD10-inv7) showed significant misexpression, suggesting that inv fragment 7 (inv7) does not function as a PRE, a few SD10-inv7 lines showed very little misexpression (data not shown). We have shown previously that the function of en regulatory DNA in this construct can sometimes be influenced through interaction with flanking genomic PREs, maintaining striped expression patterns late in development (10). Therefore, we utilized the FLP recombinase system to remove the inv fragments, leaving the remainder of the en promoter and upstream regulatory DNA intact. If the restrictive expression patterns observed for β-Gal are indeed the result of PRE activity from the inv fragments, then removal of the putative PRE should result in increased misexpression of the β-Gal reporter gene.
Embryos were collected and stained for β-Gal expression after removal of the inv DNA fragments. Fragments 1, 4, and 5 all led to increased misexpression in the ΔPRE embryos compared to the wild-type embryos (Fig. 3B and data not shown). The other inv fragment insertions, including the inv3 insertion (Fig. 3C and data not shown), produced no observable difference in β-Gal expression between the ΔPRE and wild-type embryos, further indicating that fragments 2, 3, 6, and 7 do not exhibit PRE activity.
The bxd PRE can also function with en regulatory DNA.
Our results from the β-Gal expression experiments indicated that multiple DNA fragments from the inv locus can maintain en-like expression patterns of β-Gal in a manner indicative of PRE function. These results touch on the questions of what elements are required for PRE function and whether PREs, if fundamentally similar, are interchangeable. Our results indicate that inv PREs can substitute for en PREs at the en promoter. However, evidence suggests that inv and en are functionally redundant and likely share regulatory DNA (17, 18), so it is possible that these DNA fragments have evolved to interact with one another in vivo. To explore the possibility of whether other previously identified PRE fragments could also function with the en promoter, we inserted PRED (from the bxd region of Ubx [14]) upstream of the en promoter in the SD10 construct to see if it was able to regulate β-Gal expression. The embryo stainings in Fig. 3D show that PRED is able to maintain almost perfect expression of β-Gal in en pattern-like stripes, even very late in embryonic development (data not shown). This result reaffirms not only that PRED is a strong PRE, but also that it is able to regulate the promoter of another gene, suggesting that there is some core element of these PREs that allows them to function with other promoters. PRED also appeared to be a stronger PRE than any of the inv PREs. That is, while nearly perfect maintenance of stripes was seen in the two PRED lines, all inv lines showed some misexpression between stripes (Fig. 3). Previous evidence suggests that there are several PRED subfragments that can silence mini-white, indicating that PRED is a complex element (19).
DNA consensus binding sites are not a good PRE predictor.
The data generated from the previous two experiments suggest that out of six predicted PREs in the inv locus, two acted as strong PREs and one acted as a weak PRE. Since most of the predicted fragments were chosen based on the frequency of consensus DNA binding sites in the jPREdictor program, we were interested to see if there were any clear differences in the binding site characteristics between the three identified PREs and the remaining negative fragments. In addition, other consensus binding sites that are thought to contribute to PRE function have been discovered since the 2003 study, which may aid in the development of an improved set of prediction criteria.
The DNA sequence for each predicted PRE fragment in this study was analyzed for the presence of various consensus binding sequences. In addition to the consensus sequences used in jPREdictor, we searched for DSP1 binding sites (GAAAA) (9) and Sp1 binding sites (RRGGYG) (4). Sp1 binding sites were included in the Ringrose study but were not specifically enriched in known PRE sequences versus random DNA sequences (35). DSP1 was discovered more recently in a screen for corepressors of Dorsal and was subsequently reported to aid in the recruitment of PcG proteins to the Ab-Fab PRE of Abd-B (8, 9). However, while DSP1 may aid in PRE function at some genes, it has been shown to be dispensable at others, rendering its involvement in general PRE function still uncertain (15, 25). Similarly, while the Grh protein has been shown to aid in Pho binding to the PREs, it has also been reported previously that Grh is not sufficient for PcG recruitment and also functions as an activator elsewhere in the genome (2). This observed variability in protein requirements suggests that not all PREs have the same DNA binding proteins.
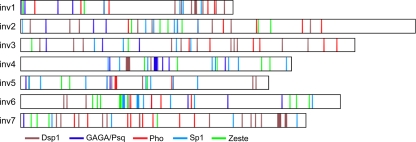
The distribution of the binding consensus sites identified in each inv fragment is shown in Fig. 4 and numerically displayed in Table 2; the same analysis was done for the well-characterized en PRE fragments for comparison. Upon first inspection, the visual comparison between the fragments seemed to show that active PRE fragments (inv1 and inv4) had clusters of binding sites, in contrast to other fragments (inv3 and inv7) in which the binding sites appeared to be more diffuse across the region. However, inv6 also had clustered binding sites, and this fragment was negative for PRE function, so binding site proximity does not seem to be a clear marker of PREs. Looking at the numbers of binding sites in Table 2, we find that inv4 does not contain any Pho binding sites. This was surprising since multiple studies have shown that Pho is an important contributor to the recruitment of PcG proteins (30, 34, 43). Analysis of the DSP1 and Sp1 sites did not show any significant enrichment in the PRE fragments compared to the negative fragments, and no Grh sites were found in any of the inv fragments.
FIG. 4.
Distribution of various DNA binding motifs within inv fragments. DNA fragments depicted in Fig. 1 were analyzed for the presence of various protein binding sites typically present in PREs. References for consensus sequences used are provided in Table 2. Each line represents a DNA consensus sequence, with thicker lines indicating multiple overlapping sites.
TABLE 2.
Occurrence of various DNA binding motifs within inv fragmentsa
| Fragment or element | No. of binding sites for: |
|||||
|---|---|---|---|---|---|---|
| GAF/Psq | Pho | Zeste | DSP1 | Sp1 | Grh | |
| inv1 | 5 | 6 | 2 | 9 | 4 | 0 |
| inv2 | 3 | 6 | 6 | 9 | 6 | 0 |
| inv3 | 3 | 6 | 2 | 11 | 4 | 0 |
| inv4 | 8 | 0 | 6 | 4 | 8 | 0 |
| inv5 | 3 | 4 | 3 | 4 | 8 | 0 |
| inv6 | 4 | 4 | 10 | 4 | 8 | 0 |
| inv7 | 5 | 6 | 2 | 9 | 4 | 0 |
| en PRE1 | 3 | 2 | 4 | 2 | 3 | 1 |
| en PRE2 | 3 | 2 | 1 | 1 | 1 | 0 |
DNA fragments depicted in Fig. 1 were analyzed for the presence of various protein binding sites typically present in PREs. DNA consensus sequences for GAF/Psq (GAGAG), Pho (GCCAT), and Zeste (YGAGYG) were the same as those used in the prediction analysis. DSP1 (GAAAA) (9), Sp1 (RRGGYG) (4), and Grh (AACYGGTYY) (12) consensus sequences were also counted. Binding sites present in the en PRE1 and PRE2 fragments were included for comparison.
Overall, it did not seem that DNA consensus sequences were useful in predicting functional PREs. This is especially obvious when comparing negative fragments with high prediction scores (like inv6) to inv4, which had strong PRE activity but virtually no PRE prediction score (Fig. 1).
Pho binds inv4 DNA fragments lacking a Pho consensus sequence in vitro.
DNA sequence analysis of the inv PRE fragments revealed that inv4 PRE did not contain a consensus Pho binding site, which is believed to be important for PRE function. However, recent ChIP-on-chip binding profiles show that Pho is bound to the regions where inv1 and inv4 are located, with stronger binding of Pho at inv1 and weaker binding at inv4 (33). This proposes an interesting question: how does the Pho protein bind a PRE that appears to lack Pho binding sites?
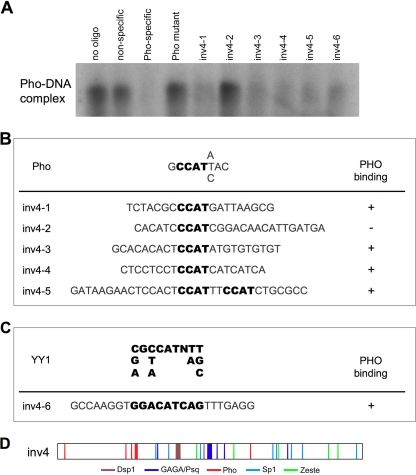
The DNA sequence required for Pho binding has been a source of much investigation. Pho is homologous to the mammalian transcription factor YY1, exhibiting 95% identity in the DNA binding domain (5). Previous studies have tried to identify a Pho-specific consensus sequence, proposing CNGCCATNDNND (28) and GCCATHWY (14) as possible motifs. However, these efforts have not yielded agreement on any sequences outside a GCCAT core sequence, similar to the YY1 consensus sequence [(C/g/a)(G/l)(C/t/a)CATN(T/a)(T/g/c), where uppercase letters indicate the preferred base] (21, 44). Since the initial analysis of the inv DNA fragments was done with the core GCCAT motif, any potential Pho sequences should have been identified, regardless of the flanking DNA sequences. However, as no Pho sites that matched this core sequence were found, we manually scanned the inv4 fragment for sequences that matched the Pho or YY1 sequence with minimal base pair mismatches. Six possible Pho binding sequences were identified, and Pho binding to these short sequences was tested using in vitro competition binding assays (Fig. 5). As expected, cold Pho-specific oligonucleotides are able to compete with radiolabeled Pho oligonucleotides for protein binding, as indicated by the lack of a radiolabeled band shift. In contrast, mutated Pho oligonucleotides lose the ability to compete for Pho binding. Interestingly, five of the six inv oligonucleotides were able to compete with the radiolabeled Pho-specific oligonucleotide, indicating that the oligonucleotide sequences have high affinities for Pho binding in vitro. We were unable to determine by further analysis of DNA sequences why the inv4-2 oligonucleotide was unable to compete for Pho binding. Thus, despite the lack of a traditional Pho consensus binding site, which resulted in our failure to computationally predict fragment 4, we find that Pho is still able to bind fragment 4 in vitro.
FIG. 5.
inv4 can compete for Pho binding in vitro. (A) Radiolabeled Pho-specific oligonucleotides were incubated with various cold oligonucleotides listed at the top of the gel. Binding of the Pho protein to the radiolabeled Pho oligonucleotide results in the visualization of a band shift. Successful competition of the cold oligonucleotide with the hot Pho-specific oligonucleotide results in the loss of a radiolabeled band shift, as seen in the Pho-specific lane. (B and C) DNA sequences of the oligonucleotides used in the band shift analysis shown in panel A. Consensus sequences for the Pho (B) (14) and YY1 (C) (21, 44) proteins are in bold and aligned among the various sequences, and their abilities to compete for the hot Pho-specific oligonucleotide as demonstrated in panel A are represented in the Pho binding column. (D) Updated schematic of inv4 DNA binding sites. Newly identified sequences that bound Pho in the gel shift assay were added to the DNA binding site map from Fig. 4. New Pho sites are represented in red.
DISCUSSION
We utilized PRE prediction programs and protein binding ChIP-on-chip data to identify new PREs at the inv gene. Two PREs were identified upstream of the inv transcription start site, and their activities were confirmed using two assays which tested their abilities to silence different reporter genes at various developmental stages. Interestingly, we found that of the two major PREs identified, only one was identified using the jPREdictor program and that both PREs tracked with the PH ChIP binding profile. The absence of a PRE prediction score for the downstream PRE was due to the lack of Pho consensus sequences, one of the strongest known predictors of PREs (35). However, biochemical data presented here as well as recently published ChIP data indicate that Pho is capable of binding to this PRE fragment despite its lack of a traditional Pho consensus sequence. A third fragment was also capable of functioning as a PRE, albeit weakly in the assays used in this study. Given that the weak PRE, fragment 5, overlaps with 485 bp of fragment 4, a stronger PRE, we anticipated that the low level of activity was attributable to the partial overlap of the two fragments, thus giving fragment 5 only some of the information needed to function fully as a PRE. However, further analysis of the overlap between fragments 4 and 5 revealed that they share only four protein binding sites—two GAF sites and two Sp1 sites. Thus, the DNA sequence alone is not enough to help us understand why fragment 4 acts as a stronger PRE than fragment 5 when fragment 5 appears to contain all of the requisite binding sites for a well-functioning PRE. While inv5 is capable of producing a low level of PRE activity on its own, its close proximity to inv4 in vivo suggests that it may work with inv4 to produce an even stronger PRE near the inv transcription start site. Thus, we feel that overall, only two major PREs have been identified at inv.
Challenges of predicting PREs.
PREs are an integral component of the cell differentiation process, ensuring that specific genes remain silenced throughout the remainder of development. Given their crucial involvement in various developmental processes, much time and effort have been devoted to gaining a better understanding of what basic components constitute a PRE. Many groups have demonstrated that elimination of more than one protein binding site is usually required to significantly affect PRE activity, indicating that PREs contain a multitude of protein binding sites that function cooperatively to recruit PcG proteins. However, our findings, as well as other published data (9, 33, 37), seem to indicate that specific clusters of binding sites are not enough to constitute a PRE. Additional unidentified binding sites or other chromatin-related modifications may be some of the missing links.
There is also the added challenge of identifying the exact sequences required for protein binding. Many protein consensus binding sequences are initially identified by using in vitro biochemical assays or by evaluating DNA sequence homology among multiple DNA loci. As research continues, additional experimental evidence often leads to further refinement of the consensus sequences with enhanced specificity, as recently demonstrated with longer consensus sequences identified for the Pho protein (26, 33). While these superstringent DNA sequences aid in more rapid identification of potential binding sites, they also pose the risk of eliminating other potential candidate sites from consideration. Our results from studying the inv DNA sequence demonstrate this pitfall, as one of our PREs (inv4) was capable of acting as a PRE in vivo and specifically binding the Pho protein in vitro despite its apparent lack of a Pho consensus sequence. Had the ChIP-on-chip data not been available to us, the lack of Pho consensus sites would have prevented us from identifying the inv4 PRE fragment. Furthermore, other DNA regions that had much stronger prediction scores, such as fragments 2 and 6, did not show any PRE activity. Thus, while computerized prediction tools can be a useful starting point, it is clear that PREs cannot be identified by DNA sequence alone, at least not at this time.
PSS and PRE activity are highly correlated.
There has been much speculation regarding the strength of the relationship between PREs and PSS. It is not completely understood at what stage PSS takes effect, but the results are manifested in adults. Our findings allow us to make a much stronger conclusion regarding the relationship between these two silencing phenomena, and we find that in each case where PSS was observed, the same fragment was also able to maintain en pattern-like stripes as seen with en PREs. The magnitude of these silencing effects was also highly correlated with PRE activity; fragments that exhibited higher frequencies of PSS also maintained sharper stripes in the en/β-Gal PRE assay, and vice versa. While this correlation is very strong in this particular case, there are reports of fragments of DNA that can act as pairing-sensitive silencers but not as PREs in embryos (reviewed in reference 22a). In these cases, it seems likely that the DNA fragments that can mediate PSS act in combination with additional, nearby DNA fragments to form fully active PREs (as described previously for iab-2 [39]). Thus, available evidence suggests that DNA fragments that mediate PSS are components of PREs.
Pho exhibits high DNA binding affinity for shorter consensus sequences.
The DNA consensus sequence for Pho was identified from sequence comparisons among various PREs, including the 181-bp en PRE fragment originally used to isolate Pho (5, 28). The original core sequence identified to be necessary for Pho binding was similar to the CCATNTT consensus sequence identified for YY1, the mammalian homolog of Pho (40). The core sequence for Pho was later honed down to the GCCAT motif, although more recent studies on the Pho protein and its target DNA binding sites have led to further refinement of the Pho consensus sequence, with the most recent submission being the 11-bp sequence G(C/A)(C/G)GCCAT(T/C)TT (33). These identified sequences are frequently used in genome-wide searches for predicting Pho binding sites and are a central component in predicting PREs. Thus, it is important to include a site that is specific enough to separate the true Pho binding sites from the false positives. However, our findings indicate that future PRE predictions should be made cautiously when DNA sequences are scoured for potential Pho binding sites, as longer, more stringent DNA consensus sequences may force many PREs to be overlooked. This is further demonstrated by reports that PcG binding profiles, as determined by ChIP-on-chip studies, coincide with an average of only 20% of the PREs predicted by jPREdictor (33, 37). The goal of computationally predicting PREs is laudable, as PREs are developmentally regulated and will vary in different tissues and, thus, detection using ChIP-on-chip may miss PREs present in only a few cells of an embryo or larva.
Finally, our data suggest that the inv PREs are weaker than the bxd and en PREs, which are all composite PREs. Since PREs mediate long-distance interactions, we suggest that in vivo, the two inv PREs interact with each other and also with the en PREs to stabilize PcG repression at the en-inv locus.
Acknowledgments
We thank Jürg Müller for sending the ChIP-on-chip data on Pho binding to the en and inv regions of the genome prior to publication and Bernd Schuettengruber and Giacomo Cavalli for sending us their tiling array data for PH and PC in the region of en and inv. We thank Kris Langlais and Yuzhong Cheng for comments on the manuscript.
This research was supported by the Intramural Research Program of the NIH, NICHD.
Footnotes
Published ahead of print on 30 November 2009.
REFERENCES
- 1.Americo, J., M. Whiteley, J. L. Brown, M. Fujioka, J. B. Jaynes, and J. A. Kassis. 2002. A complex array of DNA-binding proteins required for pairing-sensitive silencing by a polycomb group response element from the Drosophila engrailed gene. Genetics 160:1561-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blastyak, A., R. K. Mishra, F. Karch, and H. Gyurkovics. 2006. Efficient and specific targeting of Polycomb group proteins requires cooperative interaction between Grainyhead and Pleiohomeotic. Mol. Cell. Biol. 26:1434-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, J. L., C. Fritsch, J. Müller, and J. A. Kassis. 2003. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development 130:285-294. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. L., D. J. Grau, S. K. DeVido, and J. A. Kassis. 2005. An Sp1/KLF binding site is important for the activity of a Polycomb group response element from the Drosophila engrailed gene. Nucleic Acids Res. 33:5181-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, J. L., D. Mucci, M. Whiteley, M. L. Dirksen, and J. A. Kassis. 1998. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell 1:1057-1064. [DOI] [PubMed] [Google Scholar]
- 6.Coleman, K. G., S. J. Poole, M. P. Weir, W. C. Soeller, and T. Kornberg. 1987. The invected gene of Drosophila: sequence analysis and expression studies reveal a close kinship to the engrailed gene. Genes Dev. 1:19-28. [DOI] [PubMed] [Google Scholar]
- 7.Comet, I., E. Savitskaya, B. Schuettengruber, N. Negre, S. Lavrov, A. Parshikov, F. Juge, E. Gracheva, P. Georgiev, and G. Cavalli. 2006. PRE-mediated bypass of two Su(Hw) insulators targets PcG proteins to a downstream promoter. Dev. Cell 11:117-124. [DOI] [PubMed] [Google Scholar]
- 8.Decoville, M., E. Giacomello, M. Leng, and D. Locker. 2001. DSP1, an HMG-like protein, is involved in the regulation of homeotic genes. Genetics 157:237-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dejardin, J., A. Rappailles, O. Cuvier, C. Grimaud, M. Decoville, D. Locker, and G. Cavalli. 2005. Recruitment of Drosophila Polycomb group proteins to chromatin by DSP1. Nature 434:533-538. [DOI] [PubMed] [Google Scholar]
- 10.DeVido, S. K., D. Kwon, J. L. Brown, and J. A. Kassis. 2008. The role of Polycomb-group response elements in regulation of engrailed transcription in Drosophila. Development 135:669-676. [DOI] [PubMed] [Google Scholar]
- 11.DiNardo, S., J. M. Kuner, J. Theis, and P. H. O'Farrell. 1985. Development of embryonic pattern in D. melanogaster as revealed by accumulation of the nuclear engrailed protein. Cell 43:59-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dynlacht, B. D., L. D. Attardi, A. Admon, M. Freeman, and R. Tjian. 1989. Functional analysis of NTF-1, a developmentally regulated Drosophila transcription factor that binds neuronal cis elements. Genes Dev. 3:1677-1688. [DOI] [PubMed] [Google Scholar]
- 13.Fiedler, T., and M. Rehmsmeier. 2006. jPREdictor: a versatile tool for the prediction of cis-regulatory elements. Nucleic Acids Res. 34:W546-W550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fritsch, C., J. L. Brown, J. A. Kassis, and J. Müller. 1999. The DNA-binding polycomb group protein Pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development 126:3905-3913. [DOI] [PubMed] [Google Scholar]
- 15.Fujioka, M., G. L. Yusibova, J. Zhou, and J. B. Jaynes. 2008. The DNA-binding Polycomb-group protein Pleiohomeotic maintains both active and repressed transcriptional states through a single site. Development 135:4131-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gehring, W. J. 1970. A recessive lethal (l(4)29) with a homeotic effect in D. melanogaster. Drosoph. Inf. Serv. 45:103. [Google Scholar]
- 17.Goldsborough, A. S., and T. B. Kornberg. 1994. Allele-specific quantification of Drosophila engrailed and invected transcripts. Proc. Natl. Acad. Sci. U. S. A. 91:12696-12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gustavson, E., A. S. Goldsborough, Z. Ali, and T. B. Kornberg. 1996. The Drosophila engrailed and invected genes: partners in regulation, expression and function. Genetics 142:893-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horard, B., C. Tatout, S. Poux, and V. Pirrotta. 2000. Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Mol. Cell. Biol. 20:3187-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hur, M. W., J. D. Laney, S. H. Jeon, J. Ali, and M. D. Biggin. 2002. Zeste maintains repression of Ubx transgenes: support for a new model of Polycomb repression. Development 129:1339-1343. [DOI] [PubMed] [Google Scholar]
- 21.Hyde-DeRuyscher, R. P., E. Jennings, and T. Shenk. 1995. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 23:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kassis, J. A. 1994. Unusual properties of regulatory DNA from the Drosophila engrailed gene: three “pairing-sensitive” sites within a 1.6-kb region. Genetics 136:1025-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Kassis, J. A. 2002. Pairing-sensitive silencing, Polycomb group response elements, and transposon homing in Drosophila. Adv. Genet. 46:421-438. [DOI] [PubMed] [Google Scholar]
- 23.Kassis, J. A., E. P. VanSickle, and S. M. Sensabaugh. 1991. A fragment of engrailed regulatory DNA can mediate transvection of the white gene in Drosophila. Genetics 128:751-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klymenko, T., B. Papp, W. Fischle, T. Kocher, M. Schelder, C. Fritsch, B. Wild, M. Wilm, and J. Müller. 2006. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 20:1110-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozma, G., W. Bender, and L. Sipos. 2008. Replacement of a Drosophila Polycomb response element core, and in situ analysis of its DNA motifs. Mol. Genet. Genomics 279:595-603. [DOI] [PubMed] [Google Scholar]
- 26.Kwong, C., B. Adryan, I. Bell, L. Meadows, S. Russell, J. R. Manak, and R. White. 2008. Stability and dynamics of polycomb target sites in Drosophila development. PLoS Genet. 4:e1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehmann, M., T. Siegmund, K. G. Lintermann, and G. Korge. 1998. The Pipsqueak protein of Drosophila melanogaster binds to GAGA sequences through a novel DNA-binding domain. J. Biol. Chem. 273:28504-28509. [DOI] [PubMed] [Google Scholar]
- 28.Mihaly, J., R. K. Mishra, and F. Karch. 1998. A conserved sequence motif in Polycomb-response elements. Mol. Cell 1:1065-1066. [DOI] [PubMed] [Google Scholar]
- 29.Mishra, R. K., J. Mihaly, S. Barges, A. Spierer, F. Karch, K. Hagstrom, S. E. Schweinsberg, and P. Schedl. 2001. The iab-7 polycomb response element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol. Cell. Biol. 21:1311-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohd-Sarip, A., F. Venturini, G. E. Chalkley, and C. P. Verrijzer. 2002. Pleiohomeotic can link polycomb to DNA and mediate transcriptional repression. Mol. Cell. Biol. 22:7473-7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller, J., and J. A. Kassis. 2006. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr. Opin. Genet. Dev. 16:476-484. [DOI] [PubMed] [Google Scholar]
- 32.Müller, J., and P. Verrijzer. 2009. Biochemical mechanisms of gene regulation by Polycomb group protein complexes. Curr. Opin. Genet. Dev. 19:150-158. [DOI] [PubMed] [Google Scholar]
- 33.Oktaba, K., L. Gutierrez, J. Gagneur, C. Girardot, A. K. Sengupta, E. E. Furlong, and J. Muller. 2008. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev. Cell 15:877-889. [DOI] [PubMed] [Google Scholar]
- 34.Poux, S., D. McCabe, and V. Pirrotta. 2001. Recruitment of components of Polycomb group chromatin complexes in Drosophila. Development 128:75-85. [DOI] [PubMed] [Google Scholar]
- 35.Ringrose, L., M. Rehmsmeier, J. M. Dura, and R. Paro. 2003. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev. Cell 5:759-771. [DOI] [PubMed] [Google Scholar]
- 36.Saurin, A. J., Z. Shao, H. Erdjument-Bromage, P. Tempst, and R. E. Kingston. 2001. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412:655-660. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz, Y. B., T. G. Kahn, D. A. Nix, X. Y. Li, R. Bourgon, M. Biggin, and V. Pirrotta. 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 38:700-705. [DOI] [PubMed] [Google Scholar]
- 38.Schwartz, Y. B., and V. Pirrotta. 2008. Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 20:266-273. [DOI] [PubMed] [Google Scholar]
- 39.Shimell, M. J., A. J. Peterson, J. Burr, J. A. Simon, and M. B. O'Connor. 2000. Functional analysis of repressor binding sites in the iab-2 regulatory region of the abdominal-A homeotic gene. Dev. Biol. 218:38-52. [DOI] [PubMed] [Google Scholar]
- 40.Shrivastava, A., and K. Calame. 1994. An analysis of genes regulated by the multi-functional transcriptional regulator Yin Yang-1. Nucleic Acids Res. 22:5151-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soto, M. C., T. B. Chou, and W. Bender. 1995. Comparison of germline mosaics of genes in the Polycomb group of Drosophila melanogaster. Genetics 140:231-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strutt, H., G. Cavalli, and R. Paro. 1997. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 16:3621-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, L., J. L. Brown, R. Cao, Y. Zhang, J. A. Kassis, and R. S. Jones. 2004. Hierarchical recruitment of Polycomb group silencing complexes. Mol. Cell 14:637-646. [DOI] [PubMed] [Google Scholar]
- 44.Yant, S. R., W. Zhu, D. Millinoff, J. L. Slightom, M. Goodman, and D. L. Gumucio. 1995. High affinity YY1 binding motifs: identification of two core types (ACAT and CCAT) and distribution of potential binding sites within the human beta globin cluster. Nucleic Acids Res. 23:4353-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]