Abstract
Understanding inhibitory mechanisms of transforming growth factor β1 (TGF-β1) has provided insight into cell cycle regulation and how TGF-β1 sensitivity is lost during tumorigenesis. We show here that TGF-β1 utilizes a previously unknown mechanism targeting the function of prereplication complexes (pre-RCs) to acutely block S-phase entry when added to cells in late G1, after most G1 events have occurred. TGF-β1 treatment in early G1 suppresses Myc and CycE-Cdk2 and blocks pre-RC assembly. However, TGF-β1 treatment in late G1 acutely blocks S-phase entry by inhibiting activation of fully assembled pre-RCs, with arrest occurring prior to the helicase unwinding step at G1/S. This acute block by TGF-β1 requires the function of Rb in late G1 but does not involve Myc/CycE-Cdk2 suppression or transcriptional control. Instead, Rb mediates TGF-β1 late-G1 arrest by targeting the MCM helicase. Rb binds the MCM complex during late G1 via a direct interaction with Mcm7, and TGF-β1 blocks their dissociation at G1/S. Loss of Rb or overexpression of Mcm7 or its Rb-binding domain alone abrogates late-G1 arrest by TGF-β1. These results demonstrate that TGF-β1 acutely blocks entry into S phase by inhibiting pre-RC activation and suggest a novel role for Rb in mediating this effect of TGF-β1 through direct interaction with and control of the MCM helicase.
Transforming growth factor β1 (TGF-β1) is a potent inhibitor of cell proliferation. TGF-β1-induced arrest occurs during G1 and is mediated by Smad proteins, which regulate transcriptional targets, including c-myc (11, 13, 37). Downregulation of c-myc allows induction of the Cdk inhibitor (CKI) p15INK4B, which inhibits Cdk4-CycD (20, 45). The p27Kip1 inhibitor is also utilized by TGF-β1 to inhibit Cdk2-CycE (39). Cdk suppression prevents hyperphosphorylation of Rb (28), causing Rb to remain in a hypophosphorylated, growth-suppressive form. The pivotal roles for c-myc suppression and Rb are illustrated by the demonstration that ectopic c-Myc or viral tumor proteins that inactivate Rb override TGF-β1 (3, 28, 38). However, this pathway utilized by TGF-β1 has been largely derived from experimentation in which TGF-β1 is added to cells in early G1, prior to the occurrence of most G1 events, and progression into late-G1/S phase is hindered due to these mechanisms.
Several studies have raised important questions with regard to the mechanisms of TGF-β1 signaling. Cells lacking p27Kip1, p15INK4B, or p21Cip1 remain sensitive to growth arrest by TGF-β1 (24, 34, 46). Thus, CKIs are not absolutely required for TGF-β1 to arrest cells, and it has been suggested that transcriptional suppression of Cdc25A is an alternative means for TGF-β1 to suppress Cdks (24). TGF-β1 can block S-phase entry when added to cells in early G1 or late G1, including just prior to G1/S, after most G1 events have already occurred (4, 23). This ability of late-G1 TGF-β1 exposure to acutely block G1-S transit is particularly intriguing, since mammalian cells no longer require de novo mRNA synthesis in late G1 for S-phase entry (4, 9, 33) and the effectiveness of TGF-β1 arrest after exposure in late G1 is not affected by agents that block de novo mRNA synthesis (4, 23). Thus, TGF-β1 signals in late G1 acutely block S-phase entry utilizing mechanisms independent of transcriptional upregulation or downregulation. This calls into question the need for acute transcriptional control of c-Myc, Cdc25A, CKIs, or other transcriptional targets by TGF-β1 specifically in late G1 and elicits questions as to the transcription-independent and acute nature of the mechanisms by which TGF-β1 achieves arrest when added to cells in late G1.
We reasoned that TGF-β1 likely produces negative effects on the prereplication complex (pre-RC) and that understanding such effects might offer insight into TGF-β1 signaling that explains these unanswered questions. Pre-RCs assemble at future origins of DNA replication and play a pivotal role in regulating the transition to S phase (6). Each pre-RC is composed of the origin recognition complex (ORC), which recruits Cdt1, Cdc6, and the hexameric minichromosome maintenance (MCM) helicase. Initiation of DNA replication (i.e., the G1/S transition) commences after Cdc45, DNA polymerases, and PCNA (and other proteins) are recruited to the pre-RCs and MCMs are activated to melt origin DNA (6).
We show here that TGF-β1 signals do indeed target pre-RC functionality and that the effects of TGF-β1 on pre-RC dynamics provide novel explanations for these unanswered questions. TGF-β1 treatment in early G1, prior to pre-RC assembly, blocks such assembly and causes numerous other cell cycle changes, including suppression of Myc and inhibition of CycE-Cdk2. In contrast, treatment with TGF-β1 in late G1, after pre-RCs have already assembled, does not cause disassembly of pre-RCs. Instead, TGF-β1 acutely inhibits pre-RC activation and arrests cells prior to the helicase unwinding step at G1/S. This late-G1 arrest does not involve CycE-Cdk2 inhibition or Myc suppression. However, Rb is critically required for acute inhibition of pre-RC activation by TGF-β1. Rb mediates TGF-β1 arrest in late G1 via direct targeting of the MCM helicase, specifically through Mcm7. Loss of Rb or gain of Mcm7 overrides TGF-β1 arrest in late G1, indicating that the Rb-MCM interaction plays a pivotal role. These observations provide novel insight into the mechanisms of TGF-β1 arrest and suggest a role for Rb in mediating late-G1 arrest by TGF-β1 that involves regulating the MCM complex.
MATERIALS AND METHODS
Cell culture, synchronization, and drugs.
Mouse keratinocytes (BALB/MK) were maintained as described previously (3). HaCaT and SaOS-2 cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. TGF-β1 (R&D Systems) was used at 10 ng/ml. MK synchronization was achieved by culturing cells in medium lacking epidermal growth factor (EGF) for 3.5 days. HaCaT synchronization was optimized and carried out by plating cells at ∼70% density, with serum withdrawal done for 24 h to render the cells quiescent. Transfections utilized Fugene6 (Roche) or polyethylenimine (40). Hydroxyurea (Hu; 3 mM), aphidicolin (Aph; 10 μg/ml), and 5,6-dichlorobenzimidazole riboside (DRB) (40 μM) were used.
Antibodies.
Antibodies were used at 1:1,000 dilutions and derived from rabbits unless otherwise specified. Antibodies used were as follows: from Covance, mouse monoclonal (MAb) antihemagglutinin (anti-HA), MAb anti-HA-fluorescein isothiocyanate (FITC), MAb anti-glutathione S-transferase (anti-GST), and MAb anti-His6; from Cell Signaling, anti-Rb-P-Ser807/811 (1:500), anti-Cdc7 (8 μl/immunoprecipitation [IP]), and rat anti-RPA-32 (1:500, immunoblotting [IB]; 1:100, immunofluorescence [IF]); from Calbiochem, MAb anti-PCNA (1:10,000) and anti-Rb (1:500, IB); from BD Biosciences: MAb anti-Orc4, anti-Mcm2 (1:3,000), and MAb anti-Rb (1:500, IP); anti-BrdU-Alexa 594 (1:20; Invitrogen); from Santa Cruz, MAb anti-CycA (5 μg/ml, IP), anti-CycE (5 μg/ml, IP), anti-Cdc6 (1:500), MAb anti-Cdc6-P (1:500), anti-p107 (1:200), MAb anti-Mcm7, anti-Cdt1 (1:200), anti-RPA-70 (1:50), and goat anti-DNA polymerase delta (Pol delta) (1:50); MAb anti-CycA (for IB; Neomarkers); and anti-Myc (1:500; Steve Hann, Vanderbilt University). Chicken anti-Cdc45 and anti-Mcm5 were described previously (33).
IF and single-stranded-DNA (ssDNA) helicase assays.
Replicating DNA was labeled by pulsing for 30 min with 15 μM bromodeoxyuridine. For helicase assays (19), 15 μM BrdU was in the medium during the entire synchronization. BrdU detection and IF assays used standard procedures as described previously (2), except no HCl was used where indicated for the helicase assays. Two percent formaldehyde was used as fixative in all IF assays. RPA and DNA Pol delta IF assays used a 10-min preextraction at 4°C with 0.1% Triton X-100 in 10 mM HEPES, pH 7.9, 10 mM KCl, 0.5 mM dithiothreitol, and 1.5 mM MgCl2 to enrich for chromatin-bound complements/foci of each protein tested.
shRNA design and cDNAs.
Small-hairpin RNA (shRNA) was expressed using pSuperior vectors (Oligo Engine). The luciferase target sequence was 5′-CGTACGCGGAATACTTCGA-3′. Rb-shRNA-2573 and Rb-shRNA-240 are described in Results. Homo sapiens Mcm7 (HsMcm7), HsMcm7-CT, and HsMcm6 were generated by reverse transcription (RT)-PCR from HeLa mRNA. RbN was generated by PCR from a full-length HsRb cDNA. pcDNA3-HA, pET28A, and pGEX4T were used to subclone cDNAs with appropriate tags. Approaches are available upon request. The Rb-M10-glu cDNA was provided by David Goodrich (Roswell Park Cancer Center).
Bacterial expression.
BL21 bacteria were used to generate GST-tagged (Rb proteins) and His6-tagged proteins (Mcm7, Mcm7-CT, Mcm6, and Mcm2). Standard purification approaches were used, and details of purifications are available upon request.
Immunoprecipitations, immunoblots, and kinase assays.
Immunoprecipitations (IPs) used log or synchronized cells. IPs using anti-Rb, anti-CycE, anti-CycA, or anti-Cdc7 were performed in 0.1% NP-40, 50 mM Tris, pH 7.4, and 250 mM NaCl for 2 h (overnight for Rb IPs) in the cold after preclearing of samples with nonspecific IgG and 0.1% bovine serum albumin-blocked agarose beads. Washing was done using the same buffer for 3 × 10 min. For immunoblots (IB), equal numbers of cells were lysed and boiled in loading dye (for total lysates [TCE]) or were separated into detergent-resistant (P3/chromatin) or detergent-soluble fractions as described previously (32, 33). Pre-RC subunits present in the chromatin fraction are sensitive to nuclease digestion (32). TCE and chromatin samples representing equivalent cell numbers were analyzed by standard immunoblotting and enhanced chemiluminescence techniques. Histone H1 (10 μg/reaction) and GST-Rb (100 ng/reaction) kinase assays were performed as described previously (2). Cdc7 kinase assays were performed as described previously (12) using 1.8 μg His6-Mcm2 per reaction.
RESULTS
TGF-β1 treatment in early G1 blocks MCM loading.
We made use of the model line of BALB/MK cells, which are effectively synchronized by EGF deprivation and highly sensitive to TGF-β1 growth arrest (4). Quiescent MK cells display no ongoing DNA synthesis as measured by BrdU incorporation (Fig. 1A and B). Readdition of EGF leads to cell cycle entry, with the G1/S transition 12 h after release and almost all cells in S phase at 15 and 18 h (Fig. 1B). Treatment with TGF-β1 in early G1 (2 h postrelease) effectively blocks progression into S phase (Fig. 1B).
FIG. 1.
Early-G1 TGF-β1 treatment blocks MCM loading. (A) Diagram showing experimental design. (B) EGF-synchronized MK cells pulsed with BrdU at each hour to measure cell cycle progression. Effectiveness of early-G1 TGF-β1 treatment is shown below. Percentages are means from triplicate fields. (C) Immunoblot analysis of several established TGF-β1 targets after early-G1 TGF-β1 treatment. (D) Analysis of effects of TGF-β1 on CycE-associated histone H1 kinase activity. (E) Immunoblot analyses of TGF-β1 effects on total protein expression and chromatin-bound complement of multiple pre-RC subunits and of Cdc45 and PCNA.
We verified that several well-established negative effects of TGF-β1 on the cell cycle machinery were intact in MK cells. Total protein lysates were collected at time points indicated in Fig. 1. Myc expression was suppressed by TGF-β1, as was CycA (Fig. 1C). Phosphorylation of Rb in late G1, as measured using antibodies against phosphoserines 807/811, was also blocked by TGF-β1 (Fig. 1C). Ser807/811 phosphorylation is preferentially catalyzed by CycE-Cdk2 in vitro (26) and occurs around the restriction point (∼8 h in MK cells) when this kinase becomes active against Rb (17, 30, 33). Consistent with the absence of Ser807/811-P, and as shown by others (27), CycE-Cdk2 kinase was blocked from becoming active by TGF-β1 (Fig. 1D).
We next analyzed several aspects of pre-RC dynamics after TGF-β1 treatment in early G1. During pre-RC assembly, subunits become progressively bound to chromatin and are operationally defined at such time as they are ready for initiating DNA replication (6, 32). The chromatin-bound state is indicated by resistance to extraction with nonionic detergents. Parallel to experiments described above that were carried out with total cell lysates, we collected separate samples and fractionated them into detergent-sensitive and detergent-resistant lysates, defined as the nucleosolic/cytosolic and chromatin-bound pools, respectively (2, 32). For simplicity, only the total and chromatin-bound analyses are shown. Treatment with TGF-β1 in early G1 does not adversely affect the total protein levels or chromatin-bound pool of Orc4, Orc5 (data not shown), Cdt1, and Cdc6 (Fig. 1E, top). However, TGF-β1 suppresses the total protein levels of MCM subunits and thereby blocks the loading of MCM subunits onto chromatin (Fig. 1E, bottom). These results indicate that following early G1 exposure to TGF-β1, pre-RC assembly proceeds through the ORC, Cdc6, and Cdt1 loading steps but MCM hexamer formation on chromatin is blocked.
TGF-β1 treatment in early G1 targets proteins involved in MCM loading and activation.
Cdc6 is phosphorylated by CycE-Cdk2, which is required for MCM loading (31). Such phosphorylated forms of Cdc6 also interact with chromatin (1). One of our anti-Cdc6 antibodies preferentially recognizes slower-mobility forms of Cdc6 that are likely representative of phosphorylated forms of Cdc6 (called anti-Cdc6-P). Such forms of Cdc6 are efficiently blocked in accumulation and absent from chromatin after TGF-β1 exposure (Fig. 1E), consistent with the lack of CycE-Cdk2 activity (Fig. 1D). In addition, CycE-Cdk2 activity is required for Cdc7 to function in the phosphorylation of Mcm2, which is also necessary for MCM assembly (12).
Cdc45 and PCNA assemble at pre-RCs after MCMs have loaded, leading to formation of preinitiation complexes (pre-ICs) that are involved in MCM activation at G1/S (and DNA polymerase activation). TGF-β1 treatment inhibits the loading of Cdc45 and PCNA onto chromatin (Fig. 1E). Although the total protein levels of Cdc45 are blocked by TGF-β1, explaining its absence from chromatin, the total levels of PCNA are not suppressed by TGF-β1 (Fig. 1E). We conclude that early G1 exposure to TGF-β1 produces multiple negative effects on MCM loading and activation. MCM protein expression is suppressed, and the ability of any residual MCM proteins to load onto chromatin will be severely hindered due to an absence of CycE-Cdk2 activity and its necessary effects on Cdc6 and Cdc7. The loading of Cdc45 and PCNA onto chromatin is blocked, preventing activation of MCMs. Further, c-Myc binds to MCMs and functions in a nontranscriptional manner in promoting S-phase entry and replication origin activity (15), and its suppression by TGF-β1 also prevents MCM activation.
TGF-β1 treatment in late G1 arrests cells with pre-RCs fully assembled.
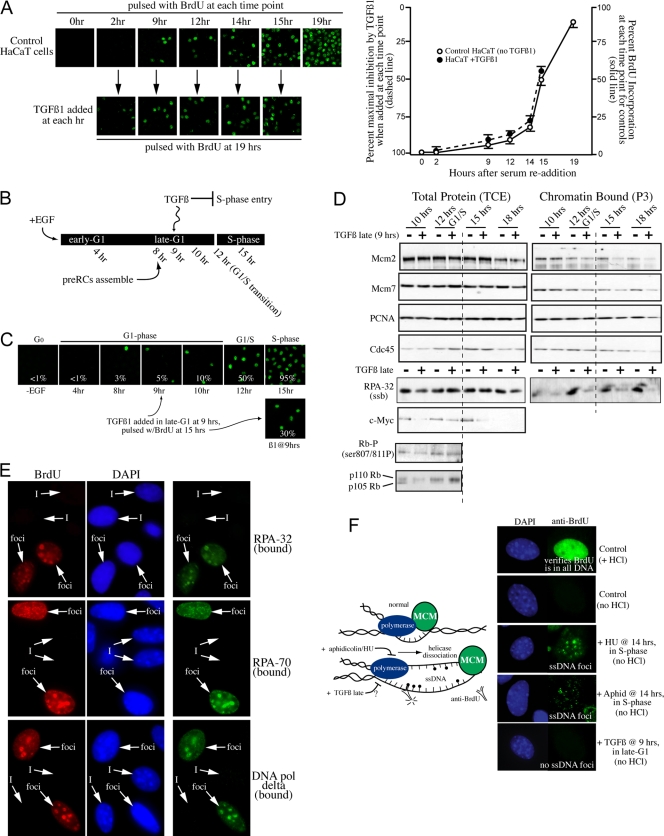
TGF-β1 can also block entry into S-phase when added to cells in late G1. Although this is true for MK (4, 38) and mink lung cells (23, 28), one report exists suggesting that human HaCaT cells are not sensitive to TGF-β1 in late G1 (18). However, given that very few HaCaT cells were capable of reentering the cell cycle after quiescence in this study, we reasoned that the confluent cell culture conditions and 3-day serum withdrawal were not optimal for a conclusive determination of TGF-β1 sensitivity. Using optimized quiescence-release conditions in which cells are plated at a lower density and serum deprived for only 24 h (allowing almost 90% of cells to reenter the cycle), we find that HaCaT cells are also sensitive to TGF-β1 inhibition throughout G1 (Fig. 2A). Thus, all three model cell lines for TGF-β1 analysis are similarly sensitive to TGF-β1 throughout the G1 period.
FIG. 2.
Late-G1 TGF-β1 treatment arrests cells with assembled pre-RCs and prior to the helicase unwinding step. (A) Synchronized HaCaT cells are sensitive to TGF-β1 throughout G1. Control cells were pulsed with BrdU at times indicated. TGF-β1 was added to parallel plates at each time but allowed to proceed to 19 h (peak S phase), at which time BrdU pulsing determined the inhibition index. Means of triplicate fields are plotted ± 1 standard deviation (SD). (B) Experimental design for MK assays. (C) BrdU analysis as for Fig. 1B of synchronized MK cells. Effectiveness of late-G1 TGF-β1 treatment is shown below. Percentages are means from triplicate fields. (D) IB assays performed as for Fig. 1E, but after late-G1 TGF-β1 treatment. (E) Synchronized MK cells treated with TGF-β1 at 9 h (late G1), allowed to proceed to 15 h, and then pulsed with BrdU and processed by IF for BrdU and indicated replication proteins. “I” denotes examples of TGF-β1-inhibited cells; “foci” denotes noninhibited cells displaying BrdU and replication protein foci that overlap. (F) Diagram illustrating in vivo helicase assay principles and photographs of representative nuclei for the helicase assays carried out with synchronized MK cells.
MCMs, Cdc45, and PCNA normally load onto chromatin ∼2/3 of the way into G1 phase, 8 h after release in MK cells (Fig. 1E and our recent study, reference 33). Since TGF-β1 can block entry into S phase when added after this time, we next asked how late-G1 treatment with TGF-β1 affected pre-RCs once assembled. MK cells were synchronized and released into G1, and at 9 h (late G1), TGF-β1 was added, followed by collection of total and chromatin-bound protein fractions at the indicated times (diagrammed in Fig. 2B). Exposure to TGF-β1 in late G1 effectively blocks entry into S phase (Fig. 2C; also verifies population was indeed in late G1 at the time of TGF-β1 addition). Expression of Myc in late G1 is blocked by TGF-β1 (Fig. 2D), confirming that TGF-β1 signals to the nucleus are intact. Interestingly, Rb phosphorylation on serines 807/811, which occurs at ∼8 h in MK cells (Fig. 1C), is not reversed by TGF-β1 exposure 1 h later (Fig. 2D). In addition, the presence of the slower-migrating p110-Rb is not acutely reduced by late G1 TGF-β treatment.
In significant contrast to the effects of early G1 TGF-β1 exposure, late G1 treatment does not disrupt preformed pre-RCs (Fig. 2D). MCM subunits remain expressed and chromatin bound equivalently to results for control cells at what would be the G1/S transition (12 h; samples to the left of the dashed line in Fig. 2D). Cdc45 and PCNA are also unperturbed, confirming that pre-RCs are assembled through the MCM loading step. After a prolonged duration in TGF-β1, cells begin to disassemble pre-RCs to some extent, as the levels of chromatin-bound MCMs, Cdc45, and PCNA are slightly diminished at later times (Fig. 2D, 15 and 18 h).
Since the MCMs and Cdc45 are loaded but polymerases are nonfunctional (due to a lack of BrdU) (Fig. 2C), we reasoned that cells might be arrested at the helicase unwinding step upstream of DNA polymerase recruitment. We determined the state of chromatin association of RPA, which binds ssDNA at replication forks with high affinity after DNA unwinding at origins and thus serves as a surrogate measure for the helicase step in vivo (44). Intriguingly, cells arrested by late-G1 TGF-β1 exposure are significantly devoid of the RPA-32 subunit in the chromatin fraction, although RPA-32 is present (in TCE) (Fig. 2D). As further confirmation, synchronized MK cells were treated with TGF-β1 at 9 h, allowed to proceed to 15 h (normal peak S phase), and assayed by IF for the chromatin presence of RPA-32, RPA-70, and DNA Pol delta. Figure 2E shows that non-TGF-β1-inhibited cells (BrdU positive, “foci”) display noticeable focal patterns for all three proteins that overlap BrdU signals, whereas TGF-β1-inhibited cells (BrdU negative, “I”) are devoid of any such focal patterns for RPA and DNA Pol delta. Collectively, these results strongly suggest that TGF-β1 acutely arrests cells in late G1 at or just prior to the helicase unwinding step at G1/S, rendering RPA and DNA Pol delta unable to become recruited although MCMs and Cdc45 are present.
MCM helicases are not functional in cells growth arrested by late-G1 TGF-β1 treatment.
We further verified that no helicase activity could be detected in TGF-β-arrested cells. We took advantage of a novel means of assessing whether MCMs are active in cells in vivo (19). Treatment with DNA polymerase blocking agents (aphidicolin or hydroxyurea) leads to a separation of helicase/MCM activity from the halted polymerases (19, 36, 44), creating stretches of ssDNA ahead of the arrested polymerases. As diagrammed in Fig. 2F (top), the DNA (labeled with BrdU in a previous cell cycle) that is unwound by the active MCMs can be observed using anti-BrdU antibodies in the absence of the HCl denaturation step that is usually necessary for antibody access to the bromo groups (19).
During EGF− synchronization, BrdU was added to the culture medium to label all of the DNA (cells proceed through 1 to 2 cycles during such synchronization). Cells were released and allowed to progress to late G1, at which time (9 h) TGF-β1 was added to one sample. Cells were then allowed to progress into S phase (but TGF-β1-treated cells do not enter), at which time (14 h) Aph or HU was added to two separate plates for 1 h. Figure 2F shows that the nuclei were fully labeled with BrdU but that HCl was required to observe a BrdU signal in untreated cells (top two panels). Aph and HU samples both display noticeable ssDNA foci as a result of continued MCM functioning, consistent with results in other studies (19). However, cells treated with TGF-β in late G1 do not display ssDNA foci (Fig. 2F). Although a small amount of DNA unwinding would not be observable by this approach, these results are in agreement with that shown above, indicating that TGF-β1 acutely arrests cells with loaded but inactive MCM helicases.
Late-G1 block to MCM activation by TGF-β1 is not mediated by Myc suppression nor by CycE/Cdk2 or Cdc7 kinase inhibition.
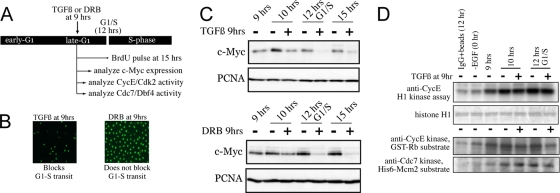
We wanted to understand the mechanism by which late-G1 treatment with TGF-β1 blocks MCM activation. The Myc protein functions in a nontranscriptional manner to stimulate S-phase entry via its interaction with MCMs (15). We asked whether acute negative effects of TGF-β1 on Myc expression, specifically in late G1, explained the failure of cells to activate the assembled pre-RCs/MCMs. Mammalian cells become insensitive to agents that block de novo mRNA synthesis in late G1, indicating that cells no longer require new mRNA production in late G1 for progression into S phase (4, 9, 33). We compared a late-G1 treatment with TGF-β1 (at 9 h), which blocks entry into S phase, to a late-G1 treatment with an effective and specific RNA polymerase II inhibitor, DRB (also at 9 h), which does not block S-phase entry (Fig. 3A and B). Both TGF-β1 and DRB acutely suppress c-myc mRNA expression during this time (33, 37). Due to the short half-life of c-myc mRNA, this acutely downregulates the Myc protein, and Fig. 3C shows that DRB and TGF-β1 display similar kinetics for loss of the Myc protein. Since DRB does not prevent MK cells from entering S phase, these results indicate that acute suppression of Myc by TGF-β1 in late G1 does not play a pivotal role in blocking S-phase entry or MCM activation at this time.
FIG. 3.
Late-G1 TGF-β1 treatment; growth arrest is not mediated by c-Myc loss nor by inhibition of CycE-Cdk2. (A) Experimental design. (B) BrdU analysis performed at 15 h, verifying that TGF-β1 treatment at 9 h blocks S-phase entry while DRB treatment does not. (C) Immunoblots on total lysates showing kinetics of c-Myc loss after TGF-β1 or DRB treatment at 9 h. (D) Analysis of effects of late-G1 TGF-β1 treatment on CycE-associated kinase measured against H1 or GST-Rb and anti-Cdc7 kinase measured against His6-Mcm2.
CycE-Cdk2 kinase becomes active prior to late G1 and is blocked from becoming active by early-G1 TGF-β1 treatment (Fig. 1D). We determined if CycE-Cdk2 kinase was acutely inactivated by late-G1 TGF-β1 treatment after it had already become active. MK cells were synchronized and released, followed by TGF-β1 treatment at 9 h and analysis of CycE-associated kinase activity 1 and 3 h later. Figure 3D shows clearly that once activated, the CycE-Cdk2 kinase is not acutely suppressed by late-G1 TGF-β1 treatment as measured against the histone H1 or GST-Rb substrate. We also found that the Cdc7-Dbf4 kinase, which is required for origin activation in yeast studies (8) and targets Mcm2 for phosphorylation (12), is not inhibited acutely by TGF-β1 (Fig. 3D). Thus, acute inactivation of CycE-Cdk2 or Cdc7 is not involved in the block to MCM activation and S-phase entry when TGF-β1 is added specifically in late G1.
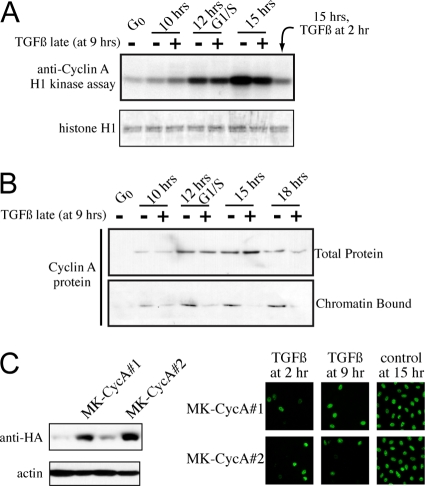
Late-G1 treatment with TGF-β1 acutely blocks CycA localization to chromatin.
We determined the effects of late-G1 TGF-β1 exposure on CycA-Cdk2 activity and on CycA expression and dynamics. Intriguingly, although the cells do not enter S phase, CycA-Cdk2 kinase activity is induced at the same time as in untreated cells and is not inhibited thereafter by late-G1 TGF-β1 treatment (Fig. 4A). Consistent with this, we also found that there are no changes in the amount of Cdk2 or p27Kip1 that is associated with CycA (data not shown). Further, late-G1 TGF-β1 treatment does not block the induction of expression of the CycA protein as it does when TGF-β1 is added in early G1 (compare Fig. 1C and 4B). However, one acute negative effect of late-G1 TGF-β1 treatment is a block to CycA localization to chromatin (Fig. 4B), where pre-RCs/MCMs are also localized but not activated. We next asked if overexpression of CycA could overcome the inhibitory effects of late-G1 treatment with TGF-β1. CycA was stably expressed in MK cells, and clones were derived, two of which are shown (Fig. 4C). In both MK-CycA clones, TGF-β1 was capable of blocking G1/S transit whether it was added to cells in early G1 or late G1 (Fig. 4C). Although the lack of CycA chromatin association may contribute to late-G1 TGF-β1 arrest, these results suggest that other unknown mechanisms, dominant to this CycA effect, mediate the late-G1 block to pre-RC activation by TGF-β1.
FIG. 4.
Late-G1 TGF-β1 treatment has no effect on CycA-associated kinase activity but blocks CycA chromatin loading. (A) Analysis of effects of late-G1 TGF-β1 treatment on CycA-associated H1 kinase activity in synchronized MK cells. (B) Immunoblot analysis performed as described for Fig. 1E, examining CycA expression and chromatin binding dynamics. (C) Immunoblot of total lysates for MK clones stably expressing elevated HA-CycA protein (left) and analysis of TGF-β1 sensitivity in early G1 and late G1 for two synchronized clones via BrdU pulsing at 15 h after release (right).
TGF-β1 block to pre-RC/MCM activation in late G1 requires Rb.
Viral tumor proteins with Rb-binding motifs abrogate TGF-β1 growth arrest (28, 38). However, viral proteins also target other cellular factors. As such, we wanted to determine conclusively if Rb alone is involved in the acute late-G1 TGF-β1 inhibition of pre-RC/MCM activation. We generated MK cells carrying stable shRNA constructs that downregulated Rb. Two shRNAs targeting two different parts of Rb were designed (Rb-shRNA-240 and Rb-shRNA-2573) and contained multiple mismatches or absent bases relative to the related p107 and p130 proteins (Fig. 5A). Three clones displaying significant downregulation of Rb [MK(Rb−)] were selected (Fig. 5B, clones #4, #5, and #13). We also generated a control cell line with an shRNA targeting luciferase (Luc-shRNA). The p107 protein was not suppressed in any of the clones (Fig. 5B).
FIG. 5.
Acute late-G1 growth arrest by TGF-β1 requires Rb, and the Rb requirement parallels kinetics of pre-RC assembly. (A) shRNA design against mouse Rb. (B) Immunoblot showing Rb depletion in three MK(Rb−) clones. (C) Immunoblot verifying Rb is depleted throughout the duration of the experiment for clone #5. Similar verifications were performed on other clones (not shown). (D) BrdU analysis at 15 h postrelease on the synchronized cell types indicated, after TGF-β1 addition in late G1. (E) Graph of results in panel D, plus results for other MK(Rb−) clones. (F) BrdU analysis (as for Fig. 1B) of synchronized MK(Rb−) clone #5, verifying cells were in late G1 when TGF-β1 was added. (G) MK(Rb−) clone #5, synchronized and tested for chromatin-bound presence of RPA-32 and Mcm7 by IB. Fractionation was done as described for Fig. 1E. (H) MK(Rb−) clones and MK-Luc-shRNA control were synchronized. TGF-β1 was added at indicated times (for +β1 samples), and such cells were allowed to progress to 15 h and BrdU labeled to determine their TGF-ß1 inhibition index (left axis). In parallel, untreated MK(Rb−) clone #5 and MK-Luc-shRNA were pulsed with BrdU at each hour to determine G1 progression kinetics (right axis). (I) Asynchronous wt-MK ± TGF-ß1 overnight were assayed by BrdU incorporation to determine amount of growth arrest (top). Asynchronous MK(Rb−) clone #5 was exposed to TGF-ß1 for ∼1 cell cycle (30 h) and ∼2 cycles (60 h) and then assayed by BrdU incorporation to determine amount of TGF-ß1 growth arrest (bottom). Percentages in panels D, E, F, and H are means of triplicate counts of ∼200 cells/field (±1 SD in graphs).
We next tested MK(Rb−) cells for their response to TGF-β1 added in late G1. Despite significant Rb reduction, all three MK(Rb−) clones could be rendered quiescent by EGF withdrawal (data not shown). Figure 5C shows that after synchronization, MK(Rb−) clone #5 contained very low levels of Rb throughout the 15-h duration of the experiment. Significantly, treatment of MK(Rb−) clone #5 with TGF-β1 in late G1 failed to block entry into S phase (Fig. 5D and E) and thus failed to block MCM activation. Similarly, MK(Rb−) clones #13 and #4 also failed to arrest following late-G1 TGF-β1 exposure (Fig. 5E). Since loss of Rb can reduce the length of G1, we were concerned that our cells were not in late G1 at the time TGF-β1 was added but instead might already be in S phase, which would serve as a simple explanation for the lack of TGF-β1 sensitivity in such experiments. However, this is not the case, since BrdU analysis of synchronized MK(Rb−) clone #5 shows that the cells are in late G1 at the time of TGF-β1 addition (Fig. 5F). Similar results were found for clones #13 and #4 (data not shown). MK(Rb−) clone #5 also regains the ability to recruit RPA-32 to chromatin (Fig. 5G). The use of two different shRNA target sites on Rb and consistent results from multiple independent clones indicate that our results are not due to off-target activity and are physiologically relevant to events following specific depletion of Rb in the MK cells. We conclude that Rb is absolutely required for TGF-β1 treatment in late G1 to block activation of MCMs and S-phase entry.
Rb is required for TGF-β1 growth arrest from the time of pre-RC/MCM assembly.
We found unexpectedly that the MK(Rb−) clones all remained sensitive to growth arrest by TGF-β1 if the inhibitor was added to cultures in early G1 (Fig. 5H). This indicated that TGF-β1 was capable of eliciting a G1 block in the absence of Rb, at least during a window of time in early G1. Consistent with this, another study has shown that Rb−/− mouse embryo fibroblasts (MEFs) remain partially sensitive to TGF-β1 arrest under low plating conditions (21). Log wild-type MK (wt-MK) cells are inhibited maximally by TGF-β1 after a 24-h exposure, which encompasses ∼1 cell cycle (and 1 G1 phase) (Fig. 5I). However, if an Rb-independent window of TGF-β1 sensitivity does exist in early G1, then log Rb-depleted MKs should be partially inhibited after one cycle of exposure to TGF-β1 but maximally inhibited after cells that were in the previous late-G1 insensitive window enter the next early-G1 window of sensitivity and become arrested. Indeed, this is the case (Fig. 5I), indicating that MK cells do not require Rb for early-G1 arrest.
The sensitivity of MK(Rb−) cells to early-G1 TGF-β1 exposure indicated that TGF-β1 signaling pathways were functional. The only exception to this was that acute late-G1 inhibition by TGF-β1 was abrogated if Rb levels were diminished. To determine when the cells transition from a lack of need for Rb in early G1 to a strict requirement for Rb in late-G1 TGF-β1 arrest, we performed an experiment in which synchronized Rb-depleted cells were compared to Rb-containing cells (Luc-shRNA control) for their sensitivity to TGF-β1 as they progressed through G1 (Fig. 5H). TGF-β1 was added at 2, 6, 8, and 9 h after release, and such cells were pulsed with BrdU at 15 h to determine the percentage of inhibition after each time of TGF-β1 exposure. In parallel, untreated control cells and MK(Rb−) clone #5 were pulsed with BrdU at indicated times to assess the flow of the synchronous populations through G1 into S phase (Fig. 5H, slashed circles at right). Comparison of the resulting inhibition and G1 progression curves reveals that relative to the G1/S transition (12 h for Luc-shRNA MK control and ∼11 h for clone #5), all three MK(Rb−) clones lose TGF-β1 sensitivity as they progress through the 7- to 8-h time period (Fig. 5H). In contrast, control cells (Luc-shRNA +β1) remain sensitive to TGF-β1 throughout G1, consistent with our previous findings for wt-MK (4). Intriguingly, this timing of desensitization to TGF-β1 in MK(Rb−) cells directly correlates with the timing of MCM assembly (∼8 h) (Fig. 1E) (33). We conclude that once MCMs load onto chromatin in late G1 and are ready for G1/S activation, TGF-β1 inhibitory mechanisms continue to function but absolutely require that Rb be present in order to successfully block such assembled pre-RCs/MCMs from becoming activated at G1/S.
Rb interacts with the MCM complex in late G1 via the Mcm7 subunit.
The acute ability of TGF-β1 to elicit a late-G1 arrest that requires Rb was intriguing. Rb becomes hyperphosphorylated at the restriction (R) point, which we showed occurs at ∼8 h in MK cells (33), shown again in Fig. 1C using Ser807/811-P, a CycE-Cdk2-catalyzed site, as a surrogate (26). Given that TGF-β1 can block entry into S phase when added to cultures even in late G1, these results collectively indicate that TGF-β1 can block entry into S phase when added after the R point. Further, our results indicate that Rb still functions in mediating G1-S transit after the R point, at least in response to TGF-β1.
Since Rb's control over E2F complexes is diminished in late G1, we hypothesized that the function of Rb in mediating TGF-β1 arrest in late G1 might arise from a direct role in binding and preventing activation of the assembled MCMs. In support of this contention, a yeast 2-hybrid study has shown that the amino terminus of Rb (RbN) can interact with the MCM complex subunit Mcm7 (43). Further, Rb can suppress in vitro DNA replication using Xenopus extracts (36, 43), and such inhibition of DNA replication by Rb derives from an ability to target and suppress MCM helicase activity via interaction with Mcm7 (36). We provide evidence here that a similar situation exists in mammalian cells that mediates the TGF-β1 inhibitory response in late G1.
The interaction of Rb and Mcm7 was shown to occur in rabbit reticulocyte extracts (43), but this did not demonstrate conclusively that Rb directly bound Mcm7. We show here that Rb does interact directly with Mcm7. Bacterially expressed His6-Mcm7 and GST-RbN coimmunoprecipitate (co-IP) when incubated together (Fig. 6A). Further, the C-terminal 137-amino-acid fragment of Mcm7 that was originally identified in the 2-hybrid assay (Mcm7-CT) also binds to RbN directly in vitro (Fig. 6A). Both Mcm7 proteins also bind to full-length Rb (data not shown). Neither Mcm7 nor Mcm7-CT binds to beads or GST alone, and a vast excess of His6-Mcm6 does not interact with RbN, indicating that the Mcm7-Rb interaction is specific.
FIG. 6.
Rb interacts with Mcm7, and Mcm7 overexpression abrogates late-G1 arrest by TGF-ß1. (A) Bacterially expressed GST or GST-RbN and His6-Mcm7, His6-Mcm7-CT, or His6-Mcm6 were incubated together and immunoprecipitated with glutathione beads. Input and IP samples were immunoblotted with anti-His or anti-GST. Images are separated due to size differences of proteins. “α” indicates antibody. (B) IP from log MK cells with anti-Rb and then IB with different anti-Rb and anti-Mcm7. (C) Co-IP performed as for panel B but probed for Mcm5. (D) Synchronized MK cells subjected to co-IP analysis as for panel B. Samples were blotted with antibodies at right. BrdU assays verified synchrony and that G1/S was at 12 h (not shown). (E) SaOS-2 cells transiently transfected with HA-Rb or HA-Rb-M10-glu for 24 h, followed by co-IP with anti-HA and IB with anti-Mcm7 or anti-Rb. (F) Log MK cells subjected to co-IP analysis as for panel B but after 24 h of TGF-ß1 treatment in one sample. (G) Synchronized MK cells subjected to co-IP analysis as for panel C but compared to TGF-ß1 treatment in late G1. BrdU verified synchrony (not shown). (H) MK cells transfected using polyethylenimine at the beginning of the synchronization. Plasmids expressed the indicated proteins (top). TGF-ß1 was added to all cultures at 9 h. Cells were allowed to proceed to 15 h and were then pulse-labeled with BrdU. Anti-HA-FITC and anti-BrdU-Alexa 594 identified transfected and replicating cells (that had entered S phase in the presence of TGF-ß1), respectively. Arrows indicate selected HA-positive (transfected) cells. (I) Quantitation of the results in panel H, performed on triplicate fields; means of such counts are plotted ± 1 SD.
Rb also binds to Mcm7 in vivo in MK cells (Fig. 6B). Immunoprecipitation with anti-Rb followed by immunoblotting for associated proteins (co-IP) revealed that Mcm7 was present in Rb-containing complexes. This also occurred with use of different anti-Rb antibodies, and transient expression of HA-tagged Mcm7 can co-IP endogenous Rb (data not shown). Further, Mcm5 co-IPs with Rb, indicating that Rb interacts with the MCM complex in vivo (Fig. 6C). Using EGF-synchronized MK cells, we found that Mcm7 and Rb interact specifically during late G1 (Fig. 6D), with the most pronounced co-IP of Mcm7 with Rb occurring near G1/S (12 h). Intriguingly, after cells entered S phase, the ability of Mcm7 to interact with Rb was diminished (Fig. 6D, 15 and 18 h). Since Mcm7 was interacting with Rb in late G1, after the R point and thus presumably after multiple phosphates have been added to Rb's C terminus (i.e., hyperphosphorylated Rb), we asked if Mcm7 could indeed interact with such an Rb molecule. The Goodrich laboratory has tested several alleles of Rb modified with glutamates at C-terminal phospho acceptor sites that mimic hyperphosphorylated forms of Rb (5). One such Rb allele, Rb-M10, carrying 10 glutamates encompassing most of the C-terminal sites (and running predictably slower in the gel), does indeed co-IP with endogenous Mcm7, similar to results with wt-Rb (Fig. 6E). Intriguingly, this particular Rb allele was found by the Goodrich group to be capable of blocking entry into S phase in SaOS-2 cells, but for unknown reasons, even though it resembles a hyperphosphorylated form of Rb (5). We conclude from these results that Rb interacts with MCMs specifically in late G1 but not after cells pass through G1/S and that the interaction occurs in a direct manner through Mcm7.
TGF-β1 treatment enhances interaction between Mcm7 and Rb.
We reasoned that TGF-β1 inhibitory signals in late G1 might be mediated through this Rb-Mcm7 interaction, resulting in a failure to activate the assembled MCMs due to a novel inhibitory function of the associated Rb protein. One prediction of this model was that TGF-β1 exposure would potentiate the Mcm7-Rb interaction, and treatment of log MK cells for 24 h with TGF-β1 revealed that the interaction between Mcm7 and Rb did become more robust than that in untreated cells (Fig. 6F). In addition, treatment of EGF-synchronized MK cells with TGF-β1 in late G1 resulted in a sustained interaction of Mcm7 and Rb from 12 to 15 h, whereas control cells again displayed a reduced Mcm7-Rb interaction at 15 h after entering S phase (Fig. 6G). We conclude from these results that TGF-β1 treatment of MK cells prevents the dissociation of the Rb-Mcm7 interaction that is present in late G1 and enhances the efficiency of the Rb-Mcm7 interaction after longer periods of time.
Overexpression of Mcm7 or its Rb-binding domain alone abrogates late-G1 arrest by TGF-β1.
If the Mcm7-Rb interaction played a functional role in TGF-β1 late-G1 arrest, then overexpression of Mcm7 should interrupt Rb's ability to target the MCM complex, thus allowing MCM activation, G1/S transit, and abrogation of TGF-β1 arrest. We show here that this is indeed the case. HA-Mcm7, HA-Mcm7-CT, or HA-Mcm6 was transfected into MK cells at the start of the EGF− synchronization and released into G1 phase with each protein overexpressed. BrdU pulsing verified synchronization and that S-phase entry was at 12 h (data not shown). Transfected cells were treated with TGF-β1 in late G1 and were allowed to progress to 15 h and then pulsed with BrdU to assess the percentage of transfected cells for each condition that had entered S phase in the presence of TGF-β1. Transfected cells were identified using anti-HA IF staining, and all three proteins were expressed similarly in anti-HA immunoblots (data not shown).
As can clearly be seen in Fig. 6H, overexpression of Mcm7 or the Rb-binding domain of Mcm7 (Mcm7-CT) abrogated the ability of TGF-β1 to acutely block entry into S phase, resulting in BrdU-positive cells. This indicated that MCMs were now functional, since cells had now initiated DNA synthesis. Cells expressing Mcm7 or Mcm7-CT had not prematurely entered S phase at the time of TGF-β1 addition (9 h; data not shown), which might have served as a simple explanation for why TGF-β1 could no longer inhibit their progression. Overexpression of Mcm6 (Fig. 6H) or Cdt1 (data not shown) was not capable of overriding TGF-β1. Neighboring nontransfected cells remained largely BrdU negative, verifying that TGF-β1 could acutely inhibit late-G1 progression in the absence of ectopic proteins. Quantitation of the results observed for multiple fields is shown in Fig. 6I. We conclude that the acute late-G1 block to MCM activation and S-phase entry by TGF-β1 can be overcome by either a loss of Rb or a gain of Mcm7 and that the Rb-binding region of Mcm7 is sufficient for this effect. Collectively, our results indicate that the Mcm7-Rb interaction is functionally involved in the mechanisms mediating late-G1 arrest by TGF-β1.
DISCUSSION
Early-G1 control of MCM assembly by TGF-β1.
TGF-β1 elicits many negative effects on the cell cycle machinery and pre-RCs when added to cells in early G1, prior to pre-RC assembly. In particular, pre-RCs fail to recruit MCMs. TGF-β1 does not block the chromatin association of pre-RC components that serve as the assembly platform (ORC) or are involved in loading MCMs (Cdc6 and Cdt1). To a great extent, the inability to load MCMs derives from TGF-β1 blocking accumulation of MCM proteins. While we do not know the mechanism for this, we do know that mRNA expression for MCM subunits is not suppressed by TGF-β1 (P. Mukherjee and M. G. Alexandrow, unpublished results).
TGF-β1 treatment in early G1 also induces changes in the cell cycle machinery that prevent residual MCMs from loading or functioning. The activity of the CycE-Cdk2 kinase is required to phosphorylate Cdc6 for MCM loading (31) and to promote association of pre-RCs with Cdc7, which phosphorylates Mcm2 during loading (12). TGF-β1-induced inhibition of CycE-Cdk2 thus indirectly blocks MCM complex assembly. Myc plays a positive role in MCM activation and G1-S transit independently of its transcriptional functions (15). Thus, Myc suppression by TGF-β1, while allowing CKI induction for CycE-Cdk2 inhibition (45), also prevents activation of any MCMs that do load. TGF-β1 also blocks the loading of Cdc45 and PCNA, Cdc45 being required for activation of MCMs at G1/S (36). In sum, TGF-β1 inhibitory signals in early G1 use multiple mechanisms to ensure that pre-RCs cannot form or become activated. Interestingly, suppression of MCM levels by TGF-β1 is sufficient to render cells unable to enter S phase, suggesting that inhibition of CycE-Cdk2 may be a secondary event that prevents loading if MCMs become available. Such a situation offers one explanation for why CKIs are not necessarily required for TGF-β1 arrest (24, 34, 46).
Late-G1 control by TGF-β1 involves mechanisms that block pre-RC/MCM activation.
TGF-β1 treatment in late G1, after pre-RC assembly has occurred, not only acutely blocks entry into S phase but does so without dissociating intact pre-RCs/MCMs that are loaded onto chromatin along with the MCM cofactor Cdc45. Since RPA and DNA Pol delta are not recruited to chromatin under these conditions, our results strongly indicate that cells are TGF-β1 arrested at the MCM helicase DNA unwinding step that produces ssDNA necessary for RPA recruitment. The lack of MCM functionality is likely due to TGF-β1 targeting critical activities necessary to stimulate their function and cause G1/S transit. Two well-established TGF-β1 targets that influence MCM loading and functionality are the CycE-Cdk2 kinase and Myc. However, our results clearly show that in late G1, neither acute inhibition of CycE-Cdk2 nor suppression of Myc by TGF-β1 explains why cells fail to activate the MCMs. Interestingly, the absence of acute CycE-Cdk2 inhibition and the lack of inhibition of CycA-Cdk2 activity described below offer another explanation for CKI-independent TGF-β1 cell cycle arrest. The lack of involvement of Myc in late-G1 control over G1-S transit and as a pivotal late-G1 target of TGF-β1 appears in opposition to data from an earlier study showing that phosphorothioate oligonucleotide suppression of Myc near G1/S blocks entry into S phase (38). One possible explanation for these differing results is that such oligonucleotides may have had off-target effects unrelated to Myc that blocked G1/S transit. Furthermore, late-G1 activation of MycER cannot override TGF-β1 arrest (3).
An Rb-MCM interaction mediates late-G1 arrest by TGF-β1.
We show here that Rb is specifically required for TGF-β1 to block S-phase entry when the inhibitor is added to cells in late G1. Although Rb is conventionally thought to regulate transcription in late G1, we and others have shown that transcriptional regulation per se in late G1 is not rate limiting for G1-S transit under normal conditions or after TGF-β1 treatment in late G1 (4, 9, 33). Thus, the Rb requirement for acute late-G1 arrest by TGF-β1 must extend beyond transcriptional regulation. Intriguingly, we show that Rb is not required for early-G1 arrest by TGF-β1, indicating that an Rb-independent window of TGF-β1 sensitivity exists in early G1. Indeed, partial Rb-independent sensitivity to TGF-β1 in MEFs has been shown by another group (21). The lack of a need for Rb in the TGF-β1 response appears in opposition to studies showing that viral oncoproteins that inactivate Rb abrogate TGF-β1 signaling (28, 38). One explanation may be that viral oncoproteins also target SMAD proteins (29, 35), which complicates assessment of such studies. Our approach, however, focused specifically on Rb.
A novel target of Rb in mediating TGF-β1 late-G1 arrest is the loaded, but nonactivated, MCM complex itself. Rb physically binds the MCM complex directly via Mcm7 in MK cells. Rb interacts with Mcm7 during late G1, overlapping the time when cells are sensitive to arrest by TGF-β1. TGF-β1 treatment in late G1 prevents the dissociation of Rb and Mcm7 that is normally seen as cells progress into S phase, and Rb and Mcm7 interact more efficiently after longer TGF-β1 durations. Overexpression of Mcm7 can specifically and effectively abrogate the ability of TGF-β1 to acutely block cells in late G1. Importantly, the Rb-binding C-terminal domain of Mcm7 is sufficient for this. Thus, Rb and Mcm7 form complexes in late G1 that are targeted by TGF-β1 signals, and perturbation of the Rb-Mcm7 interaction by loss of Rb or overexpression of Mcm7 leads to disruption of TGF-β1's ability to block late-G1 transit into S phase. Intriguingly, Mcm7 is an oncogene (22, 41), and our results indicate that one reason for its oncogenicity involves its ability to override TGF-β1.
Model for Rb function in MCM regulation and TGF-β1 arrest.
We propose a model for late-G1 arrest by TGF-β1 that incorporates multiple findings in the literature with those shown here and explains how TGF-β1 can elicit an Rb-dependent arrest in late G1 that is transcriptionally independent (4). Rb interacts with the metazoan MCM helicase (36, 43) and inhibits such helicase activity in vitro (36), and Rb can negatively control metazoan origin function directly through binding the pre-RC (7, 42). We show here that the mammalian MCM complex, which is loaded in late G1 but inactive until G1/S, is bound by Rb during late G1 but dissociates from Rb once cells transit G1/S. We propose that Rb functions in a negative manner to modulate G1-S transit in mammalian cells by preventing MCM/origin activation in late G1 (in addition to Rb's other roles). At G1/S, the interaction of Rb with the MCM complex and Rb's negative effects on MCMs/origins are relieved by a trigger, possibly involving further phosphorylation of Rb known to occur after the R point (14). Although the identity of such a trigger is unknown, the CycA-Cdk2 kinase is an attractive candidate. Homologues of this kinase in yeast trigger MCM and origin activation (16, 47); CycA-Cdk2 can phosphorylate Rb (26), and it is present and required at mammalian replication sites (2, 10). Consistent with this model, Rb is present during G1 phase at future origin sites, at least in primary cells, but not in S phase (25).
We currently do not know how Rb achieves pre-RC/MCM control biochemically or molecularly. Although an understanding of the exact mechanism of Rb control over MCM function is beyond the scope of this study, there are several possibilities worth testing in the future. Clearly, direct control of MCM helicase activity is one potential mechanism (36), which may be relieved by specific phosphorylations on Rb and/or MCM subunits, such as Mcm7. Alternatively, Rb may regulate MCMs via steric mechanisms that control recruitment of regulatory factors necessary for G1/S activation of MCMs/origins or other pre-RC components.
TGF-β1 treatment in late G1 prevents dissociation of Rb from the MCM complex, possibly due to inhibition of a trigger(s), resulting in the MCM complex remaining in an inactive state. The inactivity of the loaded MCMs is supported by the fact that RPA and DNA Pol delta fail to be recruited to chromatin, indicative of cells being arrested at the helicase unwinding step that produces ssDNA for RPA recruitment. TGF-β1 also prevents the CycA-Cdk2 kinase from binding to chromatin (but does not block its kinase activity), where pre-RCs/MCMs are located and awaiting activation. Although the effect of TGF-β1 on CycA-Cdk2 may comprise part of the block to the trigger that dissociates Rb from the MCMs/origins, the validity of this requires further investigation. Given that Rb targets the MCM complex through Mcm7, another prediction of this model, demonstrated here experimentally, is that either a loss of Rb or Mcm7 overexpression (which would interrupt Rb function) will succeed at abrogating this negative control of MCMs by Rb and render TGF-β1 incapable of blocking entry into S phase. In sum, our results demonstrate that Rb plays a necessary negative role specifically in the late-G1 response to TGF-β1, and this involves a functional and novel role for Rb in binding and likely regulating the MCM complex.
Acknowledgments
This work was supported by the National Functional Genomics Center at the Moffitt Cancer Center, an ACS-IRG award, and a grant from the Florida Department of Health and James and Esther King Foundation to M.G.A. (award 06-NIR-01).
Footnotes
Published ahead of print on 30 November 2009.
REFERENCES
- 1.Alexandrow, M. G., and J. L. Hamlin. 2004. Cdc6 chromatin affinity is unaffected by serine-54 phosphorylation, S-phase progression, and overexpression of cyclin A. Mol. Cell. Biol. 24:1614-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandrow, M. G., and J. L. Hamlin. 2005. Chromatin decondensation in S-phase involves recruitment of Cdk2 by Cdc45 and histone H1 phosphorylation. J. Cell Biol. 168:875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandrow, M. G., M. Kawabata, M. Aakre, and H. L. Moses. 1995. Overexpression of the c-Myc oncoprotein blocks the growth-inhibitory response but is required for the mitogenic effects of transforming growth factor b1. Proc. Natl. Acad. Sci. U. S. A. 92:3239-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandrow, M. G., and H. L. Moses. 1995. Transforming growth factor beta 1 inhibits mouse keratinocytes late in G1 independent of effects on gene transcription. Cancer Res. 55:3928-3932. [PubMed] [Google Scholar]
- 5.Barrientes, S., C. Cooke, and D. W. Goodrich. 2000. Glutamic acid mutagenesis of retinoblastoma protein phosphorylation sites has diverse effects on function. Oncogene 19:562-570. [DOI] [PubMed] [Google Scholar]
- 6.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 7.Bosco, G., W. Du, and T. L. Orr-Weaver. 2001. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol. 3:289-295. [DOI] [PubMed] [Google Scholar]
- 8.Bousset, K., and J. F. X. Diffley. 1998. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 12:480-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campisi, J., and A. B. Pardee. 1984. Post-transcriptional control of the onset of DNA synthesis by an insulin-like growth factor. Mol. Cell. Biol. 4:1807-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardoso, M. C., H. Leonhardt, and B. Nadal-Ginard. 1993. Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell 74:979-992. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C. R., Y. Kang, P. M. Siegel, and J. Massague. 2002. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell 110:19-32. [DOI] [PubMed] [Google Scholar]
- 12.Chuang, L. C., L. K. Teixeira, J. A. Wohlschlegel, M. Henze, J. R. Yates, J. Mendez, and S. I. Reed. 2009. Phosphorylation of Mcm2 by Cdc7 promotes pre-replication complex assembly during cell-cycle re-entry. Mol. Cell 35:206-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffey, R. J., Jr., C. C. Bascom, N. J. Sipes, R. Graves-Deal, B. E. Weissman, and H. L. Moses. 1988. Selective inhibition of growth-related gene expression in murine keratinocytes by transforming growth factor beta. Mol. Cell. Biol. 8:3088-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeCaprio, J. A., Y. Furukawa, F. Ajchenbaum, J. D. Griffin, and D. M. Livingston. 1992. The retinoblastoma-susceptibility gene product becomes phosphorylated in multiple stages during cell cycle entry and progression. Proc. Natl. Acad. Sci. U. S. A. 89:1795-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dominguez-Sola, D., C. Y. Ying, C. Grandori, L. Ruggiero, B. Chen, M. Li, D. A. Galloway, W. Gu, J. Gautier, and R. Dalla-Favera. 2007. Non-transcriptional control of DNA replication by c-Myc. Nature 448:445-451. [DOI] [PubMed] [Google Scholar]
- 16.Donaldson, A. D., M. K. Raghuraman, K. L. Friedman, F. R. Cross, B. J. Brewer, and W. L. Fangman. 1998. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol. Cell 2:173-182. [DOI] [PubMed] [Google Scholar]
- 17.Ezhevsky, S. A., A. Ho, M. Becker-Hapak, P. K. Davis, and S. F. Dowdy. 2001. Differential regulation of retinoblastoma tumor suppressor protein by G(1) cyclin-dependent kinase complexes in vivo. Mol. Cell. Biol. 21:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geng, Y., and R. A. Weinberg. 1993. Transforming growth factor β effects on expression of G1 cyclins and cyclin-dependent protein kinases. Proc. Natl. Acad. Sci. U. S. A. 90:10315-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groth, A., A. Corpet, A. J. Cook, D. Roche, J. Bartek, J. Lukas, and G. Almouzni. 2007. Regulation of replication fork progression through histone supply and demand. Science 318:1928-1931. [DOI] [PubMed] [Google Scholar]
- 20.Hannon, G. J., and D. Beach. 1994. p15INK4B is a potential effector of TGF-β-induced cell cycle arrest. Nature 371:257-261. [DOI] [PubMed] [Google Scholar]
- 21.Herrera, R. E., T. P. Makela, and R. A. Weinberg. 1996. TGF beta-induced growth inhibition in primary fibroblasts requires the retinoblastoma protein. Mol. Biol. Cell 7:1335-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honeycutt, K. A., Z. Chen, M. I. Koster, M. Miers, J. Nuchtern, J. Hicks, D. R. Roop, and J. M. Shohet. 2006. Deregulated minichromosomal maintenance protein MCM7 contributes to oncogene driven tumorigenesis. Oncogene 25:4027-4032. [DOI] [PubMed] [Google Scholar]
- 23.Howe, P. H., G. Draetta, and E. B. Leof. 1991. Transforming growth factor beta1 inhibition of p34cdc2 phosphorylation and histone H1 kinase activity is associated with G1S-phase growth arrest. Mol. Cell. Biol. 11:1185-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iavarone, A., and J. Massague. 1997. Repression of the Cdk activator Cdc25A and cell cycle arrest by cytokine TGF-β in cells lacking the Cdk inhibitor p15. Nature 387:417-422. [DOI] [PubMed] [Google Scholar]
- 25.Kennedy, B. K., D. A. Barbie, M. Classon, N. Dyson, and E. Harlow. 2000. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 14:2855-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitagawa, M., H. Higashi, H. K. Jung, I. Suzuki-Takahashi, M. Ikeda, K. Tamai, J. Kato, K. Segawa, E. Yoshida, S. Nishimura, and Y. Taya. 1996. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 15:7060-7069. [PMC free article] [PubMed] [Google Scholar]
- 27.Koff, A., M. Ohtsuki, K. Polyak, J. M. Roberts, and J. Massagué. 1993. Negative regulation of G1 in mammalian cells: inhibition of cyclin E-dependent kinase by TGF-β. Science 260:536-539. [DOI] [PubMed] [Google Scholar]
- 28.Laiho, M., J. A. DeCaprio, J. W. Ludlow, D. M. Livingston, and J. Massague. 1990. Growth inhibition by TGF-beta linked to suppression of retinoblastoma protein phosphorylation. Cell 62:175-185. [DOI] [PubMed] [Google Scholar]
- 29.Lee, D. K., B. C. Kim, I. Y. Kim, E. A. Cho, D. J. Satterwhite, and S. J. Kim. 2002. The human papilloma virus E7 oncoprotein inhibits transforming growth factor-beta signaling by blocking binding of the Smad complex to its target sequence. J. Biol. Chem. 277:38557-38564. [DOI] [PubMed] [Google Scholar]
- 30.Lundberg, A. S., and R. A. Weinberg. 1998. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol. Cell. Biol. 18:753-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mailand, N., and J. F. Diffley. 2005. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell 122:915-926. [DOI] [PubMed] [Google Scholar]
- 32.Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukherjee, P., T. V. Cao, S. L. Winter, and M. G. Alexandrow. 2009. Mammalian MCM loading in late-G(1) coincides with Rb hyperphosphorylation and the transition to post-transcriptional control of progression into S-phase. PLoS One 4:e5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakayama, K., N. Ishida, M. Shirane, A. Inomata, T. Inoue, N. Shishido, I. Horii, D. Y. Loh, and K.-I. Nakayama. 1996. Mice lacking p27Kip1 display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell 85:707-720. [DOI] [PubMed] [Google Scholar]
- 35.Nishihara, A., J. Hanai, T. Imamura, K. Miyazono, and M. Kawabata. 1999. E1A inhibits transforming growth factor-beta signaling through binding to Smad proteins. J. Biol. Chem. 274:28716-28723. [DOI] [PubMed] [Google Scholar]
- 36.Pacek, M., and J. C. Walter. 2004. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 23:3667-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pietenpol, J. A., J. T. Holt, R. W. Stein, and H. L. Moses. 1990. Transforming growth factor β-1 suppression of c-myc gene transcription: role in inhibition of keratinocyte proliferation. Proc. Natl. Acad. Sci. U. S. A. 87:3758-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pietenpol, J. A., R. W. Stein, E. Moran, P. Yaciuk, R. Schlegel, R. M. Lyons, M. R. Pittelkow, K. Munger, P. M. Howley, and H. L. Moses. 1990. TGF-β1 inhibition of c-myc transcription and growth in keratinocytes is abrogated by viral transforming proteins with pRB binding domains. Cell 61:777-785. [DOI] [PubMed] [Google Scholar]
- 39.Polyak, K., J. Kato, M. J. Solomon, C. J. Sherr, J. Massague, J. M. Roberts, and A. Koff. 1994. p27Kip1, A cyclin-Cdk inhibitor, links transforming growth factor-β and contact inhibition to cell cycle arrest. Genes Dev. 8:9-22. [DOI] [PubMed] [Google Scholar]
- 40.Reed, S. E., E. M. Staley, J. P. Mayginnes, D. J. Pintel, and G. E. Tullis. 2006. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J. Virol. Methods 138:85-98. [DOI] [PubMed] [Google Scholar]
- 41.Ren, B., G. Yu, G. C. Tseng, K. Cieply, T. Gavel, J. Nelson, G. Michalopoulos, Y. P. Yu, and J. H. Luo. 2006. MCM7 amplification and overexpression are associated with prostate cancer progression. Oncogene 25:1090-1098. [DOI] [PubMed] [Google Scholar]
- 42.Royzman, I., R. J. Austin, G. Bosco, S. P. Bell, and T. L. Orr-Weaver. 1999. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev. 13:827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sterner, J. M., S. Dew-Knight, C. Musahl, S. Kornbluth, and J. M. Horowitz. 1998. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol. Cell. Biol. 18:2748-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walter, J., and J. Newport. 2000. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell 5:617-627. [DOI] [PubMed] [Google Scholar]
- 45.Warner, B. J., S. W. Blain, J. Seoane, and J. Massague. 1999. Myc downregulation by transforming growth factor beta required for activation of the p15Ink4b G1 arrest pathway. Mol. Cell. Biol. 19:5913-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfraim, L. A., T. M. Walz, Z. James, T. Fernandez, and J. J. Letterio. 2004. p21Cip1 and p27Kip1 act in synergy to alter the sensitivity of naive T cells to TGF-beta-mediated G1 arrest through modulation of IL-2 responsiveness. J. Immunol. 173:3093-3102. [DOI] [PubMed] [Google Scholar]
- 47.Zegerman, P., and J. F. Diffley. 2007. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445:281-285. [DOI] [PubMed] [Google Scholar]