Abstract
ATF-2 is a member of the ATF/CREB family of transcription factors and is activated by stress-activated protein kinases, such as p38. To analyze the physiological role of ATF-2 family transcription factors, we have generated mice with mutations in Atf-2 and Cre-bpa, an Atf-2-related gene. The trans-heterozygotes of both mutants were lean and had reduced white adipose tissue (WAT). ATF-2 and CRE-BPa were required for bone morphogenetic protein 2 (BMP-2)-and p38-dependent induction of peroxisome proliferator-activated receptor γ2 (PPARγ2), a key transcription factor mediating adipocyte differentiation. Since stored fat supplies have been recognized as a possible target for antiobesity treatments, we tested whether inhibition of the p38-ATF-2 pathway suppresses adipocyte differentiation and leads to reduced WAT by treating mice with a p38 inhibitor for long periods of time. High-fat diet (HFD)-induced obesity was significantly reduced in mice fed the p38 inhibitor. Furthermore, the p38 inhibitor alleviated HFD-induced insulin resistance. In p38 inhibitor-treated mice, macrophage infiltration into WAT was reduced and the tumor necrosis factor alpha (TNF-α) levels were lower than control mice. Thus, p38 inhibitors may provide a novel antiobesity treatment.
ATF-2 (originally called CRE-BP1) is a member of the ATF/CREB family of transcription factors, which possess a b-ZIP DNA-binding domain (33, 15). ATF-2 forms homodimers as well as heterodimers with c-Jun and binds to the cyclic AMP response element ([CRE] 5′-TGACGTCA-3′) (14, 36). The ATF-2 subfamily contains three members: ATF-2, CRE-BPa, and ATF-7 (originally known as ATF-a) (41, 10). These proteins possess a trans-activation domain that consists of a zinc finger motif and phosphorylation sites for stress-activated protein kinases (SAPKs) such as p38 and Jun N-terminal protein kinase (JNK) (40). SAPKs are activated by various extracellular stresses, including hypoxia and inflammatory cytokines (28). In response to various stresses, p38/JNK phosphorylates ATF-2 and enhances its trans-activating capacity (13, 31, 55). The transforming growth factor β (TGF-β)-TAK1 pathway also induces phosphorylation of ATF-2 by p38, and phosphorylated ATF-2 (P-ATF-2) interacts with Smad3/Smad4 to synergistically activate transcription in response to TGF-β stimulation (49). Additionally, p38 activation occurs via TAK1 in response to bone morphogenetic protein (BMP) stimulation (25, 39).
The physiological role of ATF-2 has been examined using Atf-2 mutants. Atf-2 null mutant mice die immediately after birth due to respiratory defects that appear to be caused by impaired proliferation of cytotrophoblasts in the mutant placenta (32). Atf-2 heterozygotes are highly prone to mammary tumors after long periods of latency, due to dramatic reductions in the expression of Maspin, a mammary tumor suppressor, and Gadd45α, which is induced by hypoxic stress (35, 34). Analysis of Atf-2 hypomorphic mutant mice, which express an ATF-2 fragment, revealed that ATF-2 is required for skeletal and central nervous system development (44). ATF-2 is also expressed in liver and white adipose tissue (WAT), which are critical components in metabolic homeostasis. ATF-2 activates the transcription of the phosphoenolpyruvate carboxykinase-cytosolic (PEPCK-C) gene via the p38 pathway (3, 29). PEPCK catalyzes the first committed step in hepatic gluconeogenesis and is a rate-limiting enzyme in gluconeogenesis. PEPCK is also a key enzyme in glyceroneogenesis, which is the pathway required for triglyceride synthesis. Interestingly, Drosophila ATF-2 controls triglyceride stores via regulation of PEPCK expression (42). These observations suggest that the ATF-2 family transcription factors play a role in adipocyte differentiation and fat storage.
Diet and lifestyle changes are contributing to the rapidly increasing incidence of obesity (defined as having a body mass index [BMI] greater than 30 kg per m2) in virtually all societies of the world (7). Obesity is associated with an increased risk of type 2 diabetes mellitus, cancer, and heart disease (6, 23). The development of antiobesity drugs that do not produce side effects could significantly reduce the occurrence of these diseases. Despite major progress in our understanding of the molecular mechanisms that lead to obesity, only a few agents that control abnormal fat accumulation are currently available. Most antiobesity drugs developed thus far are appetite-suppressing compounds, activators of energy expenditure, or inhibitors of fat absorption through the gastrointestinal tract (20, 57, 5, 4, 2). However, even the most effective agents can reduce weight by up to only 5%, and strict dieting is needed for further weight loss.
Stored fat supplies have been recognized as a possible target for antiobesity treatment. WAT plays an important role in storing triacylglycerol and releasing free fatty acids and glycerol. Lean mice have been produced by a genetic manipulation that blocked the formation of mature adipocytes. However, mice lacking functional mature adipocytes have fatty livers and elevated circulating triglyceride levels and are usually insulin resistant (46, 50, 38). Furthermore, thiazolidinediones, which are now widely used for treating type 2 diabetes (48), usually enhance weight gain (8). The thiazolidinediones improve insulin sensitivity by stimulating adipocyte differentiation via activation of the peroxisome proliferator-activated receptor γ (PPARγ), a key transcription factor for adipocyte differentiation. These results suggest that leanness without functional adipocytes is incompatible with maintaining insulin sensitivity. In contrast, PPARγ heterozygous mutant mice were much more sensitive to insulin and were more resistant to high-fat diet (HFD)-induced obesity than were wild-type (WT) mice (27). Similarly, mice with mutations in the gene encoding Schnurri-2, which is required for BMP-2-dependent PPARγ2 expression, exhibited reduced WAT and increased insulin sensitivity (22). These results suggest that an appropriate degree of pharmacological interference with adipocyte differentiation, for instance, inhibition of PPARγ expression or activity, could possibly constitute an approach for antiobesity and anti-insulin resistance treatments.
Here, we report that the p38-ATF-2 pathway is involved in adipocyte differentiation and that p38 inhibitors may potentially be used as novel antiobesity and anti-insulin resistance treatments.
MATERIALS AND METHODS
Animals and histological analysis.
Generation of Cre-bpa−/− mice will be described elsewhere. All Atf-2+/− Cre-bpa+/− and control WT littermate mice had a 94% C57BL/6J and 6% CBA genetic background. Visceral fat and liver were removed, and the specimens were fixed in 4% paraformaldehyde and embedded in paraffin. Serial sections (4 μm) were mounted on slides and stained with hematoxylin and eosin (H&E).
Energy expenditure.
Energy expenditure was quantified by measuring oxygen consumption using an open-air circuit indirect calorimetry system (Oxymax Equal Flow; Columbus Instruments). Mice were placed in calorimetric chambers with free access to food and water for approximately 24 h. Expired gases from each chamber were collected and analyzed for CO2 and O2 content every 4 min. Room air was also sampled every 4 min as a reference, and airflow through the chambers was 650 ml/min. Oxygen consumption was expressed as ml of O2/g of body weight/h.
Glucose and insulin tolerance tests.
For glucose tolerance tests, animals were fasted for 6 h, from 9:30 to 15:30. Animals were injected intraperitoneally with a bolus of glucose (1.5 g/kg of body weight), and then blood glucose levels were measured using Glutest PRO R (Sanwa Kagaku). For insulin tolerance tests, animals were fasted for 4 h, from 9:30 to 13:30. Mice were injected intraperitoneally with a 0.8 U of recombinant human insulin/kg (Eli Lilly), and blood glucose levels were measured as described above.
Real-time RT-PCR.
Total RNA was isolated from mouse embryonic fibroblasts (MEFs), livers, visceral fats, and muscles using Trizol (Gibco BRL). Real-time reverse transcription-PCR (RT-PCR) was performed using an ABI 7500 Real-Time PCR Instrument and the QuantiTect Probe RT-PCR Kit (Qiagen), according to the manufacturer's instructions. The PCR conditions were 50°C for 30 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. The primer sequences are available on request. The relative level of mRNA expression was normalized against the amount of 18S rRNA in each sample using rRNA Control Reagents (Applied Biosystems).
In vitro adipogenic differentiation.
MEFs were prepared from 14.5-day-old embryos (Atf-2+/−Cre-bpa+/−, Atf-2−/−Cre-bpa−/−, and WT), which had a 94% C57BL/6J and 6% CBA genetic background, and cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Induction of adipogenic differentiation was carried out essentially as described by Sottile and Seuwen (51), with minor modifications. Briefly, 4 × 105 MEFs were seeded in 60-mm plates and grown to confluence for 3 to 4 days. Cultures were then treated with 1 μM BRL 49653, 300 ng/ml BMP-2, 0.2 mM 3-isobutyl-1-methylxanthine (IBMX), 10 mM β-glycerophosphate, and 50 μM ascorbic acid phosphate. Cultures were maintained under differentiating conditions for up to 8 days, and the medium was changed every 3 days. The presence of mature adipocytes was assessed by oil red O staining. To examine the effect of p38 inhibitors on adipocyte differentiation, 10 ng/ml SB203580 (Calbiochem) or FR167653 was added 8 days before oil red staining. To examine the effect of PPARγ2 or C/EBPα expression on adipocyte differentiation, cells were infected with a PPARγ2-or C/EBPα-containing retrovirus and exposed to differentiation medium. The PPARγ2 virus vector (27) was kindly provided by T. Kitamura. The C/EBPα virus vector was generated using the pMSCV-IRES-puro vector (where MSCV is murine stem cell virus and IRES is internal ribosome entry site). In the control experiments, cells were infected with virus carrying empty vector.
Luciferase reporter assays.
3T3-L1 fibroblasts were differentiated to adipogenic cells by cultivation for 2 days in DMEM containing 0.25 μM dexamethazone, 0.5 mM IBMX, and 1 μg/ml insulin and then by cultivation for an additional 12 days in DMEM containing 1 μg/ml insulin. To examine the effect of ATF-2 and CRE-BPa on luciferase expression driven by the PPARγ promoter, differentiated 3T3-L1 cells were transfected with 1 μg of PPARγ-luciferase (nucleotides −255 or −235 to +64), 1 μg each of pcDNASmad1 and pcDNASmad4, 0.1 μg of pEF-C/EBPα, and 1 μg of pact-ATF-2 or pact-CRE-BPa or the control empty vector, and 0.5 μg of pRL-SV40 (where SV40 is simian virus 40) (Promega) as an internal control, using the Nucleofector Kit L (program A-33; Amaxa), according to the manufacturer's protocol. At 48 h postnucleofection, luciferase activity was measured and normalized for nucleofection efficiency against Renilla luciferase activity. Immediately after nucleofection, 10 ng/ml SB203580 or FR167653 was added. The cells were treated with a second dose of the p38 inhibitors 24 h before harvesting.
Gel mobility shift assay.
Gel mobility shift assays were performed essentially as described previously (34). Briefly, nuclear extracts of 293T cells that were transfected with pact-ATF-2 were incubated for 15 min at 25°C with a 32P-labeled oligonucleotide in a 20-μl solution containing 10 mM Tris-HCl (pH 7.9), 50 mM KCl, 1 mM dithiothreitol (DTT), 0.04% NP-40, 1 μg of poly(dI-dC), and 5% glycerol. Subsequently, 2 μg of anti-ATF-2 (C19; Santa Cruz) or control rabbit IgG was added, and the extracts were incubated for 30 min. The reaction mixture was separated by electrophoresis using a 4% polyacrylamide gel in 0.25× TBE buffer (22 mM Tris-borate, 22 mM boric acid, and 0.5 mM EDTA), followed by autoradiography. The oligonucleotides probes used were5′-GAATGTGTGGGTCACTGGCGAGA-3′ and 5′-TGTCTCGCCAGTGACCCACACATT-3′ (the CRE-like sequence is underlined).
Western blotting to detect P-ATF-2.
3T3-L1 cells were cultured in DMEM supplemented with 0.2% FBS for 24 h. The cells were then transferred to DMEM containing 0.2% FBS and 300 ng/ml BMP-2 and cultured for various periods of time. The p38 inhibitor SB203580 or FR167653 (10 ng/ml) was added 2 h before BMP-2 stimulation. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1% sodium desoxycholate, 0.1% SDS, 50 mM NaF, 2 mM NaVO4, 1 μM okadaic acid, and protease inhibitor cocktail). The lysates were subjected to SDS-PAGE, followed by Western blotting using anti-ATF-2 (C19; Santa Cruz) or anti-p71-ATF-2 (9221; Cell Signaling) and ECL Detection Reagents (Amersham).
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described previously (22). Briefly, 3T3-L1 fibroblasts were cultured in differentiation medium for 7 days in the presence or absence of SB203580 (20 μM) to inhibit differentiation. The medium was changed every third day. On day 7, cells were cross-linked in 1.5% formaldehyde for 15 min at room temperature. After addition of glycine to a final concentration of 0.125 M to quench the cross-linking reaction, the chromatin was solubilized, extracted with lysis buffer, and sheared to 400-to 600-bp fragments by sonication. Immunoprecipitation was performed overnight at 4°C using anti-ATF-2 raised against recombinant ATF-2 lacking the N-terminal 107 amino acids (Δ107ATF-2) or anti-GST-CRE-BPa (where GST is glutathione S-transferase) raised against recombinant GST-CRE-BPa. Preimmune rabbit serum was used as a negative control. The immunocomplexes were washed and incubated at 65°C in 100 ml of immunoprecipitation elution buffer (1% SDS, 0.1 M NaHCO3, 250 mM NaCl, 200 mg/ml proteinase K, 10 mM DTT) to release proteins. The precipitated DNA was further purified using a QIAquick PCR Purification Kit (Qiagen) and eluted in 30 μl of elution buffer. Eluted DNA samples were used for real-time PCR (7500 Real Time PCR System; Applied Biosystems). The primers and TaqMan probe (Nippon EGT Co., Ltd.) used for amplification were 5′-TGGGTCACTGGCGAGACA-3′, 5′-CTTTGGCAAGACTTGGTACATTACA-3′, and 5′(FAM)-TGTAGCAACGTTTTCC-(TAMRA)3′ (where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine), respectively.
Diet study and physiological measurements.
Mice (8-week-old female C57BL/6J mice) were divided at random into two groups. One group was fed an HFD (400 g of beef fat, 100 g of cornstarch, 90 g of glucose, 40 g of AIT-76TM mineral mixture, AIT-76TM vitamin mixture, and 360 g of casein per kilogram of diet; Oriental Yeast Ltd.), while the other was fed the same diet supplemented with 0.08% FR167653 for at least 8 months. Food consumption and body weight were measured twice a week. In a second experiment, 4-month-old Atf-2+/− Cre-bpa+/− female mice and age-matched WT female mice were fed the HFD for 7 weeks. Rectal temperature was monitored using an electronic thermistor (BDT-100; Bio Research Center) equipped with a rectal probe (RET-3; Physitemp).
Blood analysis.
Blood glucose was measured using the glucose oxidase method (Sanwa Kagaku). Serum free fatty acid (FFA) and cholesterol levels were determined using a nonesterified fatty acid C-test and a cholesterol Etest (Wako), respectively. Serum triglyceride levels were determined using a serum triglyceride determination kit (TR0100; Sigma). Plasma leptin, adiponectin, and tumor necrosis factor alpha (TNF-α) were assayed using a Leptin enzyme-linked immunosorbent assay (ELISA) kit (B-Bridge International), an ELISA-based Quantikine adiponectin immunoassay kit (R&D systems), and an ELISA-based Quantikine TNF-α immunoassay kit (R&D systems), respectively, according to the manufacturer's instructions. Plasma insulin levels were measured using an ultrasensitive mouse insulin ELISA kit (Morinaga Institute of Biological Science, Inc., Yokohama, Japan).
Detection of macrophage infiltration into adipose tissue.
Adipose tissues were fixed in 4% paraformaldehyde for 24 h. Sections (10 μm) prepared from tissue frozen in OCT (optimal cutting temperature compound; Tissue-Tek) were stained with the macrophage-specific anti-F4/80 (sc-71088; Santa Cruz) and Alexa Fluor 488-conjugated goat anti-rat IgG (H+L). Nuclei were stained with propidium iodide (50 μg/ml). Confocal images were obtained using an LSM510 laser scanning microscope (Zeiss).
Statistical analysis.
Results are presented as the mean ± standard deviation (SD) or mean ± standard error of the mean (SEM).Differences between groups were examined for statistical significance using a Student's t test or analysis of variance (ANOVA) with Fisher's probable least squares difference (PLSD) test.
RESULTS
Atf-2+/−Cre-bpa+/− mice have reduced WAT.
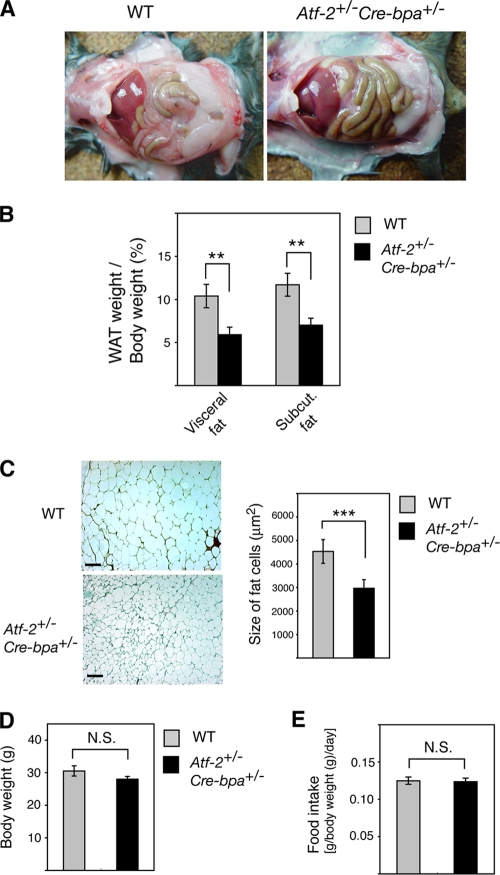
We generated Cre-bpa−/− mice and found that, like Atf-2−/− mice, they die immediately after birth (data not shown). These results suggest that the functions of ATF-2 and CRE-BPa may redundant. To identify novel functions of these ATF-2 family transcription factors, we generated Atf-2+/−Cre-bpa+/− double heterozygotes. The mice appeared to be healthy, but the WAT weight was significantly lower than that of WT mice (Fig. 1A). The weight of visceral and subcutaneous WAT relative to the total body weight of Atf-2+/− Cre-bpa+/− mice was 43% and 40% lower, respectively, than that of WT mice (Fig. 1B). No other tissues had gross abnormalities, and the weights were similar to those of WT tissues (data not shown). There was also no significant difference in brown adipose tissue (BAT) between Atf-2+/− Cre-bpa+/− and WT mice (data not shown). Adipocytes of Atf-2+/− Cre-bpa+/− mice exhibited a marked reduction in size (Fig. 1C), suggesting that the reduced WAT weight was due to fewer mature adipocytes. There was no difference in body weight and cumulative food intake between WT and Atf-2+/− Cre-bpa+/− mice (Fig. 1D and E).
FIG. 1.
Reduction in adipose tissue in Atf-2+/− Cre-bpa+/− mice. (A) Defects in adipose tissue development. Peritoneal cavities of 8-month-old WT and Atf-2+/− Cre-bpa+/− mice are shown. (B) Reduced WAT weight in Atf-2+/− Cre-bpa+/− mice. WAT weight/body weight of 8-month-old WT and Atf-2+/−Cre-bpa+/− mice is shown. Each bar represents the mean ± SEM (n = 4 to 5). **, P < 0.01. (C) Histology of adipose tissue using H&E staining of visceral fat sections (left). Bar, 100 μm. For the graph of the size of fat cells (right), the total numbers of cells examined were 660 to 1,260 from three different mice for each group, and each bar represents the mean ± SEM. ***, P < 0.001. (D) Body weight of 8-month-old WT and Atf-2+/− Cre-bpa+/− mice. Each bar represents the mean ± SEM (n = 4 to 5). (E) Relative food intake during a 3-week period. Each bar represents the mean ± SEM (n = 4 to 5). N.S., no significant difference.
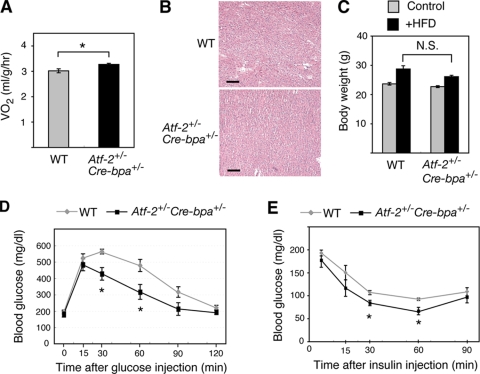
Since food intake appeared to be similar but fat tissue was reduced in Atf-2+/− Cre-bpa+/− mice, the whole-animal oxygen consumption rate was measured. Atf-2+/− Cre-bpa+/− mice exhibited a significant increase in whole-animal oxygen consumption (Fig. 2A). No lipid accumulation was observed in the livers of Atf-2+/− Cre-bpa+/− mice (Fig. 2B). To examine the response to an HFD, WT and Atf-2+/− Cre-bpa+/− mice were fed an HFD for 7 weeks. There was no difference in the degree of weight increase between WT and Atf-2+/− Cre-bpa+/− mice (Fig. 2C). In intraperitoneal glucose tolerance tests, however, Atf-2+/− Cre-bpa+/− mice exhibited less HFD-induced hyperglycemia (Fig. 2D). The hypoglycemic response to insulin was also greater in Atf-2+/− Cre-bpa+/− mice than in WT mice (Fig. 2E). Thus, the loss of one copy of Atf-2 and Cre-bpa inhibited the development of insulin resistance associated with dietary obesity.
FIG. 2.
Energy expenditure and insulin sensitivity of Atf-2+/− Cre-bpa+/− mice. (A) Oxygen consumption (VO2) in Atf-2+/− Cre-bpa+/− and WT mice (n = 4 in each group). *, P < 0.05. (B) H&E-stained sections of liver from Atf-2+/− Cre-bpa+/− and WT mice. Bar, 100 μm. (C) Increased body weight in Atf-2+/− Cre-bpa+/− and WT mice fed an HFD for 7 weeks (n = 6 for each group). N.S., no significant difference. (D) Plasma glucose levels during a glucose tolerance test were examined in 5-month-old Atf-2+/− Cre-bpa+/− or WT mice fed an HFD for 7 weeks. Values indicate the mean ± SEM (n = 5 for each group). *, P < 0.05. (E) Plasma glucose levels during an insulin tolerance test were examined using 5-month-old Atf-2+/− Cre-bpa+/− and WT mice fed an HFD for 7 weeks. Values indicate the mean ± SEM (n = 5 for each group). *, P < 0.05.
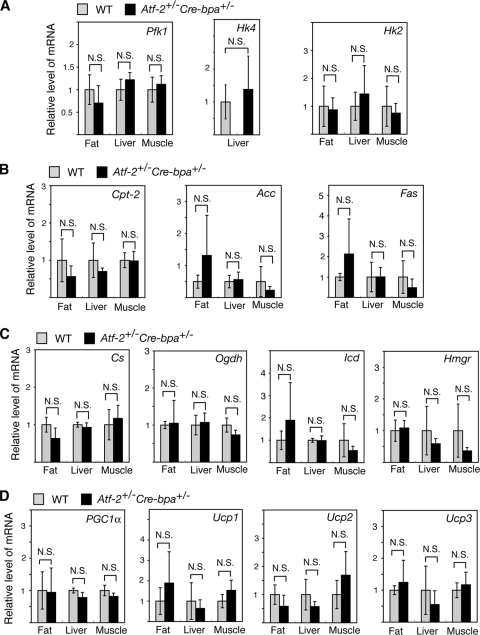
To analyze the difference in metabolism between WT and Atf-2+/− Cre-bpa+/− mice, the expression of key genes that play important roles in metabolism was compared. There were no significant differences between WT and Atf-2+/− Cre-bpa+/− mice in the expression of 14 genes that play key roles in the metabolism of glucose, fatty acids, cholesterol, and energy expenditure in the key metabolic tissues, fat, liver, and muscle (Fig. 3).
FIG. 3.
Comparison in Atf-2+/− Cre-bpa+/− and WT mice of the mRNA levels of genes playing a key role in metabolism. The mRNA levels of the following genes were determined: genes that contribute to glucose metabolism, i.e., Pfk1 (phosphofructo-1-kinase), Hk4 (hexokinase 4), and Hk2 (hexokinase 2) (A); genes involved in fatty acid metabolism, i.e., Cpt-2 (carnitine palmitoyltransferase-2), Acc (acetyl-CoA carboxylase), and Fas (fatty acid synthetase) (B); genes involved in the TCA cycle and cholesterol synthesis, i.e., Cs (citrate synthetase), Icd (isocitrate dehydrogenase), Ogdh (2-oxo-glutarate dehydrogenase), and Hmgr (HMG-CoA [3-hydroxy-3-methylglutaryl-coenzyme A] reductase) (C); and genes involved in energy expenditure, i.e., PGC-1α and Ucp1, Ucp2, and Ucp3 (uncoupling proteins 1, 2, and 3) (D). Total RNA was prepared from the fat, liver, or muscle of Atf-2+/− Cre-bpa+/− or WT mice, and the mRNA levels were measured by real-time RT-PCR. The levels (mean ± SD; n = 3 to 6) are shown relative to WT. N.S., no significant difference.
ATF-2 is required for BMP-and p38-dependent transcription of PPARγ2.
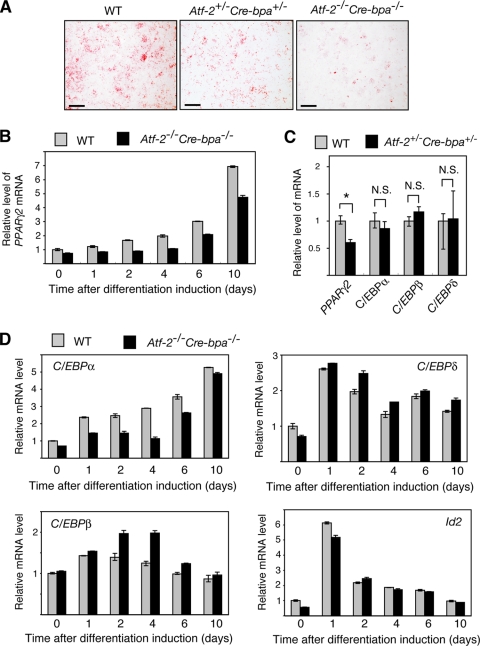
Adipogenesis is stimulated by BMP-2 (51, 16, 22), while the TGF-β/BMP family of ligands activate ATF-2 via the TAK1-p38 signaling pathway (49, 39, 25). In MEF in vitro adipocyte differentiation assays using a BMP-2-containing differentiation medium, Atf-2+/− Cre-bpa+/− MEFs exhibited significantly less adipocyte differentiation than WT cells. Atf-2−/− Cre-bpa−/− MEFs exhibited even less adipocyte differentiation than Atf-2+/− Cre-bpa+/− cells (Fig. 4A). Several independently isolated populations of MEFs exhibited similar results, indicating that this difference is not due to variability in the adipogenic potential of individual primary MEFs.
FIG. 4.
ATF-2 is required for maximal PPARγ2 transcription. (A) Adipocyte differentiation assay. MEFs from WT, Atf-2+/− Cre-bpa+/−, or Atf-2−/− Cre-bpa−/− mice were induced to differentiate into adipocytes using differentiation medium containing BMP-2. Oil-Red O staining shows fat accumulation in cells at 8 days after differentiation induction. Bar, 5 mm. (B) Reduced levels of PPARγ2 mRNA in Atf-2−/− Cre-bpa−/− MEFs. The levels of PPARγ2 mRNA during adipocyte differentiation of WT and Atf-2−/− Cre-bpa−/− MEFs were measured by real-time RT-PCR. The levels (mean ± SD; n = 3) are shown relative to preinduction levels of WT MEFs. (C) Reduced levels of PPARγ2 mRNA in adipose tissues of Atf-2+/−Cre-bpa+/− mice. The levels of PPARγ2, C/EBPα, C/EBPβ, and C/EBPδ mRNAs in adipose tissues of Atf-2+/− Cre-bpa+/− and WT mice were measured by real-time RT-PCR and are shown relative to WT levels (mean ± SD; n = 3). *, P < 0.05; N.S., no significant difference. (D) Key transcription factor mRNA levels. The levels of C/EBPα, C/EBPβ, C/EBPδ, and Id mRNAs during adipocyte differentiation of WT and Atf-2−/− Cre-bpa−/− MEFs were measured by real-time RT-PCR. The levels (mean ± SD; n = 3) are shown relative to preinduction levels of WT MEFs.
We examined the expression of transcription factors that play important roles in adipocyte differentiation (45). In Atf-2−/− Cre-bpa−/− cells, the PPARγ2 mRNA level was approximately 30% to 45% lower than that of WT cells during the first 10 days after induction of differentiation (Fig. 4B). A significant decrease in the PPARγ2 mRNA level (∼40%) was also detected in the adipocytes of Atf-2+/− Cre-bpa+/− mice (Fig. 4C). Additionally, the C/EBPα mRNA level in Atf-2−/− Cre-bpa−/− cells was approximately 30% to 60% lower than that of WT cells during the first 6 days after induction of differentiation although there was almost no difference by 10 days after induction (Fig. 4D). The reduction in C/EBPα mRNA levels in Atf-2−/− Cre-bpa−/− cells may be due to a decrease in the PPARγ2 mRNA level because PPARγ mutant mice cannot develop adipocytes, and they express C/EBPα poorly (27). The mRNA levels of C/EBPβ, C/EBPδ, and Id2 in Atf-2−/− Cre-bpa−/− cells were not significantly reduced after induction of differentiation compared to WT cells (Fig. 4D). There was also no difference in the levels of C/EBPα, C/EBPβ, or C/EBPδ between the adipocytes from WT or Atf-2+/− Cre-bpa+/− mice (Fig. 4C).
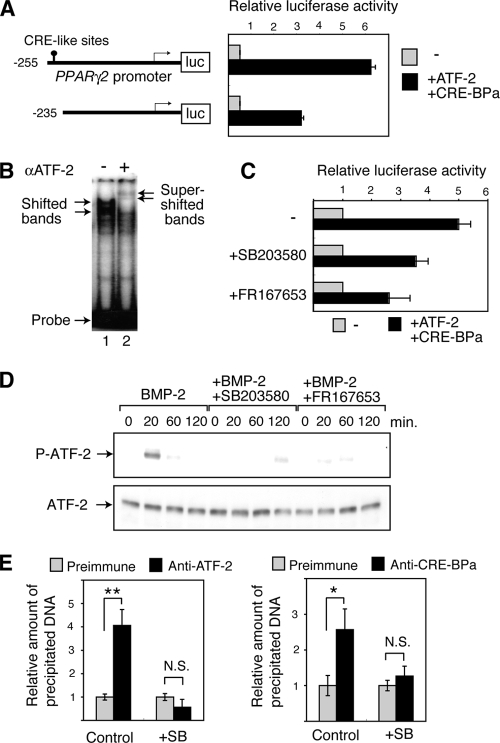
Since ATF-2 has been shown to activate transcription of PPARγ2 (30), we examined the regulation of PPARγ2 transcription by ATF-2 in detail. We used two luciferase reporter constructs fused to different fragments of the PPARγ2 promoter, the −255 and −235 promoter constructs, which contain the region between −255 and +64 and the region between −235 and +64 of the PPARγ2 promoter, respectively. The region between −255 and −235 contains one CRE-like sequence (Fig. 5A). When ATF-2 and CRE-BPa were expressed in WT MEFs with Smad1, Smad4, and C/EBPα, which activate the PPARγ2 promoter, ATF-2/CRE-BPa activated luciferase expression from the −255 promoter construct by approximately 6.1-fold (Fig. 5A). In contrast, ATF-2/CRE-BPa activated luciferase expression from the −235 promoter construct by only 3-fold. The region between −255 and −235 contains one CRE-like sequence (Fig. 5A). In gel mobility shift assays using a DNA probe containing the −255 to −235 sequence, retarded bands were detected in the presence of ATF-2 and were further supershifted by inclusion of an anti-ATF2 antibody (Fig. 5B). Taken together, these data indicate that ATF-2 directly binds to the PPARγ2 promoter and activates transcription. The ATF-2-induced expression of luciferase by the PPARγ2 promoter was reduced by treating the transfected cells with the p38 inhibitors SB203580 or FR167653 (53) (Fig. 5C). Treatment of mouse 3T3-L1 preadipocyte cells with BMP-2 transiently induced phosphorylation of ATF-2, and this phosphorylation was abrogated by the p38 inhibitors (Fig. 5D). To confirm the in vivo binding of ATF-2 family transcription factors to the PPARγ2 promoter, we performed ChIP assays using 3T3-L1 cells. DNA containing the CRE-like site of the PPARγ2 promoter was precipitated by anti-ATF-2 and anti-CRE-BPa (Fig. 5E). Furthermore, treatment of 3T3-L1 cells with the p38 inhibitor SB203580 significantly reduced precipitation of the PPARγ2 promoter by either antibody. These results indicate that both ATF-2 and CRE-BPa bind to the promoter region of PPARγ2 and that their binding depends on p38 activity.
FIG. 5.
The BMP-p38-ATF-2 signaling pathway regulates adipocyte differentiation via PPARγ2 transcription. (A) Activation of the PPARγ2 promoter by ATF-2/CRE-BPa. WT MEFs were transfected with a luciferase reporter containing different fragments of the PPARγ2 promoter (shown at left) and a vector to express Smad1, Smad4, C/EBPα, ATF-2, or CRE-BPa. The relative luciferase activity is shown (mean ± SD; n = 3). (B) Gel mobility shift assays. Nuclear extracts of 293T cells that were transfected with pact-ATF-2 were incubated with a DNA probe containing the CRE-like sequence (nucleotides −255 to −235) from the PPARγ2 promoter and separated by gel electrophoresis, followed by autoradiography. In lane 2, anti-ATF-2 antibodies were added (+). (C) p38 inhibitors attenuate the ATF-2-dependent activation of the PPARγ2 promoter. MEFs were transfected with the PPARγ2 promoter-luciferase reporter and the ATF-2/CRE-BPa expression vectors or the control empty vectors, as described above. The relative luciferase activity is shown (mean ± SD; n = 3). (D) BMP-2 induces p38-dependent phosphorylation of ATF-2. 3T3-L1 cells were treated with BMP-2 at various time points. Phosphorylated ATF-2 (P-ATF-2) and total ATF-2 were detected by Western blot analysis using specific antibodies. 3T3-L1 cells were treated with BMP-2 and p38 inhibitor SB203580 or FR167653. (E) ChIP assays. Soluble chromatin was prepared from SB203580 (+SB) treated 3T3-L1 cells or untreated cells (Control) and immunoprecipitated with anti-ATF-2, anti-CRE-BPa or preimmune serum. The final DNA extraction was amplified by real-time PCR using primers that cover the PPARγ2 promoter region containing the CRE-like site. The relative densities of the bands are indicated, and each bar represents the mean ± SD (n = 3). **, P < 0.01; *, P < 0.05. N.S., no significant difference.
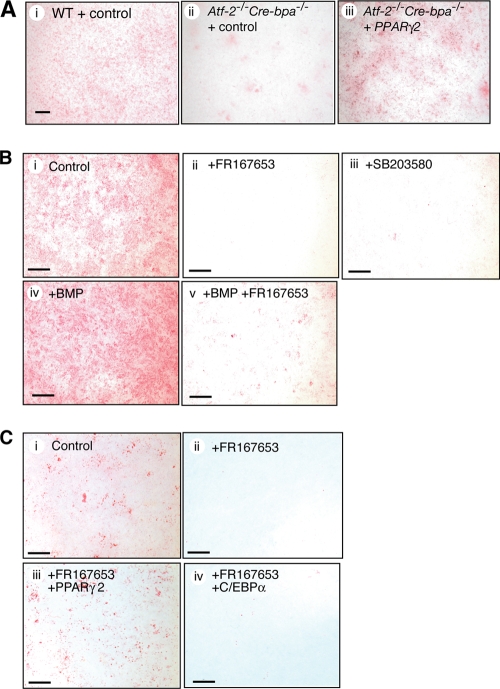
The reduced capacity of the Atf-2−/− Cre-bpa−/− MEFs to differentiate into adipocytes, compared to WT cells, was rescued by overexpression of PPARγ2 (Fig. 6A). This result further supports the hypothesis that PPARγ2 is a key target of the ATF-2 family transcription factors in regulating adipocyte differentiation. Adipocyte differentiation from MEFs was inhibited by p38 inhibitors using a differentiation medium in the presence or absence of BMP-2 (Fig. 6B). In the absence of added BMP-2, p38 inhibitors may block p38 activation in response to BMP present in the serum component of the medium. Inhibition of adipocyte differentiation by FR167653 suggests that the p38-ATF-2 signaling pathway regulates adipocyte differentiation by activating PPARγ2 transcription. Thus, p38 inhibitors severely inhibited adipocyte differentiation from MEFs while the inhibitors only partially inhibited luciferase expression from the PPARγ2 promoter (Fig. 5C). In luciferase reporter assays, many copies of the PPARγ2 gene promoter-luciferase reporter and ATF-2/CRE-BPa exist in the nucleus, so there may be basal luciferase expression, which is not regulated by p38 and is, thus, insensitive to p38 inhibitors. Inhibition of adipocyte differentiation by the p38 inhibitor FR167653 was rescued by overexpression of PPARγ2 but not by overexpression of C/EBPα (Fig. 6C). These results suggest that the p38-ATF-2 signaling pathway controls adipocyte differentiation by inducing PPARγ2 transcription in response to BMP.
FIG. 6.
Adipocyte differentiation is positively regulated by p38 via PPARγ2 expression. (A) Rescue of adipocyte differentiation of Atf-2−/− Cre-bpa−/− MEFs by overexpression of PPARγ2. Adipocyte differentiation of Atf-2−/− Cre-bpa−/− and WT MEFs was induced using differentiation medium containing BMP-2. PPARγ2 was expressed using a viral expression vector, while control cells were infected with virus harboring the empty vector. Bar, 2 μm. (B) Inhibition of adipocyte differentiation by p38 inhibitors. WT MEFs were induced to differentiate into adipocytes using differentiation medium with (frames iv and v) or without (frames i to iii) BMP-2. In some cases, p38 inhibitor FR167653 or SB203580 was added. Oil-Red O staining shows fat accumulation in cells at 8 days after induction of differentiation. Bar, 5 mm. (C) The effect of the p38 inhibitor FR167653 on adipocyte differentiation is blocked by overexpression of PPARγ2. Adipocyte differentiation of WT MEFs was induced using differentiation medium without BMP, as described above. In some cases, the p38 inhibitor FR167653 was added, or PPARγ2 or C/EBPα was expressed using a viral expression vector. Control cells were infected with virus harboring the empty vector. Bar, 5 mm.
Inhibition of p38 exerts an antiobesity effect and reduces insulin resistance.
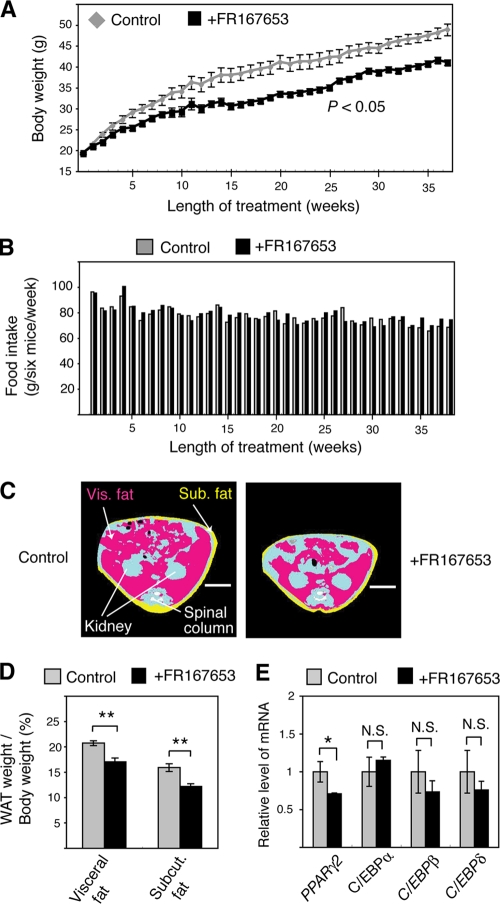
The above results suggest that p38 inhibitors have an antiobesity effect by regulating adipocyte differentiation. To test this hypothesis, WT mice were fed an HFD in the presence or absence of the p38 inhibitor FR167653. Oral administration of FR167653 inhibited development of HFD-induced obesity by 14% to 19%, as measured from 10 to 38 weeks after administration (Fig. 7A). Food intake over the course of the experiment was not affected by FR167653 (Fig. 7B). Microcomputed tomography (micro-CT) scan analysis revealed that FR167653 treatment reduced the size of adipocytes (Fig. 7C). The weights of visceral and subcutaneous fat were reduced by 18% and 23%, respectively, by FR167653 treatment (Fig. 7D). As in Atf-2+/− Cre-bpa+/− mice, a significant decrease in the PPARγ2 mRNA level (∼30%) was detected in the adipocytes of FR167653-treated mice while no decrease in the mRNA level of C/EBPα, C/EBPβ, or C/EBPδ was observed (Fig. 7E). FR167653 had no adverse effects on the major organs of the mice and did not alter blood cell counts (data not shown). These results suggest that the antiobesity effect of FR167653 is not due to systemic toxicity or adverse side effects of the drug.
FIG. 7.
FR167653 prevents HFD-induced obesity. (A) Body weight change during dietary administration of FR167653. Each bar represents the mean ± SEM (n = 6). (B) Food intake during dietary administration of FR167653. Food intake by six mice during 1 week is shown. (C) Computer tomography (CT)-based body composition analysis. Representative CT-images taken of mice treated with FR167653 for 38 weeks and of control mice. Pink, yellow, and blue areas represent visceral (Vis) fat, subcutaneous (Sub) fat, and lean mass, respectively. Bar, 1 cm. (D) Reduced WAT weight in FR167653-treated mice. WAT weight/body weight of 8-month-old WT mice treated with FR167653 for 38 weeks and of control mice over the same period is shown. Each bar represents the mean ± SEM (n = 6). **, P < 0.01. (E) Reduced levels of PPARγ2 mRNA in adipose tissues of FR167653-treated mice. The mRNA levels of PPARγ2, C/EBPα, C/EBPβ, and C/EBPδ in adipose tissues of FR167653-treated and control mice were measured by real-time RT-PCR. The levels (mean ± SD; n = 3) are shown relative to WT. *, P < 0.05; N.S., no significant difference.
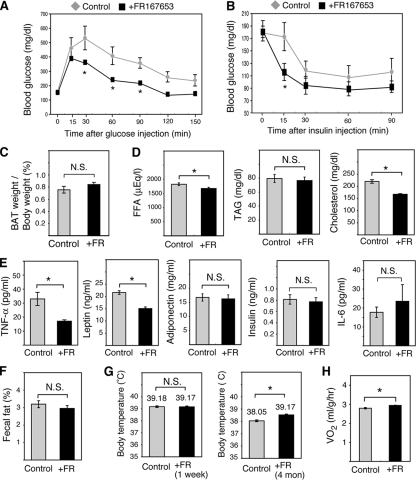
Intraperitoneal glucose tolerance tests revealed that FR167653 administration reduced HFD-induced hyperglycemia (Fig. 8A). The hypoglycemic response to insulin was also more robust in mice treated with FR167653 than in control mice (Fig. 8B). Thus, FR167653 administration blocked the development of insulin resistance associated with dietary obesity. The BAT weight per total body weight of FR167653-treated mice was similar to that of control mice (Fig. 8C). Compared with control mice, the FR167653-treated mice displayed a progressive reduction in the level of serum free fatty acids (12% decrease at 38 weeks) and cholesterol (24% decrease at 38 weeks) (Fig. 8D). However, serum triglyceride concentrations were only marginally reduced by FR167653 administration (Fig. 8D).
FIG. 8.
FR167653 attenuates HFD-induced insulin resistance, hyperlipidemia, and TNF-α overexpression. (A) Plasma glucose levels during glucose tolerance tests were examined using mice treated with FR167653 for 32 weeks or control, untreated mice. Values indicate the mean ± SEM (n = 6 for each group). *, P < 0.05. (B) Plasma glucose levels during insulin tolerance tests were examined using mice treated with FR167653 for 32 weeks or control, untreated mice. Values indicate the mean ± SEM (n = 6 for each group). *, P < 0.05. (C) BAT weight/body weight of mice treated with FR167653 for 38 weeks and of control untreated mice is shown. Each bar represents the mean ± SEM (n = 6). N.S., no significant difference. (D) Serum concentration of free fatty acids, triacylglycerol (TAG), and cholesterol after treatment with FR167653 for 38 weeks. Values indicate the mean ± SEM (n = 6 for each group). *, P < 0.05. (E) Serum concentration of TNF-α, leptin, adiponectin, insulin, and IL-6 after treatment with FR167653 for 38 weeks. Values indicate the mean ± SEM (n = 6 for each group). *, P < 0.05. (F) Fecal fat in mice treated with FR167653 for 36 weeks and control mice. Values indicate the mean ± SEM (n = 6 for each group). The results are expressed as the percentage of fat in the total stool dry weight. (G) Rectal temperature. Average rectal temperature after FR167653 treatment for 1 week or 4 months is shown (mean ± SEM; n = 6 for each group). *, P < 0.05. (H) Oxygen consumption in mice (VO2) treated with FR167653 for 36 weeks and in control untreated mice. Values indicate the mean ± SEM (n = 4 for each group). *, P < 0.05.
Inhibition of p38 suppresses HFD-induced TNF-α expression.
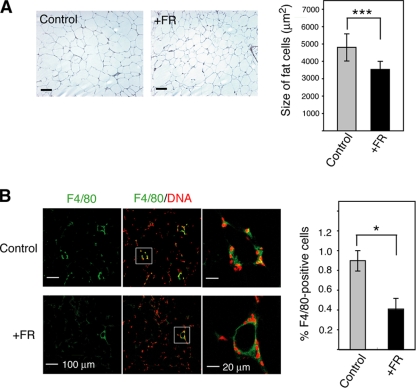
Excess TNF-α is produced by macrophages recruited to HFD-induced hypertrophic adipocytes and by adipocytes themselves (11). Furthermore, a physiological role for TNF-α in modulating insulin responses is well recognized since knockouts of TNF-α and of TNF-α receptors have improved insulin sensitivity (54). The serum TNF-α levels in FR167653-treated mice were significantly lower (48%) than levels in control mice (Fig. 8E). Adipocytes in the visceral WAT of FR167653-treated mice were significantly smaller than those of control mice (Fig. 9A). This result suggests that the decrease in TNF-α in FR167653-treated mice is correlated with the reduction in hypertrophy of the adipocytes. Furthermore, macrophage infiltration into adipose tissue was also inhibited by FR167653 administration (Fig. 9B). The significant reduction (30%) in serum leptin levels (Fig. 8E) is consistent with the decrease in the number of mature adipocytes. FR167653 administration did not affect the serum adiponectin level (Fig. 8E). Recently, p38δ knockout mice were reported to display improved glucose tolerance due to enhanced insulin secretion (52). However, serum insulin levels were not affected by F167653 administration (Fig. 8E). Furthermore, p38δ is not blocked by pyrinyl imidazoles (21) such as FR167653, suggesting that protection from HFD-induced insulin resistance by FR167653 is not due to p38δ inhibition. Recently, knockout of Jnk1, another SAPK, was shown to block HFD-induced insulin resistance by downregulating interleukin-6 (IL-6) expression (47). FR167653 administration did not affect serum IL-6 levels (Fig. 8E), suggesting that protection from HFD-induced insulin resistance by FR167653 is not due to inhibition of JNK1.
FIG. 9.
Effect of the p38 inhibitor FR167653 on HFD-induced enlargement of adipocytes and macrophage infiltration into WAT. (A) FR167653 prevented the HFD-induced enlargement of adipocytes. (Left) Sections from fat around ovary of mice treated with FR167653 for 32 weeks and control untreated mice were stained with H&E. Bar, 100 μm. (Right) Average size of adipocytes in visceral WAT is shown (mean ± SEM). The total number of cells examined was 760 to 1,350 from three different mice for each group. ***, P < 0.001. Control, untreated mice; +FR, FR167653-treated mice. (B) FR167653 prevented HFD-induced macrophage infiltration into WAT. (Left) Sections from fat around ovary were stained with anti-F4/80 antibodies (green). Cell nuclei were identified by staining DNA using propidium iodide (red). The sections were examined by laser confocal microscopy, and representative images are presented. In the middle panels, staining with anti-F4/80 was merged with DNA staining. The white box indicates a subregion of each image that is also presented at higher magnification in the right-hand frames. (Right) Averaged ratio of F4/80-positive cells is shown by bar graph with SEM. The ratio of F4/80-expressing cells was calculated as the sum of the number of nuclei of F4/80-expressing cells divided by the total number of nuclei. The total number of cells examined was 1,800 to 3,960 from three different mice for each group. *, P < 0.05. Control, untreated mice; +FR, FR167653-treated mice.
The amount of fecal fat in FR167653-treated mice was similar to that in control mice (Fig. 8F), suggesting that FR167653 administration did not affect the gastrointestinal absorption of fat. The body temperature of FR167653-treated mice was similar to that of control mice (39.18°C versus 39.17°C) during the first week after treatment (Fig. 8G), suggesting that FR167653 administration did not directly affect metabolism. Obese mice fed the HFD for 4 months had a lower body temperature than mice fed the HFD for 1 week (38.05°C versus 39.18°C) (Fig. 8G). The body temperature of FR167653-treated mice fed the HFD for 4 months was higher than that of control mice (39.17°C versus 38.05°C) (Fig. 8G). R167653-treated mice exhibited a significant increase in whole-animal oxygen consumption (Fig. 8H). Thus, in the FR167653-treated mice, the reduced amount of adipose tissue appears to be correlated with an increase in energy expenditure although the underlying mechanism is not clear.
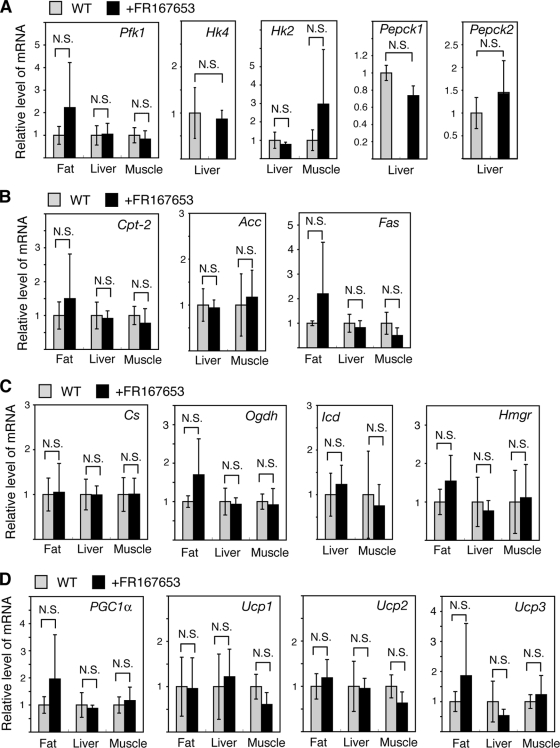
To analyze the differences in metabolism between control and FR167653-treated mice, the expression levels of key genes that play important roles in metabolism were measured and compared. There were no significant differences in the expression levels of 16 genes that have key roles in glucose, fatty acid, and cholesterol metabolism, as well as energy expenditure, in fat, liver, or muscle between control and FR167653-treated mice (Fig. 10). ATF-2 can activate PEPCK transcription (3), which raises the possibility that FR167653 may block insulin resistance and lipid storage by altering metabolism through the inhibition of PEPCK expression. However, the level of Pepck mRNA from the liver of FR167653-treated mice was similar to that of control mice (Fig. 10A), indicating that this possibility is unlikely.
FIG. 10.
Comparison of the mRNA levels of genes playing key roles in metabolism between mice treated with FR167653 for 36 weeks and control mice. The mRNA levels were measured and are presented as described in the legend of Fig. 3. Note that the level of some mRNAs, such as the Acc mRNA, was undetectable in fat tissue and is not shown.
DISCUSSION
This study demonstrated a role for the p38-ATF-2 signaling pathway in adipocyte differentiation and suggested an approach for treatment of HFD-induced obesity and insulin resistance using a p38 inhibitor. A decreased PPARγ2 mRNA level was observed in the adipose tissues of both Atf2+/− Cre-bpa+/− mice and the FR167653-treated mice. Furthermore, no significant difference in the mRNA levels of key genes that play important roles in metabolism was observed. These results strongly suggest that the effect of p38 inhibitor is mediated by a decreased PPARγ2 mRNA level, which was caused by the inhibition of activity of ATF-2 family transcription factors, although we cannot completely exclude the possibility that p38 inhibitor suppresses the HFD-induced obesity via unknown target(s).
Oral administration of the p38 inhibitor FR167653 reduced HFD-induced obesity and improved glucose tolerance and insulin resistance. These observations are reminiscent of the previously reported phenotypes of PPARγ-heterozygous mutant mice and Shn-2 mutant mice, both of which exhibited reduced WAT and increased insulin sensitivity compared to WT mice (27, 22). Furthermore, a similar lean phenotype with higher insulin sensitivity was also reported for mice with apoptosis-inducing peptide injected into the adipose tissue (26). In contrast, inhibiting adipose tissue formation by ectopically expressing the nSREBP-1c or A-ZIP/F toxins increased plasma levels of FFAs and caused an inappropriate deposition of lipids in liver and skeletal muscle, which led to insulin resistance (46, 50, 38). Why, then, does a reduction in WAT have the opposite effect on insulin resistance? In this study, fat ablation may be relatively slow and incomplete in mice treated with the p38 inhibitors or in mice treated with the apoptosis-inducing peptide compared to the rate of fat ablation in transgenic mice expressing nSREBP-1c or A-ZIP/F. Furthermore, fat ablation in PPARγ heterozygotes and Shn-2 mutant mice is also incomplete compared to that in transgenic mice expressing the nSREBP-1c or A-ZIP/F toxins. Thus, relatively slow and incomplete fat ablation could result in the absence of dyslipidemia and retention of insulin sensitivity because such conditions lead to a normal body habitus with normal amounts of body fat.
Despite the similar phenotypes, the mechanism by which insulin resistance is blocked in the p38 inhibitor-treated mice appears to differ from the mechanism in PPARγ heterozygotes or mice injected with apoptosis-inducing peptide in the adipose tissue. Overexpression of leptin was observed in both the PPARγ heterozygotes and in mice treated with the apoptosis-inducing peptide (27, 26), whereas serum leptin levels in mice treated with the p38 inhibitor were reduced compared to control mice. Furthermore, the TNF-α levels were greatly reduced in mice treated with the p38 inhibitor while a smaller decrease in TNF-α levels was observed in the PPARγ heterozygotes (56). Since knockouts of TNF-α and of TNF-α receptors resulted in improved insulin sensitivity (54), a physiological role for TNF-α in modulating insulin responses is well recognized. Thus, a reduction in the TNF-α level upon treatment with a p38 inhibitor may prevent HFD-induced insulin resistance. Although the mechanisms by which TNF-α levels are modulated in mice treated with p38 inhibitor or in PPARγ heterozygotes are unknown, differences in the amount of inhibition of adipocyte differentiation may be correlated with the reduction in insulin resistance.
Although p38 has been shown to regulate metabolism by multiple mechanisms, it is unlikely that the p38 inhibitor blocks HFD-induced obesity and insulin resistance by directly regulating the expression of genes such as PEPCK. Expression of PEPCK, a key enzyme in gluconeogenesis and glyceroneogenesis, is regulated by p38 via ATF-2 (3, 29). However, the level of hepatic PEPCK mRNA in mice treated with p38 inhibitor was similar to that of untreated mice, suggesting that the p38 inhibitor does not increase insulin sensitivity by modulating PEPCK expression. Phosphorylation of the transcriptional PPARγ coactivator-1 (PGC-1) by p38 has been reported to stimulate energy expenditure (43). This observation suggests that inhibition of p38 activity may result in obesity by lowering overall energy expenditure, which contradicts the observations in this study that mice treated with the p38 inhibitor FR167653 were resistant to HFD-induced obesity.
JNK plays a central role in HFD-induced obesity and insulin resistance (17, 47), and a cell-permeable JNK-inhibitory peptide is a novel therapeutic agent for diabetes (24). JNK and p38 belong to the same superfamily of SAPKs, and some inhibitors act on both of these kinases. A series of pyridinyl imidazole compounds, including SB20350 and FR167653, inhibit the p38 pathway by competing with ATP at the p38 ATP-binding site (19, 53, 9, 12). Although SB203580 inhibits not only p38 but also JNK (1), FR167653 appears to be highly selective and inhibits p38 without affecting kinases, such as JNK, protein kinase A, and protein kinase C, or cyclooxygenases (53). Continuous treatment of mice with FR167653 actually inhibits in vivo activation of p38 (18, 37). Thus, it is unlikely that FR167653 affects HFD-induced obesity and insulin resistance by inhibiting JNK activity.
After extended treatments with FR167653, mice appeared to be healthy and suffered no obvious side effects, such as inflammation or abnormal behavior. These data suggest that the p38 inhibitor does not cause significant side effects. However, we cannot completely exclude the possibility that p38 inhibitors may cause side effects, for instance, in the immune system, because these mice were maintained under pathogen-free conditions. Despite the remaining obstacles, the p38 inhibitor described here may be a useful tool for the treatment of human obesity.
Acknowledgments
We are grateful to the Fujisawa Pharmaceutical Co., Ltd. (now the Astellas Pharmaceutical Co., Ltd.), for providing FR167653, to T. Kitamura and T. Kadowaki for the PPARγ2 virus vectors, and to members of the Experimental Animal Division of the RIKEN Tsukuba Institute for maintenance of the mice.
This work was supported in part by Grants-in-Aid for Scientific Research and a grant from the Genome Network Project from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 30 November 2009.
REFERENCES
- 1.Ammendrup, A., A. Maillard, K. Nielsen, N. Aabenhus Andersen, P. Serup, O. Dragsbaek Madsen, T. Mandrup-Poulsen, and C. Bonny. 2000. The c-Jun amino-terminal kinase pathway is preferentially activated by interleukin-1 and controls apoptosis in differentiating pancreatic β-cells. Diabetes 49:1468-1476. [DOI] [PubMed] [Google Scholar]
- 2.Bray, G. A., and L. A. Tartaglia. 2000. Medicinal strategies in the treatment of obesity. Nature 404:672-677. [DOI] [PubMed] [Google Scholar]
- 3.Cheong, J., J. E. Coligan, and J. D. Shuman. 1998. Activating transcription factor-2 regulates phosphoenolpyruvate carboxykinase transcription through a stress-inducible mitogen-activated protein kinase pathway. J. Biol. Chem. 273:22714-22718. [DOI] [PubMed] [Google Scholar]
- 4.Chiesi, M., C. Huppertz, and K. G. Hofbauer. 2001. Pharmacotherapy of obesity: targets and perspectives. Trends Pharmacol. Sci. 22:247-254. [DOI] [PubMed] [Google Scholar]
- 5.Clapham, J. C., J. R. Arch, and M. Tadayyon. 2001. Anti-obesity drugs: a critical review of current therapies and future opportunities. Pharmacol. Ther. 89:81-121. [DOI] [PubMed] [Google Scholar]
- 6.Després, J. P., and I. Lemieux. 2006. Abdominal obesity and metabolic syndrome. Nature 444:881-887. [DOI] [PubMed] [Google Scholar]
- 7.Flier, J. S. 2004. Obesity wars: molecular progress confronts an expanding epidemic. Cell 116:337-350. [DOI] [PubMed] [Google Scholar]
- 8.Fonseca, V. 2003. Effect of thiazolidinediones on body weight in patients with diabetes mellitus. Am. J. Med. 115(Suppl. 8A):42S-48S. [DOI] [PubMed] [Google Scholar]
- 9.Frantz, B., T. Klatt, M. Pang, J. Parsons, A. Rolando, H. Williams, M. J. Tocci, S. J. O'Keefe, and E. A. O'Neill. 1998. The activation state of p38 mitogen-activated protein kinase determines the efficiency of ATP competition for pyridinylimidazole inhibitor binding. Biochemistry 37:13846-13853. [DOI] [PubMed] [Google Scholar]
- 10.Gaire, M., B. Chatton, and C. Kedinger. 1990. Isolation and characterization of two novel, closely related ATF cDNA clones from HeLa cells. Nucleic Acids Res. 18:3467-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guilherme, A., J. V. Virbasius, V. Puri, and M. P. Czech. 2008. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat. Rev. Mol. Cell Biol. 9:367-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gum, R. J., M. M. McLaughlin, S. Kumar, Z. Wang, M. J. Bowe, J. C. Lee, J. L. Adam, G. P. Livi, E. J. Goldsmith, and P. R. Young. 1998. Acquisition of sensitivity of stress-activated protein kinases to the p38 inhibitor, SB 203580, by alteration of one or more amino acids within the ATP binding pocket. J. Biol. Chem. 1273:15605-15610. [DOI] [PubMed] [Google Scholar]
- 13.Gupta, S., D. Campbell, B. Derijard, and R. J. Davis. 1995. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science 267:389-393. [DOI] [PubMed] [Google Scholar]
- 14.Hai, T., and T. Curran. 1991. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl. Acad. Sci. U. S. A. 88:3720-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hai, T. W., F. Liu, E. A. Allegretto, M. Karin, and M. R. Green. 1988. A family of immunologically related transcription factors that includes multiple forms of ATF and AP-1. Genes Dev. 2:1216-1226. [DOI] [PubMed] [Google Scholar]
- 16.Hata, K., R. Nishimura, F. Ikeda, K. Yamashita, T. Matsubara, T. Nokubi, and T. Yoneda. 2003. Differential roles of Smad1 and p38 kinase in regulation of peroxisome proliferator-activating receptor gamma during bone morphogenetic protein 2-induced adipogenesis. Mol. Biol. Cell 14:545-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirosumi, J., G. Tuncman, L. Chang, C. Z. Görgün, K. T. Uysal, K. Maeda, M. Karin, and G. S. Hotamisligil. 2002. A central role for JNK in obesity and insulin resistance. Nature 420:333-336. [DOI] [PubMed] [Google Scholar]
- 18.Iwata, Y., T. Wada, K. Furuichi, N. Sakai, K. Matsushima, H. Yokoyama, and K. Kobayashi. 2003. p38 Mitogen-activated protein kinase contributes to autoimmune renal injury in MRL-Fas lpr mice. J. Am. Soc. Nephrol. 14:57-67. [DOI] [PubMed] [Google Scholar]
- 19.Jackson, J. R., B. Bolognese, L. Hillegass, S. Kassis, J. Adams, D. E. Griswold, and J. D. Winkler. 1998. Pharmacological effects of SB220025, a selective inhibitor of P38 mitogen-activated protein kinase, in angiogenesis and chronic inflammatory disease models. J. Pharmacol. Exp. Ther. 284:687-692. [PubMed] [Google Scholar]
- 20.Jandacek, R. J., and S. C. Woods. 2004. Pharmaceutical approaches to the treatment of obesity. Drug Discov. Today 9:874-880. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, Y., H. Gram, M. Zhao, L. New, J. Gu, L. Feng, F. Di Padova, R. J. Ulevitch, and J. Han. 1997. Characterization of the structure and function of the fourth member of p38 group mitogen-activated protein kinases, p38δ. J. Biol. Chem. 272:30122-30128. [DOI] [PubMed] [Google Scholar]
- 22.Jin, W., T. Takagi, S-N. Kanesashi, T. Kurahashi, T. Nomura, J. Harada, and S. Ishii. 2006. Schnurri-2 controls BMP-dependent adipogenesis via interaction with Smad proteins. Dev. Cell 10:461-471. [DOI] [PubMed] [Google Scholar]
- 23.Kahn, S. E., R. L. Hull, and K. M. Utzschneider. 2006. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 444:840-846. [DOI] [PubMed] [Google Scholar]
- 24.Kaneto, H., Y. Nakatani, T. Miyatsuka, D. Kawamori, T. A. Matsuoka, M. Matsuhisa, Y. Kajimoto, H. Ichijo, Y. Yamasaki, and M. Hori. 2004. Possible novel therapy for diabetes with cell-permeable JNK-inhibitory peptide. Nat. Med. 10:1128-1132. [DOI] [PubMed] [Google Scholar]
- 25.Kimura, N., R. Matsuo, H. Shibuya, K. Nakashima, and T. Taga. 2000. BMP2-induced apoptosis is mediated by activation of the TAK1-p38 kinase pathway that is negatively regulated by Smad6. J. Biol. Chem. 275:17647-17652. [DOI] [PubMed] [Google Scholar]
- 26.Kolonin, M. G., P. K. Saha, L. Chan, R. Pasqualini, and W. Arap. 2004. Reversal of obesity by targeted ablation of adipose tissue. Nat. Med. 10:625-632. [DOI] [PubMed] [Google Scholar]
- 27.Kubota, N., Y. Terauchi, H. Miki, H. Tamemoto, T. Yamauchi, K. Komeda, S. Satoh, R. Nakano, C. Ishii, T. Sugiyama, K. Eto, Y. Tsubamoto, A. Okuno, K. Murakami, H. Sekihara, G. Hasegawa, M. Naito, Y. Toyoshima, S. Tanaka, K. Shiota, T. Kitamura, T. Fujita, O. Ezaki, S. Aizawa, and T. Kadowaki. 1999. PPARγ mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol. Cell 4:597-609. [DOI] [PubMed] [Google Scholar]
- 28.Kyriakis, J. M., and J. Avruch. 2001. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81:807-869. [DOI] [PubMed] [Google Scholar]
- 29.Lee, M. Y., C. H. Jung, K. Lee, Y. H. Choi, S. Hong, and J. Cheong. 2002. Activating transcription factor-2 mediates transcriptional regulation of gluconeogenic gene PEPCK by retinoic acid. Diabetes 51:3400-3407. [DOI] [PubMed] [Google Scholar]
- 30.Lee, M. Y., H. J. Kong, and J. Cheong. 2001. Regulation of activating transcription factor-2 in early stage of the adipocyte differentiation program. Biochem. Biophys. Res. Commun. 281:1241-1247. [DOI] [PubMed] [Google Scholar]
- 31.Livingstone, C., G. Patel, and N. Jones. 1995. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 14:1785-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maekawa, T., F. Bernier, M. Sato, S. Nomura, M. Singh, Y. Inoue, T. Tokunaga, H. Imai, M. Yokoyama, A. Reimold, L. H. Glimcher, and S. Ishii. 1999. Mouse ATF-2 null mutants display features of a severe type of meconium aspiration syndrome. J. Biol. Chem. 274:17813-17819. [DOI] [PubMed] [Google Scholar]
- 33.Maekawa, T., H. Sakura, C. Kanei-Ishii, T. Sudo, T. Yoshimura, J. Fujisawa, M. Yoshida, and S. Ishii. 1989. Leucine zipper structure of the protein CRE-BP1 binding to the cyclic AMP response element in brain. EMBO J. 8:2023-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maekawa, T., Y. Sano, T. Shinagawa, Z. Rahman, T. Sakuma, S. Nomura, J. D. Licht, and S. Ishii. 2008. ATF-2 controls transcription of Maspin and GADD45α genes independently from p53 to suppress mammary tumors. Oncogene 27:1045-1054. [DOI] [PubMed] [Google Scholar]
- 35.Maekawa, T., T. Shinagawa, Y. Sano, T. Sakuma, S. Nomura, K. Nagasaki, Y. Miki, F. Saito-Ohara, J. Inazawa, T. Kohno, J. Yokota, and S. Ishii. 2007. Reduced levels of ATF-2 predispose mice to mammary tumors. Mol. Cell. Biol. 27:1730-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuda, S., T. Maekawa, and S. Ishii. 1991. Identification of the functional domains of the transcriptional regulator CRE-BP1. J. Biol. Chem. 266:18188-18193. [PubMed] [Google Scholar]
- 37.Matsuoka, H., T. Arai, M. Mori, S. Goya, H. Kida, H. Morishita, H. Fujiwara, I. Tachibana, T. Osaki, and S. Hayashi. 2002. A p38 MAPK inhibitor, FR-167653, ameliorates murine bleomycin-induced pulmonary fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 283:L103-112. [DOI] [PubMed] [Google Scholar]
- 38.Moitra, J., M. M. Mason, M. Olive, D. Krylov, O. Gavrilova, B. Marcus-Samuels, L. Feigenbaum, E. Lee, T. Aoyama, M. Eckhaus, M. L. Reitman, and C. Vinson. 1998. Life without white fat: a transgenic mouse. Genes Dev. 12:3168-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monzen, K., Y. Hiroi, S. Kudoh, H. Akazawa, T. Oka, E. Takimoto, D. Hayashi, T. Hosoda, M. Kawabata, K. Miyazono, S. Ishii, Y. Yazaki, R. Nagai, and I. Komuro. 2001. Smads, TAK1, and their common target ATF-2 play a critical role in cardiomyocyte differentiation. J. Cell Biol. 153:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagadoi, A., K. Nakazawa, H. Uda, K. Okuno, T. Maekawa, S. Ishii, and Y. Nishimura. 1999. Solution structure of the transactivation domain of ATF-2 comprising a zinc finger-like subdomain and a flexible subdomain. J. Mol. Biol. 287:593-607. [DOI] [PubMed] [Google Scholar]
- 41.Nomura, N., Y. L. Zu, T. Maekawa, S. Tabata, T. Akiyama, and S. Ishii. 1993. Isolation and characterization of a novel member of the gene family encoding the cAMP response element-binding protein CRE-BP1. J. Biol. Chem. 268:4259-4266. [PubMed] [Google Scholar]
- 42.Okamura, T., H. Shimizu, T. Nagao, R. Ueda, and S. Ishii. 2007. ATF-2 regulates fat metabolism in Drosophila. Mol. Biol. Cell 18:1519-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puigserver, P., J. Rhee, J. Lin, Z. Wu, J. C. Yoon, C. Y. Zhang, S. Krauss, V. K. Mootha, B. B. Lowell, and B. M. Spiegelman. 2001. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Mol. Cell 8:971-982. [DOI] [PubMed] [Google Scholar]
- 44.Reimold, A. M., M. J. Grusby, B. Kosaras, J. W. Fries, R. Mori, S. Maniwa, I. M. Clauss, T. Collins, R. L. Sidman, M. J. Glimcher, and L. H. Glimcher. 1996. Chondrodysplasia and neurological abnormalities in ATF-2-deficient mice. Nature 379:262-265. [DOI] [PubMed] [Google Scholar]
- 45.Rosen, E. D., C. J. Walkey, P. Puigserver, and B. M. Spiegelman. 2000. Transcriptional regulation of adipogenesis. Genes Dev. 14:1293-1307. [PubMed] [Google Scholar]
- 46.Ross, S. R., R. A. Graves, and B. M. Spiegelman. 1993. Targeted expression of a toxin gene to adipose tissue: transgenic mice resistant to obesity. Genes Dev. 7:1318-1324. [DOI] [PubMed] [Google Scholar]
- 47.Sabio, G., M. Das, A. Mora, Z. Zhang, J. Y. Jun, H. J. Ko, T. Barrett, J. K. Kim, and R. J. Davis. 2008. A stress signaling pathway in adipose tissue regulates hepatic insulin resistance. Science 322:1539-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saltiel, A. R., and J. M. Olefsky. 1996. Thiazolidinediones in the treatment of insulin resistance and type II diabetes. Diabetes 45:1661-1669. [DOI] [PubMed] [Google Scholar]
- 49.Sano, Y., J. Harada, S. Tashiro, R. Gotoh-Mandeville, T. Maekawa, and S. Ishii. 1999. ATF-2 is a common nuclear target of Smad and TAK1 pathways in transforming growth factor-β signaling. J. Biol. Chem. 274:8949-8957. [DOI] [PubMed] [Google Scholar]
- 50.Shimomura, I., R. E. Hammer, J. A. Richardson, S. Ikemoto, Y. Bashmakov, J. L. Goldstein, and M. S. Brown. 1998. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev. 12:3182-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sottile, V., and K. Seuwen. 2000. Bone morphogenetic protein-2 stimulates adipogenic differentiation of mesenchymal precursor cells in synergy with BRL 49653 (rosiglitazone). FEBS Lett. 475:201-204. [DOI] [PubMed] [Google Scholar]
- 52.Sumara, G., I. Formentini, S. Collins, I. Sumara, R. Windak, B. Bodenmiller, R. Ramracheya, D. Caille, H. Jiang, K. A. Platt, P. Meda, R. Aebersold, P. Rorsman, and R. Pricci. 2009. Regulation of PKD by the MAPK p38δ in insulin secretion and glucose homeostasis. Cell 136:235-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi, S., Y. Keto, T. Fujita, T. Uchiyama, and A. Yamamoto. 2001. FR167653, a p38 mitogen-activated protein kinase inhibitor, prevents Helicobacter pylori-induced gastritis in Mongolian gerbils. J. Pharmacol. Exp. Ther. 296:48-56. [PubMed] [Google Scholar]
- 54.Uysal, K. T., S. M. Wiesbrock, N. W. Marino, and G. S. Hotamisligil. 1997. Protection from obesity-induced insulin resistance in mice lacking TNF-α function. Nature 389:610-614. [DOI] [PubMed] [Google Scholar]
- 55.van Dam, H., D. Wilhelm, I. Herr, A. Steffen, P. Herrlich, and P. Angel. 1995. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 14:1798-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamauchi, T., J. Kamon, H. Waki, K. Murakami, K. Motojima, K. Komeda, T. Ide, N. Kubota, Y. Terauchi, K. Tobe, H. Miki, A. Tsuchida, Y. Akanuma, R. Nagai, S. Kimura, and T. Kadowaki. 2001. The mechanisms by which both heterozygous peroxisome proliferator-activated receptor γ (PPARγ) deficiency and PPARγ agonist improve insulin resistance. J. Biol. Chem. 276:41245-41254. [DOI] [PubMed] [Google Scholar]
- 57.Yanovski, S. Z., and J. A. Yanovski. 2002. Obesity. N. Engl. J. Med. 346:591-602. [DOI] [PubMed] [Google Scholar]