Abstract
Estrogen receptor alpha (ER) and the insulin-like growth factor I receptor (IGF-IR) pathways are engaged in a functional cross talk in breast cancer, promoting tumor progression and increased resistance to anticancer treatments and radiotherapy. Here, we introduce new mechanisms through which proteins of the IGF-I/IGF-IR signaling pathway may regulate ER function in the absence of ligand. Our results indicate that in ER-positive breast cancer cells, Akt2 modulates ER transcriptional activity at multiple levels, including (i) the regulation of ER expression and its nuclear retention and (ii) the activation of one of its downstream targets, the Forkhead transcription factor FoxO3a. FoxO3a colocalizes and coprecipitates with ER in the nucleus, where it binds to Forkhead-responsive sequences on the ER target pS2/TFF-1 promoter; in addition, FoxO3a silencing leads to an increase of ER transcriptional activity, suggesting a repressive role of the Forkhead transcription factor in ER function. Moreover, 17β-estradiol upregulates FoxO3a levels, which could represent the basis for an ER-mediated homeostatic mechanism. These findings provide further evidence of the importance of mediators of the growth factor signaling in ER regulation, introducing the Akt2/FoxO3a axis as a pursuable target in therapy for ER-positive breast cancer.
Ovarian steroids are essential for the development, proliferation, and differentiation of normal human breast tissue (2). Cell response to 17β-estradiol (E2) is mostly mediated through estrogen receptor alpha (ER) (14), although E2 can elicit physiological events that are independent of ER (57, 60). ER is expressed at low levels in normal human mammary epithelial cells and is absent in stromal cells. However, during breast cancer development, the number of cells expressing ER and the abundance of this receptor tend to increase (48). The causative role of ER in the development of breast cancer has been substantiated by numerous in vivo and in vitro studies that documented the ability of estrogens to stimulate proliferation and differentiation in normal and cancerous mammary epithelium (24, 42). The analysis of clinical samples indicated that more than 60% of breast tumors express ER (13, 25). ER expression (i) has been defined as a marker for breast cancer diagnosis and prognosis (50), (ii) is correlated with a higher degree of tumor differentiation (35, 38), (iii) increases disease-free survival (41), and (iv) is a target for antiestrogen therapy and prevention.
In breast cancer, the expression and/or activity of specific growth factor receptors, such as the insulin-like growth factor receptor or epidermal growth factor receptor family members, including EGFR and Her-2/neu (6, 29, 44), is inversely related to ER expression and activity (16, 27, 59) and confers E2-independent growth properties (23) and antiestrogen resistance (21, 26).
Growth factors have been shown to enhance the transcriptional activity of ER in a ligand-independent manner through activation of mitogen-activated protein kinase (MAPK) or the phosphatidylinositol 3-kinase (PI3-K)/Akt pathway (23, 53, 55). In human breast cancers, PI3-K/Akt signaling is frequently deregulated either by loss of the suppressor protein PTEN or by the expression of active isoforms of PI3-K or downstream elements, such as Akt and mTOR (7).
Akt is known to play an important role in controlling cell proliferation, survival, and inhibition of apoptosis (22). Akt is a serine/threonine kinase belonging to the AGC superfamily. The Akt family is composed of three closely related isoforms, Akt1, Akt2 and Akt3, which are expressed at the mRNA level by virtually all normal human tissues (21, 64).
Since tumorigenesis has been reported not to involve a dramatic change in the RNA expression patterns of the three AKT isoforms, it has been proposed that differences in the Akt1, -2, and-3 kinase activities may be more important in clinical disease (64). For instance, elevated Akt1 kinase activity has been detected in primary tumors of the breast, prostate, and ovary (52, 56); sustained Akt2 kinase activity has been reported in breast and ovarian carcinomas (52, 55, 61); while the expression levels of Akt3 have been shown to be upregulated in ER-negative breast cancer tumors (40).
Recently, the Forkhead box class O (FoxO) family members transcription factors FoxO1a, FoxO3a, FoxO4 (formerly FKHR, FKHRL1, and AFX, respectively), and the more recent FoxO6 (20) have been identified as targets of the PI-3K/Akt pathway (8). Some reports showed evidence that the overexpression of any of these Forkhead transcription factors induced either cell cycle arrest or apoptosis (1, 8, 39, 45). Activation of PI-3K controls cell cycle entry by inactivating FoxO factors, which have been shown to regulate expression of p27/Kip1 (36), cyclin D1, and cyclin E (46). FoxO transcription factors have been shown to be functional in mammary cells and to be regulated by Akt (19); in fact, Akt-phosphorylated FoxO binds to 14-3-3 proteins, and the complex is translocated from the nucleus to the cytoplasm (8). When hypophosphorylated, Forkhead proteins are released from 14-3-3 and translocate into the nucleus, where they transactivate specific proapoptotic target genes (36, 47, 54). Recently, several reports have suggested a functional interaction between ER and FoxO members. E2 has been noted to determine ER binding to FKHR, FKHRL1, and AFX (49, 62, 65) and to induce FKHR phosphorylation in breast cancer cells (34). Particularly, E2-dependent ER binding to FKHR seems to be involved in ER nucleocytoplasmic shuttling, since site-directed mutagenesis of the ER nuclear export sequence inhibits FKHR nuclear export, the estradiol-induced cytoplasmic relocalization of receptor, and DNA synthesis (33). In transient-transfection experiments, FoxO members seemed also to regulate ER-mediated transcription, showing either coactivator or corepressor functions on estrogen-responsive element (ERE) sites, depending on the cellular model (49, 62, 65). Additionally, FoxO3a has recently been reported to suppress cell growth and tumorigenesis in breast cancer cells and in an orthotopic mouse model of breast cancer (65).
The aim of this study was to better elucidate the molecular mechanisms underlying the regulation exerted by the Akt/FoxO axis on ER in human breast cancer cells.
MATERIALS AND METHODS
Cell culture, conditions, and treatments.
The ER-positive human breast cancer epithelial cell line MCF-7 was maintained in monolayer culture in Dulbecco's modified Eagle's/Ham's F-12 medium (1:1) (DMEM/F-12), supplemented with 5% fetal bovine serum (FBS) and 1% Eagle's nonessential amino acids. Human cervical cancer (HeLa) cells (ATCC, United Kingdom) were grown in modified Eagle's medium (MEM) containing 10% calf serum, and human breast cancer (SKBR3) cells (ATCC, United Kingdom) were cultured in RPMI medium plus 10% FBS. Additionally, culture media were supplemented with 100 IU/ml penicillin, 100 μg/ml streptomycin, and 0.2 mM l-glutamine (all media and reagents were purchased from Sigma-Aldrich, United Kingdom).
For experimental purposes, cells were synchronized in phenol red-free and serum-free DMEM/F-12 (PRF-SFM) (Sigma-Aldrich, United Kingdom) for 24 h in the absence of steroids and growth factors. Following starvation, the cells were pretreated or not with 1 μM API-2 (triciribine; Calbiochem), an inhibitor of Akt activity, for 1 h and then treated with 100 nM E2 (Sigma) for 5 min (for Western blotting [WB]) or 45 min (for chromatin immunoprecipitation [ChIP] and DNA affinity precipitation assays) and 16 h (gene reporter assay and reverse transcription [RT]-PCR).
Plasmids, transfections, and ERE-luciferase assay.
The following plasmids were used: the reporter plasmid XETL, driving the expression of firefly luciferase by an ERE from the Xenopus vitellogenin promoter (9); pS2/TFF-1-ERE (pS2-ERE) containing a 1,050-bp fragment of the pS2 promoter preceding the luciferase reporter of pGL3 (5); pSG5-HeG0, a simian virus 40 (SV40) promoter-based pSG5 vector encoding wild-type ER (HeG0) (56a); the Renilla reniformis luciferase expression vector pRL-Tk, used to assess transfection efficiency (Promega); and pECE-HA-FoxO3a (HA-FoxO3a) WT, encoding wild-type FoxO3a (Addgene plasmid 1787).
To monitor the effect of API-2 on ER transactivation, MCF-7, HeLa, and SKBR3 cells (density, 5 × 104) were plated on 24-well plates, grown in culture medium to an approximate confluence of 70 to 80%, and then switched to PRF-SFM and cotransfected with XETL and pRL-Tk (MCF-7) or XETL, pRL-Tk, and HeG0 (HeLa and SKBR3). All the transfections were carried out using Fugene 6 (Roche) (DNA/Fugene ratio, 3:1). After 6 h, the medium was replaced with fresh PRF-SFM, and the cells were pretreated for 1 h with 1 μM API-2 (triciribine; Calbiochem) and then treated for 16 h with 100 nM E2 (Sigma-Aldrich, United Kingdom).
To evaluate the effect of FoxO3a on ER transactivation, FoxO3a was either silenced for 24 h with FoxO3a small interfering RNA (siRNA) (Invitrogen) or overexpressed using hemagglutinin (HA)-FoxO3a WT and then cotransfected with pRL-Tk and XETL or pRL-Tk and pS2-ERE and exposed to 100 nM E2 for 16 h. Luciferase activity was measured using the dual-luciferase assay system (Promega), normalized to pRL-Tk activity, and expressed as fold induction over the control. Cell extracts were also processed by Western blot analysis and/or RT-PCR. Similar experiments were conducted to evaluate the effect of FoxO3a on ER gene transcription, using a pGL3 plasmid bearing the full ER promoter (fragment E, containing both promoters A and B), pGL3-ERprom(E), mapping from −4,100 to +212 bp from the first transcription start site (15).
Real-time reverse transcription-PCR.
Before the experiments, cells were serum starved for 24 h and then treated for the indicated times. Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions and treated with DNase I (Ambion). Two μg of total RNA was reverse transcribed with the ImProm-II reverse transcription system kit (Promega) using random primers; cDNA was diluted 1:3 in nuclease-free water, and 5 μl was analyzed in triplicate by real-time PCR in an iCycler iQ Detection System (Bio-Rad) using SYBR green Universal PCR Master Mix (Bio-Rad) with 0.1 μmol/liter of each primer in a total volume of 30 μl of reaction mixture. The primers used for the amplification were based on published sequences for human Akt 2, ER, FoxO3a, cyclin D1 (CD1), and pS2 (data available on request). The PCR conditions were 95°C for 3 min and 40 cycles of 95°C for 30 s, Ta (data available on request) for 30 s, and 72°C for 30 s; negative controls contained water instead of first-strand cDNA. Each sample was normalized on its 18S rRNA content. The 18S quantification was done using a TaqMan rRNA reagent kit (Applied Biosystems) following the manufacturer's instructions. The relative gene expression levels were normalized to a calibrator that was chosen to be the basal, untreated sample. The final results were expressed as n-fold differences in gene expression relative to 18S rRNA and the calibrator, calculated using the ΔΔCT method as follows: n-fold = 2−(ΔCTsample − ΔCTcalibrator), where the ΔCT values of the sample and calibrator were determined by subtracting the average CT value of the 18S rRNA reference gene from the average CT value of the different genes analyzed.
Chromatin immunoprecipitation.
MCF-7 cells were grown in 100-mm plates. Subconfluent cultures (70%) were shifted to PRF-SFM for 24 h, pretreated with API-2 for 1 h, and then treated with 100 nM E2 or left untreated for 45 min. Alternatively, growing cells were switched to PRF-SFM, transfected with HA-FoxO3a using Fugene 6 (Fugene 6/plasmid ratio, 3:1), and treated the following day with E2 for 45 min. ChIP methodology was performed as described previously (37). The precleared chromatin was precipitated for 16 h with anti-ER monoclonal antibody (MAb) (Santa Cruz) for ER, anti-FoxO3a polyclonal antibody (PAb) (Cell Signaling) for FoxO3a, and anti-polymerase II PAb (Santa Cruz) for Pol II. Normal rabbit IgG and normal mouse IgG (Santa Cruz) were used instead of primary Abs as negative controls. Immunoprecipitated DNA was analyzed in triplicate by real-time PCR using 5 μl of the diluted (1:3) template DNA as described above, and the the pS2 promoter region (pS2-prom) was amplified using specific primers (data available on request).
Real-time PCR data were normalized with respect to unprocessed lysates (input DNA). Input DNA quantification was performed by using 5 μl of the diluted (1/50) template DNA. The relative antibody-bound fractions were normalized to a calibrator that was chosen to be the basal, untreated sample. The final results were expressed as fold differences with respect to the relative inputs.
Immunoprecipitation and Western blotting.
Protein expression and complex formation were assessed by Western blotting (WB) or immunoprecipitation (IP), followed by WB using total protein lysates, cytoplasmic protein lysates, or fractionated proteins, where appropriate. MCF-7 cells were serum starved for 24 h and treated with 100 nM E2 and/or the Akt inhibitor API-2 (1 μM) for different times, depending on the experiment. Cytoplasmic proteins were obtained using lysis buffer containing 50 mmol/liter HEPES (pH 7.5), 150 mmol/liter NaCl, 1% Triton X-100, 1.5 mmol/liter MgCl2, 10 mmol/liter EGTA (pH 7.5), 10% glycerol, and inhibitors (0.1 mmol/liter Na3VO4, 1% phenylmethylsulfonyl fluoride, and 20 mg/ml aprotinin). After the collection of cytoplasmic proteins, the nuclei were lysed with nuclear buffer containing 20 mmol/liter HEPES (pH 8), 0.1 mmol/liter EDTA, 5 mmol/liter MgCl2, 0.5 mol/liter NaCl, 20% glycerol, 1% NP-40, and inhibitors (as described above). For total protein extracts, RIPA buffer was used (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.25% Na deoxycholate, plus inhibitors). The protein content was determined using Bradford dye reagent (Bio-Rad). For WB, 50 μg of lysates was separated on an 11% polyacrylamide denaturing gel (SDS-PAGE) and transferred to nitrocellulose membranes. Proteins of interest were detected with specific Abs, recognized by peroxidase-coupled secondary Abs, and developed using the ECL Plus Western Blotting detection system (Amersham Pharmacia Biotech, United Kingdom). For IP, 500 μg of protein lysates was precleared for 1 h with protein A/G-agarose (Santa Cruz) for either MAb or PAbs, incubated with primary Abs at 4°C for 18 h in HNTG buffer (20 mmol/liter HEPES, pH 7.5, 150 mmol/liter NaCl, 0.1% Triton X-100, 10% glycerol, and 0.1 mmol/liter Na3VO4), and then the antigen-Ab complexes were precipitated with protein A/G agarose for 2 h in HNTG buffer. In control samples, the primary immunoprecipitating Abs were replaced with normal rabbit IgG (Santa Cruz Biotechnology). The immunoprecipitated proteins were washed three times with HNTG buffer, separated on SDS-PAGE, and processed by WB. The images were acquired by using an Epson Perfection scanner (Epson, Japan) using Photoshop software (Adobe). The optical densities of the spots were analyzed by using ImageJ software (NIH; http://rsb.info.nih.gov/IJ).
Antibodies for Western blotting and immunoprecipitation.
Total and phosphorylated Akt isoforms were detected by WB with specific Abs: anti-Akt1/2 (H-136) PAb, anti-Akt1 (G5) MAb, anti p-Akt1/2/3 (Ser473)-R (Santa Cruz), and anti-Akt2 (5B5) MAb (Cell Signaling). ER and FoxO3a were assessed by WB and IP with anti-ER F-10 MAb (Santa Cruz) and anti-FoxO3a PAb (Cell Signaling). pS2 was probed with anti-pS2 PAb (Santa Cruz), phosphorylated GSK-3β with anti-GSK-3β (Ser9) PAb (Cell Signaling), total GSK-3β with anti-GSK-3β MAb (Cell Signaling), CD1 with anti-cyclin D1 PAb (Santa Cruz), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and lamin B were assessed by WB as controls for loading and purity of lysates with the anti-GAPDH (FL-335) PAb and the anti-lamin B (C-20) PAb (Santa Cruz), respectively. Normal rabbit IgG and normal mouse IgG (Santa Cruz) were used instead of primary Abs for IP negative controls. All Abs were used at concentrations recommended by the manufacturers.
siRNA-mediated RNA interference.
Custom-synthesized siRNA (Invitrogen)-annealed duplexes (25-bp double-stranded RNA [dsRNA]) were used for effective depletion of Akt1, Akt2, and FoxO3a genes. A scrambled siRNA (Invitrogen) that lacked identity with known gene targets was used as a control for non-sequence-specific effects. Cells were trypsinized; transfected in suspension with Lipofectamine 2000 (Invitrogen), as suggested by the manufacturer; and then plated in six-well plates (3.5 × 105 cells per well). Briefly, the cells were transfected with 100 pmol siRNAs (siAkt1, siAkt2, siFoxO3a, or nonspecific siRNA) in 3 ml total growing medium. The siRNAs were diluted in 500 μl of PRF-SFM without antibiotic; after 5 min, Lipofectamine 2000 was added to the mixture and incubated at room temperature for 20 min. The siRNA-Lipofectamine 2000 complex was then added to the cells and incubated at 37°C for 6 h, switched to PRF-SFM, and further incubated for an additional 18 h. Then, the cells were treated with E2 (100 nM) for 6 h or 24 h before analysis, depending on the experiment. For time course purposes, cells were harvested after 24 h, 48 h, or 72 h.
Confocal laser scanning microscopy (CLSM).
MCF-7 cells were transfected in suspension with Akt2 siRNA as described previously, plated on coverslips, serum starved, and then treated with 100 nM E2 for 6 h. After incubation, the cells were fixed with 3% paraformaldehyde and permeabilized with 0.2% Triton X-100, and nonspecific sites were blocked with bovine serum albumin (BSA) (3% for 30 min). The blocked samples were incubated for 1 h with a mixture of primary antibodies (2 mg/ml each) recognizing ER (MAb; Santa Cruz) and FoxO3a (PAb; Cell Signaling), washed with phosphate-buffered saline (PBS) (Gibco), and incubated with a mixture of fluorescein-conjugated goat anti-rabbit IgG and rhodamine-conjugated goat anti-mouse IgG (Santa Cruz) secondary Abs. The cellular localization of the two proteins was examined under a Leica TCS SP2 confocal laser scanning microscope at ×400 magnification. The optical sections were taken at the central plane. The fluorophores were imaged separately to ensure there was no excitation/emission wavelength overlap. The optical densities of stained proteins were analyzed by ImageJ software.
DAPA.
The binding of nuclear FoxO3a to Forkhead-responsive elements on the pS2 promoter was assessed in vitro using a modified version of the DNA affinity precipitation assay (DAPA) protocol of Zhu et al. (63). Briefly, nuclear-protein extracts were obtained from starved cells pretreated with API-2 (1 μM) for 1 h and stimulated with E2 (100 nM) for 45 min. One hundred μg of nuclear proteins was mixed with 2 μg of specific biotinylated DNA probes (see below) in 400 μl of buffer D (20 mM HEPES, pH 7.9, 10% glycerol, 50 mM KCl, 0.2 mM EDTA, 1.5 mM MgCl2, 10 μM ZnCl2, 1 mM dithiothreitol, and 0.25% Triton X-100) and then incubated on ice for 45 min. After that, 20 μl of streptavidin-agarose beads (Promega) was added, and the samples were incubated under rotation for 2 h at 4°C. Next, the agarose bead-protein complexes were collected by brief centrifugation and washed twice in buffer D. Proteins were uncoupled from DNA probes by the addition of 40 μl of 2× Laemmli's sample buffer and by heating them at 96°C for 10 min. The beads were removed by centrifugation, and the supernatants were analyzed by WB for the presence of FoxO3a. The DNA motif probes (pS2/FKH) were prepared by annealing a 5′-biotinylated sense oligonucleotide bearing a Forkhead consensus sequence (5′-Bio-ACGCTCTTTAAGCAAACAGAGCCTGCCCTA-3′) with a nonbiotinylated antisense oligonucleotide (5′-TAGGGCAGGCTCTGTTTGCTTAAAGAGCGT-3′). A labeled probe with the consensus sequence [pS2/FKH(−)], underlined above, deleted was used as a negative control (forward, 5′-Bio-ACGCTCTTTAAACAGAGCCTGCCCTA-3′; reverse, 5′-TAGGGCAGGCTCTGTTTAAAGAGCGT-3′). The optical densities of the spots were analyzed by ImageJ software.
Proliferation assay.
MCF-7 cells were transfected in suspension with either HA-FoxO3a or siFoxO3a (the empty vector or scrambled siRNA, respectively, was used as a control) in growth medium without antibiotic and plated in 12-well plates at a concentration of 105 cells/plate. After 6 h, the cells were starved for 18 h (day zero) and then treated or not with E2 (100 nM) for 1, 2, and 3 days (the hormone was refreshed every day to maintain constant levels in the medium). At each time point, the cells were harvested by trypsinization and counted in a hemocytometer using the trypan blue exclusion assay.
Statistical analysis.
All data were expressed as the means ± standard deviations (SD) of at least three independent experiments. Statistical significances were tested using Student's t test.
RESULTS
Akt inhibition decreases ER transcriptional activity.
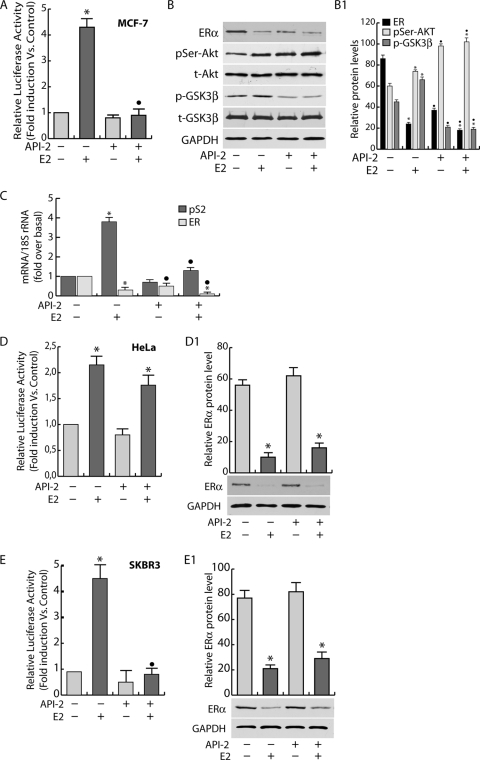
It has been well established that Akt is involved in the control of cell survival and that its inhibition is responsible for growth retardation; in MCF-7 cells, this effect is paralleled by a decrease of ER transcriptional activity (32, 53). This assumption was confirmed through transactivation experiments using an ER-responsive 1× ERE-Luc construct (XETL). In our experimental system, E2 treatment, as expected, determined a 4-fold induction of luciferase activity (Fig. 1A). The inhibition of Akt kinase function by API-2 pretreatment completely abrogated ER transcriptional activity in response to E2 administration (Fig. 1A). Similar results were obtained in two additional ER-positive cell lines, T47D and ZR75 (data not shown). Hormone treatment caused an increase of Akt phosphorylation in either untreated or API-2-treated samples (Fig. 1B). Interestingly, API-2 treatment resulted in pSer-Akt accumulation, while Akt kinase activity was dramatically reduced, since its downstream target, GSK-3β, was no longer phosphorylated (Fig. 1B). Notably, the inhibition of ER transactivation is consistent with a reduction of the ER-mediated transcription, since, under the same experimental conditions, a dramatic decrease in mRNA levels of the ER target gene pS2 was observed both under basal conditions and after E2 treatment (Fig. 1C). This event was paralleled by a significant decrease in ER protein and mRNA levels (Fig. 1B and C). Interestingly, the inhibition of Akt kinase function did not alter either Akt or GSK-3β protein expression (Fig. 1B), while it did induce a 40% reduction of ER protein and mRNA levels in untreated samples, as well as further emphasizing the ligand-induced downregulation of the receptor (Fig. 1B and C).
FIG. 1.
Akt inhibition decreases ER transcriptional activity. (A) MCF-7 cells were transfected with a mixture of XETL (1 μg/well) and pRL-Tk (50 ng/well) plasmids for 6 h. The medium was then replaced with fresh PRF-SFM, and the cells were pretreated with 1 μM API-2 for 1 h or left untreated and then stimulated with 100 nM E2 for an additional 16 h. Firefly luciferase was detected and expressed as relative luciferase activity with respect to the untreated samples (fold induction versus the control). (B) The whole-cell lysates obtained were then collected and analyzed by WB using specific Abs. (C) MCF-7 cells were treated as in panel A; total RNA was extracted, and the abundance of pS2 and ER mRNAs was detected in real time, as described in Materials and Methods. Each sample was normalized to its 18S rRNA content. (D and E) HeLa and SKBR3 cells were transfected with a mixture of XETL (0.5 μg/well), HeG0 (0.5 μg/well), and pRL-Tk (50 ng/well) plasmids for 6 h and then treated as in panel A. (D1 and E1) The ER content in total lysates. (B1, D1, and E1) Densitometric analysis of protein levels reported as means ± SD of samples normalized over GAPDH. In all experiments, significance values were as follows: *, P < 0.05 versus non-E2-treated samples; •, P < 0.05 versus the corresponding non-API-2-treated samples.
To assess cell specificity, a gene reporter assay was conducted in two ER-negative cell lines, HeLa and SKBR3, ectopically expressing ER. In both cell systems, ER presence was responsible for E2-dependent transactivation (Fig. 1D and E). Interestingly, in HeLa cells, no significant difference in luciferase induction was observed in API-2-treated samples compared to nontreated samples, while in SKBR3 cells, the response to the same treatment paralleled that observed in MCF-7 cells (Fig. 1D and E). In both cell lines, API-2 did not affect the ER protein content (Fig. 1, D1 and D2).
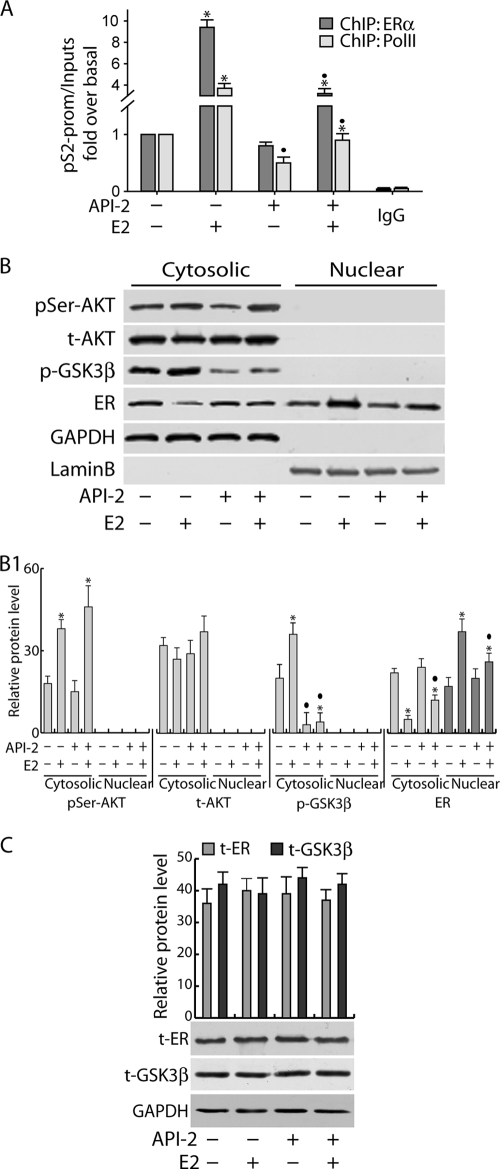
To clarify the role of Akt in ER transcriptional activity, chromatin-bound ER and Pol II were precipitated from MCF-7 cell nuclear extracts. As shown in Fig. 2A, strong inhibition of ER and Pol II recruitment on the estrogen-responsive sequence of the pS2 promoter was observed in cells pretreated with API-2 for 1 h and exposed to E2 for 45 min. The reduced occupancy of ER on the pS2 promoter was related to a decrease in ER nuclear content. Indeed, 1 h of exposure to API-2 was able to interfere with ligand-dependent retention of ER in the nucleus (Fig. 2B) without affecting total ER and GSK-3β protein amounts (Fig. 2C).
FIG. 2.
Short-term Akt inhibition affects ER retention in the nucleus. (A) MCF-7 cells synchronized in PRF-SFM for 24 h were pretreated with API-2 for 1 h and then treated with 100 nM E2 or left untreated for 45 min. Then, samples were processed for ChIP analysis as described in Materials and Methods. The ERE-containing pS2 promoter region was amplified using a specific pair of primers (data available on request). A mixture of normal rabbit IgG and normal mouse IgG was used as a negative control for both primary Abs to precipitate the E2-treated samples. (B) MCF-7 cells were serum starved for 24 h, pretreated for 1 h with API-2, and then treated with 100 nM E2 for 5 min or left untreated. Cytoplasmic and nuclear protein extracts were collected as described in Materials and Methods and subjected to WB (50 μg/lane) using different Abs. (B1) The relative protein levels were analyzed, and the optical density is reported. (C) MCF-7 cells were treated as in panel B, and total protein extracts were collected with RIPA buffer (see Materials and Methods) and processed by WB. Samples were normalized over input (A), GAPDH (for cytosolic proteins [B] and total proteins [C]), and lamin B (for nuclear proteins [B]). In all experiments, significance values were as follows: *, P < 0.05 versus non-E2-treated samples; •, P < 0.05 versus the corresponding non-API-2-treated samples. The error bars indicate SD.
In our system, E2 induced phosphorylation of Akt and, as a consequence, GSK-3β phosphorylation after 5 min of treatment (Fig. 2B). The efficacy of API-2 pretreatment was evidenced by the clear inhibition of GSK-3β phosphorylation while, as expected, at this time point, API-2 did not alter Akt expression or its phosphorylation on Ser473 in either the presence or absence of E2 (Fig. 2B).
Akt2 silencing reduces ER expression and function.
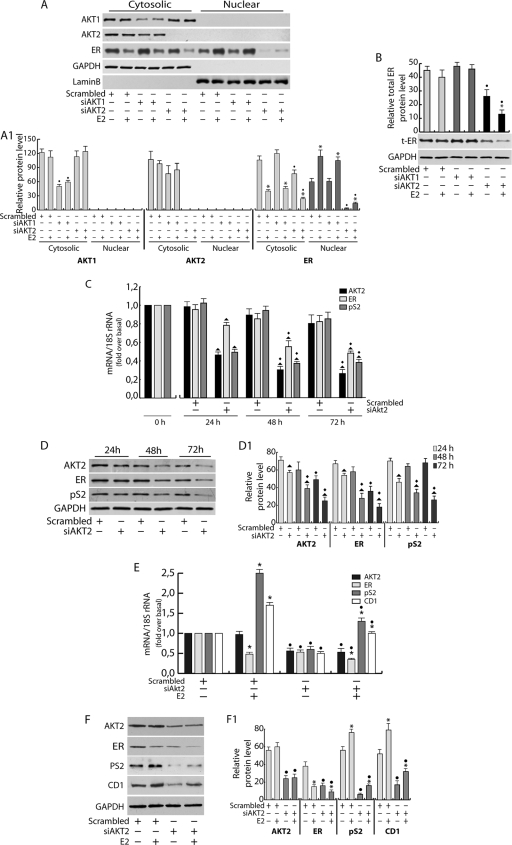
Since API-2 is not able to discriminate between the Akt isoforms, Akt1 and Akt2 mRNA transcripts (Akt3 is not expressed in MCF-7 cells [reference 21 and data not shown]) were silenced by siRNA. A strong reduction of ER total expression was observed in Akt2, but not in Akt1, silenced samples (Fig. 3A and B). Time course experiments (24 to 72 h) showed that a significant decrease in ER expression also occurs at the transcriptional level following Akt2 silencing (Fig. 3C and D). This event was reflected by a strong reduction in pS2 mRNA (Fig. 3C) and protein (Fig. 3D) expression at all investigated time points. The reduction of Akt2 and ER cytosolic content observed after 72 h in scrambled-siRNA samples (Fig. 3D) could simply be ascribed to a general phenomenon in response to the prolonged starvation. As mentioned above, siAkt1 did not alter either ER (Fig. 3A and B) or pS2 (data not shown) expression, supporting the hypothesis that ER functional disruption specifically depends on Akt2 inhibition. A comparable trend was maintained in the presence of E2 in Akt2 silenced samples. In fact, E2 treatment did not alter the inhibitory effect of silenced Akt2 on the mRNA and protein levels of ER and pS2, as well as an additional ER-regulated gene, CD1 (Fig. 3E and F). Similar experiments were conducted in T47D and ZR75 cells (data not shown). In both cell lines, Akt2 inhibition led to a dramatic reduction of ER, and consequently pS2, mRNA and protein levels.
FIG. 3.
Akt2 regulates ER expression. (A and B) MCF-7 cells were transfected in suspension with 100 pmol siRNAs/35-mm well (siAkt1, siAkt2, or scrambled siRNA for control samples), plated, starved for 48 h, and treated with 100 nM E2 for 6 h. Cytosolic and nuclear proteins (A) or total extracts (B) (50 μg/lane) were analyzed by WB using appropriate antibodies. The relative protein levels (A1 and B) were analyzed, and the optical density is reported. (C and D) AKT2 was silenced for 24, 48, and 72 h, as described previously. (C) Two μg RNA was reverse transcribed and subjected to real-time PCR analysis. Each sample was normalized to its 18S rRNA content and reported as fold increase over the control (0 h). (D) Fifty micrograms of cytosolic proteins was subjected to WB analysis. (D1) The relative protein levels were analyzed, and the optical density is shown. (E and F) MCF-7 cells were transfected, as described above, with siAkt2 or scrambled siRNA, starved for 24 h, and treated with 100 nM E2 for an additional 24 h. (E) Two μg RNA was reverse transcribed and subjected to real-time PCR. (F) Fifty micrograms of total proteins was processed by WB analysis, and the relative densitometric analysis is reported (F1). All data were normalized over GAPDH (cytosolic or total proteins) or lamin B (nuclear proteins). In all experiments, significance values were as follows: *, P < 0.05 versus non-E2-treated samples; •, P < 0.05 versus the corresponding treatment in scrambled siRNA; ▴, P < 0.05 versus scrambled siRNA; ·, P < 0.05 versus the corresponding sample at 24 h. The error bars indicate SD.
FoxO3a coprecipitates and colocalizes with ER in the nucleus.
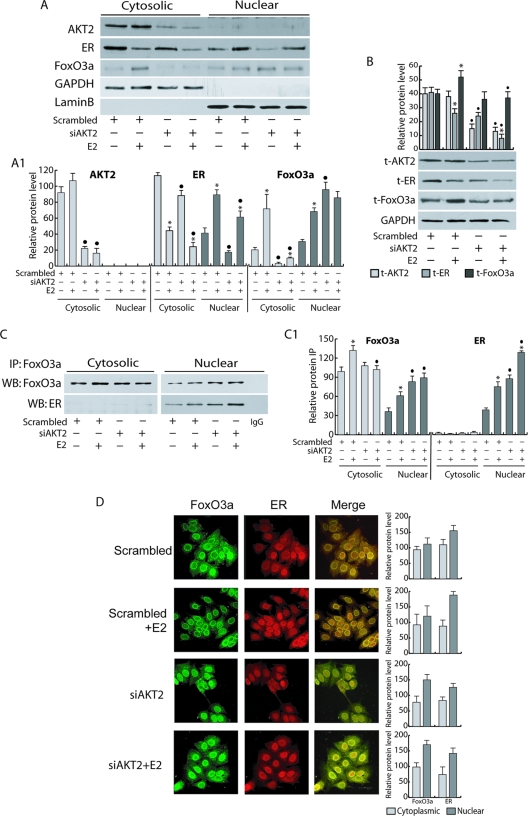
To elucidate the mechanism underlying the involvement of Akt2 on the regulation of both ER expression and function, we focused our attention on one of its downstream effectors, FoxO3a, a member of the Forkhead transcription factor family. FoxO3a is well expressed in MCF-7 cells and has been reported to functionally interact with ER (17, 62). As shown in Fig. 4A and B, an increase in total FoxO3a protein levels was observed in response to E2 stimulation. siAkt2 caused a drastic reduction of FoxO3a cytosolic content in MCF-7 cells in either the presence or absence of E2 (Fig. 4A). The latter effect was paralleled by a marked increase in FoxO3a nuclear content (Fig. 4A and D), even though it is worth noting that E2 treatment did not significantly affect siAkt2-induced FoxO3a nuclear translocation. Interestingly, Akt2 knockdown did not affect total expression of FoxO3a, while it counteracted E2-induced FoxO3a upregulation, since, as previously described, total ER expression was significantly reduced (Fig. 4B).
FIG. 4.
FoxO3a coprecipitates and colocalizes with ER in the nucleus. AKT2 silencing was carried out for 48 h, as described in Materials and Methods, followed by 100 nM E2 treatment for 6 h. Protein expression and complex formation were assessed by WB (A and B) and IP analysis (C), respectively, using fractionated protein lysates (A and C) or whole-cell lysates (B). (C) Five hundred micrograms of protein lysates was immunoprecipitated with anti-FoxO3a Ab and blotted for both FoxO3a and ER. Normal rabbit IgG was used as a negative control to precipitate E2-treated samples. (A1, B, and C1) The relative protein levels were analyzed, and the optical density is reported. Samples were normalized over GAPDH (A, cytosolic, or B, total proteins) and lamin B (A, nuclear proteins). In all experiments, significance values were as follows: *, P < 0.05 versus non-E2 treated; •, P < 0.05 versus the corresponding treatment in scrambled siRNA. (D) Cytoplasmic and nuclear distribution of ER and FoxO3a in response to AKT2 knockdown was also evaluated by confocal microscopy. To avoid fluorescence overlapping, a rabbit PAb for FoxO3a and a mouse MAb for ER were used. A mixture of fluorescein-conjugated goat anti-rabbit IgG (green) and rhodamine-conjugated goat anti-mouse IgG (red) secondary Abs was used to detect primary immune complexes of FoxO3a and ER, respectively. The merged images show FoxO3a-ER colocalization (yellow). The optical sections were taken at the central plane at ×400 magnification. The fluorophores were imaged separately to ensure there was no excitation/emission wavelength overlap. The histograms on the right show the corresponding means and SD of the densitometric analysis performed in three independent experiments.
Immunoprecipitation experiments and confocal microscopy confirmed the existence of a physical interaction (Fig. 4C) and colocalization (Fig. 4D) between ER and FoxO3a. In accordance with previously reported data in cell-free systems (49), the ER/FoxO3a interaction occurred mainly at the nuclear and perinuclear levels, where it was enhanced by E2, and became more evident in Akt2 silenced samples (Fig. 4C and D).
FoxO3a binds Forkhead-responsive elements on the pS2 promoter.
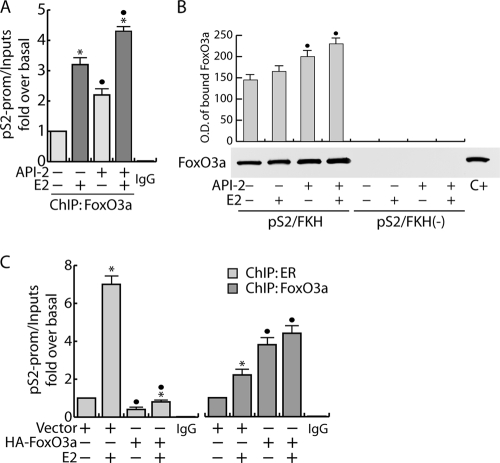
Some recently published reports evidenced an overrepresentation of TA-rich motifs, particularly Forkhead binding sites (AAG[A]TAAA[G]C[T]A), in several ER-bound regions (30-31), including the pS2 promoter (4, 11). To investigate if ER/FoxO3a interaction does exert a functional role in ER-mediated transcription, chromatin-bound FoxO3a was immunoprecipitated with a specific antibody. The presence of FoxO3a on the pS2 promoter was detected targeting the Forkhead DNA binding site close to the TATAA box (4). As shown in Fig. 5A, a constitutive association of FoxO3a with the pS2 promoter was observed, which was increased by E2 stimulation. As expected, the inhibition of Akt kinase activity by API-2 treatment increased the promoter occupancy by FoxO3a, consistent with the induced FoxO3a translocation into the nucleus (Fig. 4A). Additionally, to investigate if FoxO3a binding occurs on the Forkhead-responsive element of the pS2 promoter (pS2/FKH), a DAPA assay was conducted on nuclear extracts from MCF-7 cells. The results obtained showed that nuclear FoxO3a binds with high affinity to the pS2/FKH sequence but not to the same region partially deleted in the Forkhead-responsive element [pS2/FKH(−)] used as a negative control (Fig. 5B). These data support the hypothesis that this region is involved in pS2 regulation by the Forkhead transcription factor.
FIG. 5.
FoxO3a binds Forkhead-responsive elements on the pS2 promoter. (A) ChIP assay. Starved MCF-7 cells were pretreated with 1 μM API-2 for 1 h and then treated with 100 nM E2 or left untreated for 45 min. FoxO3a was immunoprecipitated from the precleared chromatin. Samples were analyzed through real-time PCR targeting a Forkhead-containing pS2 promoter region (see Materials and Methods). Normal rabbit IgG was used in place of the primary Ab as a negative control to precipitate E2-treated samples. Each sample was normalized to its input (*, P < 0.05 versus each non-E2-treated sample; •, P < 0.05 versus the corresponding treatment in non-API-2-treated samples). (B) DAPA assay. Cells were treated as for panel A, and FoxO3a was isolated from 100 μg of nuclear proteins using specific biotinylated DNA probes (pS2/FKH) as described in Materials and Methods and pulled down by streptavidin-agarose beads. The proteins recovered after removal of the beads were analyzed by WB for the presence of FoxO3a. Labeled probes with the consensus sequence [pS2/FKH(−)] deleted were used as negative controls. Fifty μg of cytosolic lysate (C+) was used as a positive control. The relative protein levels were analyzed, and the optical density (O.D.) is reported (•, P < 0.05 versus the corresponding treatment in non-API-2-treated samples). (C) ChIP analysis was performed on subconfluent MCF-7 cells transfected for 48 h with either HA-FoxO3a or empty vector (10 μg/100-mm petri dish). Twenty-four hours after transfection, the cells were synchronized in PRF-SFM for an additional 24 h and then treated with 100 nM E2 for 45 min or left untreated. The ERE- and FKH-containing pS2 promoter regions, precipitated with anti-ER and anti-FoxO3a Abs, respectively, were amplified using a specific pair of primers (data available on request). A mixture of normal rabbit IgG and normal mouse IgG was used as a negative control for both primary Abs to precipitate the E2-treated samples. Each sample was normalized to its input. In all experiments significance values were as follows: *, P < 0.05 versus each non-E2-treated sample; •, P < 0.05 versus the corresponding treatment in vector-transfected samples. The error bars indicate SD.
To evaluate the effects of FoxO3a recruitment to Forkhead-responsive sites on the binding of ER to ERE-containing promoters, we performed a ChIP assay transiently overexpressing FoxO3a. As shown in Fig. 5C, a significant decrease of both basal and E2-induced recruitment of ER on the pS2 promoter was observed in FoxO3a-overexpressing samples, corroborating our hypothesis of the negative role of FoxO3a in ER function.
FoxO3a inhibits ER expression and functional activity.
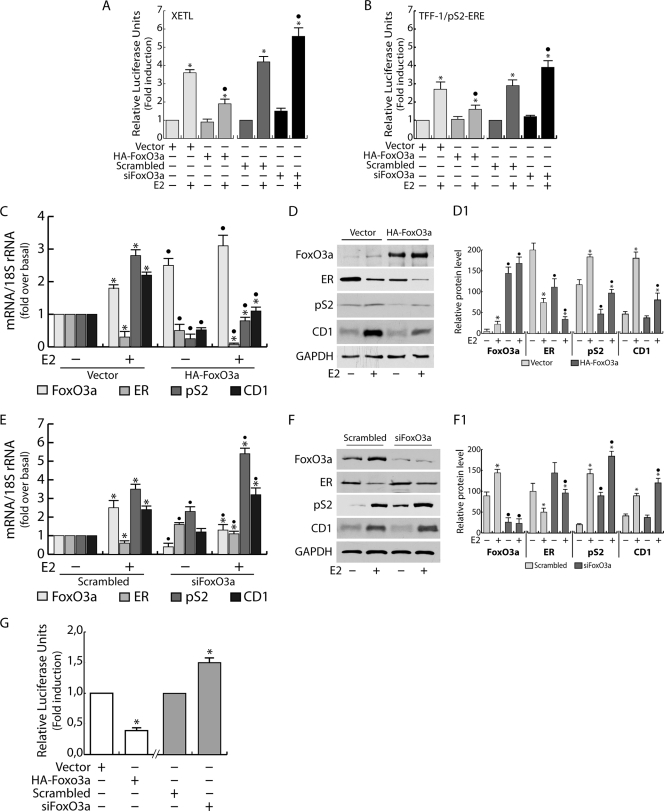
To further understand the role of FoxO3a in inhibiting ER transcription, we both overexpressed and silenced FoxO3a and performed gene reporter experiments using either XETL (Fig. 6A) or a full-length pS2 promoter construct (Fig. 6B). Our results showed that FoxO3a overexpression caused a significant decrease of ER-dependent transcription in response to E2 stimulation, while FoxO3a knockdown led to the opposite effect (Fig. 6A and B). Comparable results were obtained in T47D and ZR75 (data not shown) cell lines transfected with XETL.
FIG. 6.
FoxO3a modulates ER expression and functional activity. FoxO3a was either overexpressed or silenced in MCF-7 cells, cotransfected the day after with the XETL (A) or pS2 (B) promoter, and exposed to 100 nM E2 for 16 h (see Materials and Methods). Luciferase activity was normalized to pRL-Tk activity and expressed as relative luciferase units. (C) MCF-7 cells were transfected for 48 h in 35-mm dishes with HA-FoxO3a (1 μg) or empty vector (1 μg) and then treated for 24 h with E2 (100 nM). RNA was then extracted and reverse transcribed, and FoxO3a, ER, pS2, and CD1 transcripts were analyzed by real-time PCR. Each sample was normalized to its 18S rRNA content. (D) Fifty μg of whole-cell extracts from a duplicate set of cells treated as described above was subjected to WB analysis. (D1) The relative protein levels, normalized over GAPDH, are reported. (E) RNA extracted from FoxO3a-silenced MCF-7 cells, treated with E2 (100 nM) for 24 h, was reverse transcribed, and FoxO3a, ER, pS2, and CD1 transcripts were analyzed by real-time PCR. Each sample was normalized to its 18S rRNA content. (F) A duplicate set of cells, treated as described above, was lysed, and 50 μg of whole-cell lysates was loaded for protein expression. (F1) The relative protein levels, normalized versus GAPDH, were analyzed, and the optical density is reported. (G) MCF-7 cells were transfected with HA-FoxO3a or control vector (white bars) or FoxO3a siRNA or scrambled siRNA (gray bars) and cotransfected the day after with pGL3-ERprom(E). As described above, luciferase activity was normalized to pRL-Tk activity and expressed as relative luciferase units. In all experiments, significance values were as follows: *, P < 0.05 versus each non-E2-treated sample; •, P < 0.05 versus the corresponding treatment in scrambled siRNA or vector-transfected samples. The error bars indicate SD.
Moreover FoxO3a overexpression determined strong downregulation in ER mRNA and protein levels, both in controls and in E2-treated samples, which most likely is responsible for decreased pS2 and CD1 transcription (Fig. 6C and D). The same experiments were conducted in T47D and ZR75 (data not shown) cell lines, leading to similar results. On the other hand, a slight upregulation in ER mRNA and protein levels was observed in FoxO3a silenced samples (Fig. 6E and F), paralleled by an increase in pS2 and CD1 transcription and expression.
Preliminary data showing an ∼2.5-fold reduction in pGL3-ERprom(E) luciferase activity in FoxO3a-overexpressiong cells, as well as an ∼1.5-fold increase in FoxO3a silenced samples (Fig. 6G), confirmed these observations, leading us to hypothesize that the inhibition of ER expression exerted by FoxO3a could occur at the genomic level.
Interestingly, FoxO3a mRNA and protein were upregulated by E2 (Fig. 6C to F and 4A and B), suggesting a potential involvement of FoxO3a in a negative feedback loop aimed at downregulating ER and therefore attenuating ER function. This assumption was confirmed by proliferation assays, by either overexpressing or knocking down FoxO3a in MCF-7 cells (Fig. 7A and B). The results evidenced a dramatic inhibition of both basal and E2-stimulated cell growth in MCF-7 cells overexpressing FoxO3a (Fig. 7A), while both basal and E2-induced proliferation increased in FoxO3a-silenced cells (Fig. 7B).
FIG. 7.
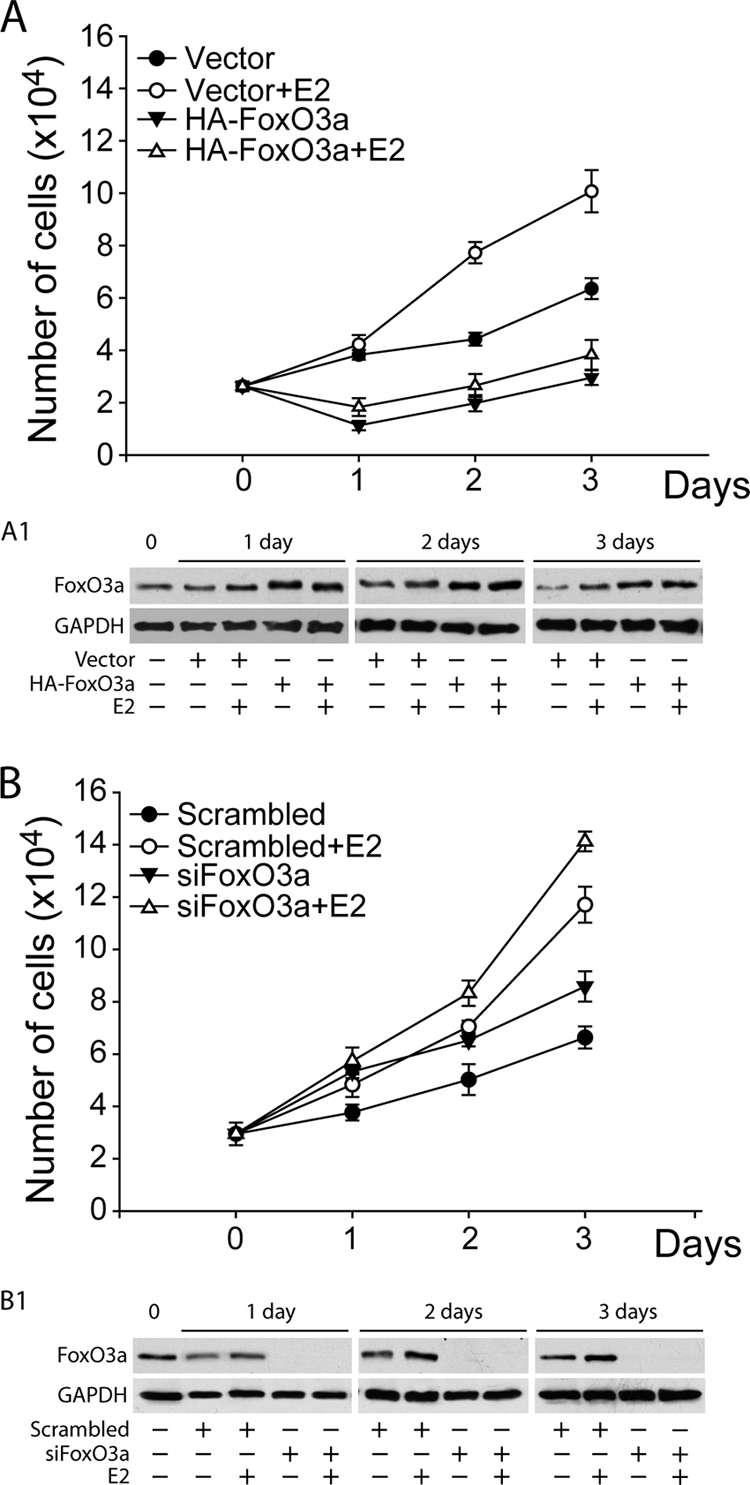
FoxO3a inhibits cell proliferation. MCF-7 cells were transfected with either HA-FoxO3a (A) or siFoxO3a (B) and treated as described in Materials and Methods. The cells were counted in a hemocytometer at 50% confluence (day 0) and 1, 2, and 3 days later. The results represent the means ± SD of three independent experiments. (A1 and B1) The FoxO3a content was analyzed by WB at each time point.
DISCUSSION
In the last 2 decades a great amount of data confirmed the existence of a functional cross talk between estrogens and growth factors and its essential role in breast cancer development and progression (51). Specifically, growth factors are known to influence the expression and activity of ER, as well as its transcriptional cofactors; conversely, ER regulates the expression of growth factor receptors and their ligands and signaling intermediates (28). For instance, several authors reported that MAPK/ERK and Akt can be activated following estrogen stimulation, independently of growth factors (12, 18, 53). The results presented here partially confirm these findings, since in MCF-7 breast cancer cells, E2 treatment increased Akt phosphorylation within 5 min, while at that time point it had no effect on MAPK/ERK activation (data not shown). Moreover, inhibition of the PI3-K/Akt pathway, either following prolonged exposure to a specific inhibitor of Akt kinase activity, API-2, or knocking down Akt2, led to a decrease in ER mRNA and protein expression, both in the presence and in the absence of ligand. Such a decrease was paralleled by a dramatic reduction of ER-regulated transcription. Our observations are in contrast to previously reported results showing ER upregulation in response to the expression of an Akt dominant negative, despite a reduction in ER-mediated transcription (53).
In particular, we observed a dual effect on ER regulation, depending on the persistence of Akt inhibition. While long-term treatment (∼16 h) with API-2 led to a decrease in both ER mRNA and protein levels, as well as in ER-mediated transcription, 1 h of treatment was not sufficient to alter ER levels but only interfered with E2-dependent ER nuclear retention, evidencing an important role for Akt in mediating the cytoplasmic-nuclear shuttling of the receptor. Ligand-induced full activation of ER seems to depend on Akt, and this event can occur, since Akt itself, in turn, undergoes phosphorylation within minutes of E2 treatment. In fact, ER can be phosphorylated on serine 167 by Akt in either an E2-dependent (12, 53, 55) or -independent manner (10, 55). Since our experiments were conducted in PRF-SFM and in the total absence of growth factors, it can be reasonably speculated that ER phosphorylation by Akt is solely dependent on E2. These observations could explain the reduced recruitment of ER and, consequently, of Pol II on the pS2 promoter following 1 h of API-2 pretreatment.
API-2 was used as an Akt inhibitor rather than the commonly used PI-3K inhibitors (i.e., wortmannin or LY294002) for several reasons: (i) we were interested in focusing on Akt's role in ER signaling and not the PI-3K/Akt axis in general; (ii) wortmannin, used at suggested concentrations, never inhibited ER function as API-2 did; and (iii) LY294002 has been suspected of having antiestrogenic properties (43), which clearly could have interfered with our experiments. However, since API-2 is unable to discriminate among Akt isoforms, small interfering RNAs were used to silence either Akt1 or Akt2.
Interestingly, only Akt2 knockdown produced effects on both ER expression and function comparable to those observed after a prolonged exposure to API-2. Indeed, an evident decrease in ER protein content and in mRNA levels, followed by a predictable decrease in estrogen-dependent pS2 expression, was evidenced. As mentioned above, these effects were obtained only by Akt2 and not by Akt1 knockdown, suggesting a pivotal role of Akt2 in both the regulation of ER function and its expression.
Although there are reports of Akt1 presence at the nuclear level, e.g., in thyroid cancer specimens and cultures (58) or in IGF-1-stimulated human embryonic kidney cells (3), in our cellular model and under our experimental conditions we never observed either Akt1 or Akt2 in the nucleus. Therefore, the genomic events deriving from Akt2 knockdown can be assumed to be mediated by one of its downstream nuclear effectors, such as the Forkhead winged-helix transcription factor FoxO3a, the most highly expressed in MCF-7 cells among the four known members of the box O family.
Forkhead members were found to bind ER in a cell-free system in the presence of ER ligand (49, 62, 65). Our data, for the first time, demonstrate that the interaction between ER and FoxO3a occurs in a whole-cell system. Indeed, ER was found to colocalize with FoxO3a at the perinuclear and nuclear levels and to coprecipitate from MCF-7 nuclear-protein extracts. E2 stimulation enhanced the binding between the two proteins, most likely as a consequence of the increased ER nuclear retention, as well as of the E2-induced FoxO3a overexpression. This ER/FoxO3a interaction became much more evident in Akt2 silenced samples, probably for a more sustained FoxO3a translocation into the nucleus. More interestingly, we found that FoxO3a was recruited to the pS2 promoter in an E2-dependent manner and that Akt2 inhibition emphasized this phenomenon. FoxO3a binding to DNA occurred on the Forkhead-responsive sequence close to the TATAA box (Fig. 5). The current literature reports controversial data regarding the Forkhead box O function on ER target promoters. Some authors state that these transcriptional factors can act as coactivators (49), while others found that they can have a bifunctional role, behaving as ER coactivators or corepressors, depending on the cellular model (62, 65). In our MCF-7 clone, FoxO3a overexpression inhibited ER-mediated transcription of E2-regulated genes, such as pS2 and CD1. This effect could most likely be ascribable to reduced recruitment of ER to ERE sites on the pS2 promoter, due to both reduced ER expression and the increased recruitment of the overexpressed negative modulator FoxO3a on its own binding motif, while FoxO3a silencing led to the opposite effect. It is worthwhile to emphasize that, unlike pS2, we cannot state at the moment whether CD1 is directly regulated by FoxO3a through its binding to Forkhead-responsive elements, though they are present on the CD1 promoter, or whether this effect is due to a reduced general ER content. In fact, as mentioned above, a strong decrease in ER expression, at both the RNA and protein levels, was observed in FoxO3a-overexpressing samples. Although our preliminary results suggest possible regulation of the ER promoter by FoxO3a, further investigations are needed to elucidate if this effect really depends on the loss of FoxO3a as a repressive element for ER gene transcription or if it is due to prolonged ER mRNA stability. Indeed, at least two functional Forkhead binding sites were identified on ER promoter B (17), but the authors demonstrateed an activating effect of constitutively active FoxO3a at this level in mouse mammary tumor-derived NF639 cells paralleled by an increase in ER protein in cells transiently overexpressing FoxO3a. These data do not support our hypothesis, nor are they in agreement with recent observations by Zou et al., who did not find significant differences in ER expression in FoxO3a-overexpressing MCF-7 stable clones (65). Furthermore, our findings also contrast with the reported decrease in ER levels in MCF-7 cells infected with dominant-negative FoxO3a-expressing adenovirus (17). The incongruence of these data could be partially explained by the different cell systems, culture conditions, and experimental procedures used, but this cannot replace further study of the possible mechanism through which FoxO3a regulates ER expression and/or stability. However, our findings were further corroborated by the growth inhibition observed in FoxO3a-overexpressing cells, as well as by the proliferative rate increase in FoxO3a silenced samples, evidencing the physiological relevance of this transcription factor in regulating the ER mitogenic signal. Moreover, we reproduced the main experiments in two additional ER-positive cell lines, T47D and ZR75, where Akt inhibition, as well as FoxO3a overexpression, led to ER downregulation and impaired transcriptional activity, suggesting how the Akt2/FoxO3a axis has a pivotal role in modulating ER function in ER-expressing breast cancer cells.
In fact, in two ER-negative cell lines, HeLa and SKBR3, that ectopically express ER, Akt inhibition did not lead to ER loss (protein and mRNA), supporting our hypothesis that the Akt pathway might control ER expression at a genomic level. Notably SKBR3 showed a significant decrease of ER transcriptional activity in response to API-2, mimicking MCF-7 behavior, while in HeLa cells, API-2 did not significantly affect ER transactivaton. The latter result could be explained by considering the different expression levels of Akt2 and Foxo3a in the two cell lines: in fact, HeLa cells express negligible amounts of both proteins, while their levels in SKBR3 cells are comparable to those found in MCF-7 cells, underlining the fact that the molecular machinery introduced here might work properly in cells expressing both Akt2 and FoxO3a.
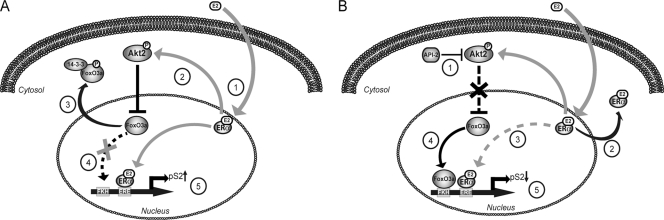
Finally, as already mentioned, E2 stimulation induced FoxO3a transcription and protein expression, which leads us to hypothesize the presence of estrogen target sequences on the FoxO3a promoter region. This observation could suggest a homeostatic control mechanism through which E2-induced FoxO3a upregulation is a required event to ensure liganded ER inactivation. The molecular basis on which E2 activates FoxO3a expression is currently under investigation in our laboratory. In conclusion, in our model, FoxO3a, transcriptionally activated by Akt inhibition, might behave as a repressor for ER-mediated transcription (i) directly, by binding to Forkhead-responsive elements on ER target gene promoters; (ii) indirectly, by the recruitment on ERE sequences of the FoxO3a/ER complex; and (iii) by inhibiting ER expression.
Taken together, the results presented here show that, among PKB members, only Akt2 seems to modulate ER activity at multiple levels, having a key role in the regulation of (i) the retention of ER in the nucleus, (ii) ER expression, and (iii) FoxO3a activation. A schematic summary of our findings is shown in Fig. 8. Our data point out the importance of the Akt/FoxO axis in E2-induced ER activation and signaling, evidencing additional mechanisms that could represent novel targets in ER-positive breast cancer therapy.
FIG. 8.
Suggested mechanism through which the Akt2/FoxO3a axis modulates ER expression and activity. (A) Liganded ER (1) activates Akt2 (2), which in turn phosphorylates, and thus inhibits, the FoxO3a transcription factor, promoting its nuclear exclusion and retention in the cytoplasm, where it binds to 14-3-3 proteins (3). FoxO3a is no longer recruited on the pS2 promoter (4), leading to enhanced ER transactivation and pS2 transcription (5). (B) Akt2 inhibition (1) leads to ER transcriptional repression, acting at different levels: reduction of liganded-ER retention in the nucleus (2) and, consequently, of ER recruitment on the pS2 promoter (3); activation and subsequent recruitment of FoxO3a on the pS2 promoter (4), leading to pS2 downregulation (5).
Acknowledgments
We thank B. Van der Burg (Utrecht, the Netherlands) for providing MCF-7 cells, V. Giguère (McGill University, Quebec City, Canada) for the pS2/TFF-1-ERE (pS2-ERE) plasmid, and S. A. W. Fuqua (Baylor College of Medicine, Breast Center, Houston, TX) for the pGL3-ERprom(E).
Financial support for this work was received from AIRC, MIUR Ex 60%, and Regione Calabria.
We declare that there is no conflict of interest that would prejudice the impartiality of this scientific work.
Footnotes
Published ahead of print on 23 November 2009.
REFERENCES
- 1.Alvarez, B., A. C. Martinez, B. M. Burgering, and A. C. Carrera. 2001. Forkhead transcription factors contribute to execution of the mitotic programme in mammals. Nature 413:744-747. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, E., R. B. Clarke, and A. Howell. 1998. Estrogen responsiveness and control of normal human breast proliferation. J. Mammary Gland Biol. Neoplasia 3:23-35. [DOI] [PubMed] [Google Scholar]
- 3.Andjelkovic, M., D. R. Alessi, R. Meier, A. Fernandez, N. J. Lamb, M. Frech, P. Cron, P. Cohen, J. M. Lucocq, and B. A. Hemmings. 1997. Role of translocation in the activation and function of protein kinase B. J. Biol. Chem. 272:31515-31524. [DOI] [PubMed] [Google Scholar]
- 4.Beck, S., P. Sommer, E. dos Santos Silva, N. Blin, and P. Gott. 1999. Hepatocyte nuclear factor 3 (winged helix domain) activates trefoil factor gene TFF1 through a binding motif adjacent to the TATAA box. DNA Cell Biol. 18:157-164. [DOI] [PubMed] [Google Scholar]
- 5.Berry, M., A. M. Nunez, and P. Chambon. 1989. Estrogen-responsive element of the human pS2 gene is an imperfectly palindromic sequence. Proc. Natl. Acad. Sci. U. S. A. 86:1218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas, D. K., A. P. Cruz, E. Gansberger, and A. B. Pardee. 2000. Epidermal growth factor-induced nuclear factor kappa B activation: a major pathway of cell-cycle progression in estrogen-receptor negative breast cancer cells. Proc. Natl. Acad. Sci. U. S. A. 97:8542-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brader, S., and S. A. Eccles. 2004. Phosphoinositide 3-kinase signalling pathways in tumor progression, invasion and angiogenesis. Tumori 90:2-8. [DOI] [PubMed] [Google Scholar]
- 8.Brunet, A., A. Bonni, M. J. Zigmond, M. Z. Lin, P. Juo, L. S. Hu, M. J. Anderson, K. C. Arden, J. Blenis, and M. E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857-868. [DOI] [PubMed] [Google Scholar]
- 9.Bunone, G., P. A. Briand, R. J. Miksicek, and D. Picard. 1996. Activation of the unliganded estrogen receptor by EGF involves the MAP kinase pathway and direct phosphorylation. EMBO J. 15:2174-2183. [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell, R. A., P. Bhat-Nakshatri, N. M. Patel, D. Constantinidou, S. Ali, and H. Nakshatri. 2001. Phosphatidylinositol 3-kinase/AKT-mediated activation of estrogen receptor alpha: a new model for anti-estrogen resistance. J. Biol. Chem. 276:9817-9824. [DOI] [PubMed] [Google Scholar]
- 11.Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, E. V. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver, and M. Brown. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the Forkhead protein FoxA1. Cell 122:33-43. [DOI] [PubMed] [Google Scholar]
- 12.Castoria, G., A. Migliaccio, A. Bilancio, M. Di Domenico, A. de Falco, M. Lombardi, R. Fiorentino, L. Varricchio, M. V. Barone, and F. Auricchio. 2001. PI3-kinase in concert with Src promotes the S-phase entry of oestradiol-stimulated MCF-7 cells. EMBO J. 20:6050-6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark, G. M., C. K. Osborne, and W. L. McGuire. 1984. Correlations between estrogen receptor, progesterone receptor, and patient characteristics in human breast cancer. J. Clin. Oncol. 2:1102-1109. [DOI] [PubMed] [Google Scholar]
- 14.Couse, J. F., and K. S. Korach. 1999. Estrogen receptor null mice: what have we learned and where will they lead us? Endocr. Rev. 20:358-417. [DOI] [PubMed] [Google Scholar]
- 15.deGraffenried, L. A., S. G. Hilsenbeck, and S. A. Fuqua. 2002. Sp1 is essential for estrogen receptor alpha gene transcription. J. Steroid Biochem. Mol. Biol. 82:7-18. [DOI] [PubMed] [Google Scholar]
- 16.Dowsett, M. 2001. Overexpression of HER-2 as a resistance mechanism to hormonal therapy for breast cancer. Endocr. Relat. Cancer 8:191-195. [DOI] [PubMed] [Google Scholar]
- 17.Guo, S., and G. E. Sonenshein. 2004. Forkhead box transcription factor FOXO3a regulates estrogen receptor alpha expression and is repressed by the Her-2/neu/phosphatidylinositol 3-kinase/Akt signaling pathway. Mol. Cell. Biol. 24:8681-8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Improta-Brears, T., A. R. Whorton, F. Codazzi, J. D. York, T. Meyer, and D. P. McDonnell. 1999. Estrogen-induced activation of mitogen-activated protein kinase requires mobilization of intracellular calcium. Proc. Natl. Acad. Sci. U. S. A. 96:4686-4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson, J. G., J. I. Kreisberg, A. P. Koterba, D. Yee, and M. G. Brattain. 2000. Phosphorylation and nuclear exclusion of the forkhead transcription factor FKHR after epidermal growth factor treatment in human breast cancer cells. Oncogene 19:4574-4581. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs, F. M., L. P. van der Heide, P. J. Wijchers, J. P. Burbach, M. F. Hoekman, and M. P. Smidt. 2003. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J. Biol. Chem. 278:35959-35967. [DOI] [PubMed] [Google Scholar]
- 21.Jordan, N. J., J. M. Gee, D. Barrow, A. E. Wakeling, and R. I. Nicholson. 2004. Increased constitutive activity of PKB/Akt in tamoxifen resistant breast cancer MCF-7 cells. Breast Cancer Res. Treat. 87:167-180. [DOI] [PubMed] [Google Scholar]
- 22.Kandel, E. S., and N. Hay. 1999. The regulation and activities of the multifunctional serine/threonine kinase Akt/PKB. Exp. Cell Res. 253:210-229. [DOI] [PubMed] [Google Scholar]
- 23.Kato, S., H. Endoh, Y. Masuhiro, T. Kitamoto, S. Uchiyama, H. Sasaki, S. Masushige, Y. Gotoh, E. Nishida, H. Kawashima, D. Metzger, and P. Chambon. 1995. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science 270:1491-1494. [DOI] [PubMed] [Google Scholar]
- 24.Katzenellenbogen, B. S., K. L. Kendra, M. J. Norman, and Y. Berthois. 1987. Proliferation, hormonal responsiveness, and estrogen receptor content of MCF-7 human breast cancer cells grown in the short-term and long-term absence of estrogens. Cancer Res. 47:4355-4360. [PubMed] [Google Scholar]
- 25.Keen, J. C., and N. E. Davidson. 2003. The biology of breast carcinoma. Cancer 97:825-833. [DOI] [PubMed] [Google Scholar]
- 26.Knuefermann, C., Y. Lu, B. Liu, W. Jin, K. Liang, L. Wu, M. Schmidt, G. B. Mills, J. Mendelsohn, and Z. Fan. 2003. HER2/PI-3K/Akt activation leads to a multidrug resistance in human breast adenocarcinoma cells. Oncogene 22:3205-3212. [DOI] [PubMed] [Google Scholar]
- 27.Konecny, G., G. Pauletti, M. Pegram, M. Untch, S. Dandekar, Z. Aguilar, C. Wilson, H. M. Rong, I. Bauerfeind, M. Felber, H. J. Wang, M. Beryt, R. Seshadri, H. Hepp, and D. J. Slamon. 2003. Quantitative association between HER-2/neu and steroid hormone receptors in hormone receptor-positive primary breast cancer. J. Natl. Cancer Inst. 95:142-153. [DOI] [PubMed] [Google Scholar]
- 28.Lanzino, M., C. Morelli, C. Garofalo, M. L. Panno, L. Mauro, S. Ando, and D. Sisci. 2008. Interaction between estrogen receptor alpha and insulin/IGF signaling in breast cancer. Curr. Cancer Drug Targets 8:597-610. [DOI] [PubMed] [Google Scholar]
- 29.Lee, C. S., A. deFazio, C. J. Ormandy, and R. L. Sutherland. 1996. Inverse regulation of oestrogen receptor and epidermal growth factor receptor gene expression in MCF-7 breast cancer cells treated with phorbol ester. J. Steroid Biochem. Mol. Biol. 58:267-275. [DOI] [PubMed] [Google Scholar]
- 30.Lin, C. Y., V. B. Vega, J. S. Thomsen, T. Zhang, S. L. Kong, M. Xie, K. P. Chiu, L. Lipovich, D. H. Barnett, F. Stossi, A. Yeo, J. George, V. A. Kuznetsov, Y. K. Lee, T. H. Charn, N. Palanisamy, L. D. Miller, E. Cheung, B. S. Katzenellenbogen, Y. Ruan, G. Bourque, C. L. Wei, and E. T. Liu. 2007. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 3:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, Y., H. Gao, T. T. Marstrand, A. Strom, E. Valen, A. Sandelin, J. A. Gustafsson, and K. Dahlman-Wright. 2008. The genome landscape of ERalpha- and ERbeta-binding DNA regions. Proc. Natl. Acad. Sci. U. S. A. 105:2604-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lobenhofer, E. K., G. Huper, J. D. Iglehart, and J. R. Marks. 2000. Inhibition of mitogen-activated protein kinase and phosphatidylinositol 3-kinase activity in MCF-7 cells prevents estrogen-induced mitogenesis. Cell Growth Differ. 11:99-110. [PubMed] [Google Scholar]
- 33.Lombardi, M., G. Castoria, A. Migliaccio, M. V. Barone, R. Di Stasio, A. Ciociola, D. Bottero, H. Yamaguchi, E. Appella, and F. Auricchio. 2008. Hormone-dependent nuclear export of estradiol receptor and DNA synthesis in breast cancer cells. J. Cell Biol. 182:327-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mazumdar, A., and R. Kumar. 2003. Estrogen regulation of Pak1 and FKHR pathways in breast cancer cells. FEBS Lett. 535:6-10. [DOI] [PubMed] [Google Scholar]
- 35.McCarty, K. S., Jr., T. K. Barton, B. F. Fetter, B. H. Woodard, J. A. Mossler, W. Reeves, J. Daly, W. E. Wilkinson, and K. S. McCarty, Sr. 1980. Correlation of estrogen and progesterone receptors with histologic differentiation in mammary carcinoma. Cancer 46:2851-2858. [DOI] [PubMed] [Google Scholar]
- 36.Medema, R. H., G. J. Kops, J. L. Bos, and B. M. Burgering. 2000. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404:782-787. [DOI] [PubMed] [Google Scholar]
- 37.Morelli, C., C. Garofalo, D. Sisci, S. del Rincon, S. Cascio, X. Tu, A. Vecchione, E. R. Sauter, W. H. Miller, Jr., and E. Surmacz. 2004. Nuclear insulin receptor substrate 1 interacts with estrogen receptor alpha at ERE promoters. Oncogene 23:7517-7526. [DOI] [PubMed] [Google Scholar]
- 38.Mossler, J. A., K. S. McCarty, Jr., and W. W. Johnston. 1981. The correlation of cytologic grade and steroid receptor content in effusions of metastatic breast carcinoma. Acta Cytol. 25:653-658. [PubMed] [Google Scholar]
- 39.Nakamura, N., S. Ramaswamy, F. Vazquez, S. Signoretti, M. Loda, and W. R. Sellers. 2000. Forkhead transcription factors are critical effectors of cell death and cell cycle arrest downstream of PTEN. Mol. Cell. Biol. 20:8969-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakatani, K., D. A. Thompson, A. Barthel, H. Sakaue, W. Liu, R. J. Weigel, and R. A. Roth. 1999. Up-regulation of Akt3 in estrogen receptor-deficient breast cancers and androgen-independent prostate cancer lines. J. Biol. Chem. 274:21528-21532. [DOI] [PubMed] [Google Scholar]
- 41.Osborne, C. K. 1998. Steroid hormone receptors in breast cancer management. Breast Cancer Res. Treat. 51:227-238. [DOI] [PubMed] [Google Scholar]
- 42.Osborne, C. K., K. Hobbs, and G. M. Clark. 1985. Effect of estrogens and antiestrogens on growth of human breast cancer cells in athymic nude mice. Cancer Res. 45:584-590. [PubMed] [Google Scholar]
- 43.Pasapera Limon, A. M., J. Herrera-Munoz, R. Gutierrez-Sagal, and A. Ulloa-Aguirre. 2003. The phosphatidylinositol 3-kinase inhibitor LY294002 binds the estrogen receptor and inhibits 17beta-estradiol-induced transcriptional activity of an estrogen sensitive reporter gene. Mol. Cell Endocrinol. 200:199-202. [DOI] [PubMed] [Google Scholar]
- 44.Perou, C. M., T. Sorlie, M. B. Eisen, M. van de Rijn, S. S. Jeffrey, C. A. Rees, J. R. Pollack, D. T. Ross, H. Johnsen, L. A. Akslen, O. Fluge, A. Pergamenschikov, C. Williams, S. X. Zhu, P. E. Lonning, A. L. Borresen-Dale, P. O. Brown, and D. Botstein. 2000. Molecular portraits of human breast tumours. Nature 406:747-752. [DOI] [PubMed] [Google Scholar]
- 45.Reagan-Shaw, S., and N. Ahmad. 2006. RNA interference-mediated depletion of phosphoinositide 3-kinase activates forkhead box class O transcription factors and induces cell cycle arrest and apoptosis in breast carcinoma cells. Cancer Res. 66:1062-1069. [DOI] [PubMed] [Google Scholar]
- 46.Rena, G., S. Guo, S. C. Cichy, T. G. Unterman, and P. Cohen. 1999. Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J. Biol. Chem. 274:17179-17183. [DOI] [PubMed] [Google Scholar]
- 47.Samatar, A. A., L. Wang, A. Mirza, S. Koseoglu, S. Liu, and C. C. Kumar. 2002. Transforming growth factor-beta 2 is a transcriptional target for Akt/protein kinase B via forkhead transcription factor. J. Biol. Chem. 277:28118-28126. [DOI] [PubMed] [Google Scholar]
- 48.Santini, D., C. Ceccarelli, M. Taffurelli, S. Pileri, and D. Marrano. 1996. Differentiation pathways in primary invasive breast carcinoma as suggested by intermediate filament and biopathological marker expression. J. Pathol. 179:386-391. [DOI] [PubMed] [Google Scholar]
- 49.Schuur, E. R., A. V. Loktev, M. Sharma, Z. Sun, R. A. Roth, and R. J. Weigel. 2001. Ligand-dependent interaction of estrogen receptor-alpha with members of the forkhead transcription factor family. J. Biol. Chem. 276:33554-33560. [DOI] [PubMed] [Google Scholar]
- 50.Sisci, D., C. Morelli, C. Garofalo, F. Romeo, L. Morabito, F. Casaburi, E. Middea, S. Cascio, E. Brunelli, S. Ando, and E. Surmacz. 2007. Expression of nuclear insulin receptor substrate 1 in breast cancer. J. Clin. Pathol. 60:633-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sisci, D., and E. Surmacz. 2007. Crosstalk between IGF signaling and steroid hormone receptors in breast cancer. Curr. Pharm. Des. 13:705-717. [DOI] [PubMed] [Google Scholar]
- 52.Stal, O., G. Perez-Tenorio, L. Akerberg, B. Olsson, B. Nordenskjold, L. Skoog, and L. E. Rutqvist. 2003. Akt kinases in breast cancer and the results of adjuvant therapy. Breast Cancer Res. 5:R37-R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoica, G. E., T. F. Franke, M. Moroni, S. Mueller, E. Morgan, M. C. Iann, A. D. Winder, R. Reiter, A. Wellstein, M. B. Martin, and A. Stoica. 2003. Effect of estradiol on estrogen receptor-alpha gene expression and activity can be modulated by the ErbB2/PI 3-K/Akt pathway. Oncogene 22:7998-8011. [DOI] [PubMed] [Google Scholar]
- 54.Suhara, T., H. S. Kim, L. A. Kirshenbaum, and K. Walsh. 2002. Suppression of Akt signaling induces Fas ligand expression: involvement of caspase and Jun kinase activation in Akt-mediated Fas ligand regulation. Mol. Cell. Biol. 22:680-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun, M., J. E. Paciga, R. I. Feldman, Z. Yuan, D. Coppola, Y. Y. Lu, S. A. Shelley, S. V. Nicosia, and J. Q. Cheng. 2001. Phosphatidylinositol-3-OH kinase (PI3K)/AKT2, activated in breast cancer, regulates and is induced by estrogen receptor alpha (ERalpha) via interaction between ERalpha and PI3K. Cancer Res. 61:5985-5991. [PubMed] [Google Scholar]
- 56.Sun, M., G. Wang, J. E. Paciga, R. I. Feldman, Z. Q. Yuan, X. L. Ma, S. A. Shelley, R. Jove, P. N. Tsichlis, S. V. Nicosia, and J. Q. Cheng. 2001. AKT1/PKBalpha kinase is frequently elevated in human cancers and its constitutive activation is required for oncogenic transformation in NIH3T3 cells. Am. J. Pathol. 159:431-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56a.Tora, L., A. Mullick, D. Metzger, M. Ponglikitmongkol, I. Park, and P. Chambon. 1989. The cloned human oestrogen receptor contains a mutation which alters its hormone binding properties. EMBO J. 8:1981-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsai, E. M., S. C. Wang, J. N. Lee, and M. C. Hung. 2001. Akt activation by estrogen in estrogen receptor-negative breast cancer cells. Cancer Res. 61:8390-8392. [PubMed] [Google Scholar]
- 58.Vasko, V., M. Saji, E. Hardy, M. Kruhlak, A. Larin, V. Savchenko, M. Miyakawa, O. Isozaki, H. Murakami, T. Tsushima, K. D. Burman, C. De Micco, and M. D. Ringel. 2004. Akt activation and localisation correlate with tumour invasion and oncogene expression in thyroid cancer. J. Med. Genet. 41:161-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Witton, C. J., J. R. Reeves, J. J. Going, T. G. Cooke, and J. M. Bartlett. 2003. Expression of the HER1-4 family of receptor tyrosine kinases in breast cancer. J. Pathol. 200:290-297. [DOI] [PubMed] [Google Scholar]
- 60.Yu, X., R. V. Rajala, J. F. McGinnis, F. Li, R. E. Anderson, X. Yan, S. Li, R. V. Elias, R. R. Knapp, X. Zhou, and W. Cao. 2004. Involvement of insulin/phosphoinositide 3-kinase/Akt signal pathway in 17 beta-estradiol-mediated neuroprotection. J. Biol. Chem. 279:13086-13094. [DOI] [PubMed] [Google Scholar]
- 61.Yuan, Z. Q., M. Sun, R. I. Feldman, G. Wang, X. Ma, C. Jiang, D. Coppola, S. V. Nicosia, and J. Q. Cheng. 2000. Frequent activation of AKT2 and induction of apoptosis by inhibition of phosphoinositide-3-OH kinase/Akt pathway in human ovarian cancer. Oncogene 19:2324-2330. [DOI] [PubMed] [Google Scholar]
- 62.Zhao, H. H., R. E. Herrera, E. Coronado-Heinsohn, M. C. Yang, J. H. Ludes-Meyers, K. J. Seybold-Tilson, Z. Nawaz, D. Yee, F. G. Barr, S. G. Diab, P. H. Brown, S. A. Fuqua, and C. K. Osborne. 2001. Forkhead homologue in rhabdomyosarcoma functions as a bifunctional nuclear receptor-interacting protein with both coactivator and corepressor functions. J. Biol. Chem. 276:27907-27912. [DOI] [PubMed] [Google Scholar]
- 63.Zhu, Y., M. A. Saunders, H. Yeh, W. G. Deng, and K. K. Wu. 2002. Dynamic regulation of cyclooxygenase-2 promoter activity by isoforms of CCAAT/enhancer-binding proteins. J. Biol. Chem. 277:6923-6928. [DOI] [PubMed] [Google Scholar]
- 64.Zinda, M. J., M. A. Johnson, J. D. Paul, C. Horn, B. W. Konicek, Z. H. Lu, G. Sandusky, J. E. Thomas, B. L. Neubauer, M. T. Lai, and J. R. Graff. 2001. AKT-1, -2, and -3 are expressed in both normal and tumor tissues of the lung, breast, prostate, and colon. Clin. Cancer Res. 7:2475-2479. [PubMed] [Google Scholar]
- 65.Zou, Y., W. B. Tsai, C. J. Cheng, C. Hsu, Y. M. Chung, P. C. Li, S. H. Lin, and M. C. Hu. 2008. Forkhead box transcription factor FOXO3a suppresses estrogen-dependent breast cancer cell proliferation and tumorigenesis. Breast Cancer Res. 10:R21. [DOI] [PMC free article] [PubMed] [Google Scholar]