Abstract
Clostridium difficile is the most common infectious cause of diarrhea in hospitalized patients. The optimal approach for the detection of toxigenic C. difficile remains controversial because no single test is sensitive, specific, and affordable. We have developed a real-time PCR method (direct stool PCR [DPCR]) to detect the tcdB gene encoding toxin B directly from stool specimens and have combined it with enzyme immunoassays (EIAs) in a three-step protocol. DPCR was performed on 699 specimens that were positive for C. difficile glutamate dehydrogenase (GDH) by Wampole C Diff Quik Chek EIA (GDH-Q) and negative for toxins A and B by Wampole Tox A/B Quik Chek EIA (AB-Q), performed sequentially. The performance of this three-step algorithm was compared with a modified “gold standard” that combined tissue culture cytotoxicity (CYT) and DPCR. A separate investigation was performed to evaluate the sensitivity of the GDH-Q as a screening test, and toxigenic C. difficile was found in 1.9% of 211 GDH-Q-negative specimens. The overall sensitivity, specificity, and positive and negative predictive values, respectively, were as follows for an algorithm combining GDH-Q, AB-Q, and DPCR: 83.8%, 99.7%, 97.1%, and 97.9%. Those for CYT alone were 58.8%, 100%, 100%, and 94.9%, respectively. In comparison, the sensitivity and specificity of DPCR were estimated to be 97.5% and 99.7%, respectively, using the same modified gold standard. Neither CYT nor toxin EIA was sufficiently sensitive to exclude toxigenic C. difficile, and combining EIAs with CYT in a three-step algorithm failed to substantially improve sensitivity. DPCR is a sensitive and specific method for the detection of toxigenic C. difficile that can provide same-day results at a cost-per-positive test comparable to those of other methods. A three-step algorithm in which DPCR is used to analyze GDH EIA-positive, toxin EIA-negative specimens provides a convenient and specific alternative with rapid results for 87.7% of specimens, although this approach is less sensitive than performing DPCR on all specimens.
Clostridium difficile is the leading cause of infectious diarrhea in hospitalized patients. New challenges have been posed by the emergence of highly virulent C. difficile strains that may be refractory to standard treatment and cause disease even in immunocompetent individuals without prior antibiotic exposure (11, 28). Rapid and accurate laboratory diagnosis is critical to reduce the morbidity from C. difficile infection (CDI) and allow the implementation of specific infection control measures.
Methods for the detection of toxigenic C. difficile have long been unsatisfactory. The tissue-culture assay for cytotoxin B (CYT) is often considered the “gold standard” for diagnosis (10, 29), but many reports have documented the failure of CYT to detect symptomatic and even life-threatening cases of CDI (14, 33, 44) and the poor sensitivity of CYT in comparison to toxigenic culture (18, 22, 39, 43). The technical complexity of CYT, as well as a requirement for 24 to 48 h of incubation, has resulted in the widespread replacement of CYT with toxin immunoassays that provide results within minutes. In a 2008 College of American Pathologists report of proficiency results, 95% of laboratories reported using an EIA kit to detect C. difficile toxin (6). C. difficile immunoassays are often adopted as stand-alone assays based on validation against CYT, but studies that have employed more sensitive gold standards have documented that rapid toxin EIAs have unacceptably low sensitivities ranging from 32 to 79% (3, 20, 23, 44, 50).
Toxigenic culture, when performed under optimal culture conditions and combined with a sensitive and specific toxin detection method, is regarded as the most sensitive method of toxigenic C. difficile detection (18, 44, 50), but complexity and a prolonged turnaround time have discouraged its routine use. An alternative approach has been to test for the glutamate dehydrogenase (GDH) antigen of C. difficile as a surrogate for culture. GDH EIAs have been reported to be highly sensitive for C. difficile detection, allowing same-day reporting of negative results, but positive results must be followed by a sensitive and specific test to differentiate between toxigenic and nontoxigenic strains (5, 57, 58). In this study, we used the Wampole C Diff Quik Chek EIA (GDH-Q), a rapid GDH assay that requires less technical time and expertise in comparison to microtiter plate immunoassays, followed by the Tox A/B Quik Chek (AB-Q) on GDH-Q-negative specimens. The choices for a second-step assay to detect toxigenic C. difficile in GDH-positive specimens have included toxigenic culture, CYT, or toxin EIAs (20, 23, 46, 57), but the insensitivity of CYT and toxin EIAs potentially leaves many cases of CDI undetected.
An alternative, highly sensitive method to detect toxigenic C. difficile is real-time PCR (12, 44, 50, 54, 60), with sensitivity values ranging from 83.6% to 93.4% and specificity from 93.9% to 98.2%, respectively, when compared to toxigenic culture (50, 54, 60). Real-time PCR can be completed on the day of specimen submission, thus providing same-day results. However, PCR techniques have not been not widely used for stool specimens, due primarily to budgetary issues, as well as the challenge of extracting nucleic acids from feces and separating template DNA from potentially interfering substances.
We have developed a sensitive and specific real-time PCR assay (direct stool PCR [DPCR]) for the C. difficile toxin B-encoding tcdB gene, which can be performed directly on stool specimens. Nucleic acids can be efficiently extracted from feces with the NucliSENS miniMAG system, a semiautomated magnetic silica bead extraction system to remove inhibitors and purify nucleic acids. Results were compared with the total yield of positive specimens detected by CYT and DPCR. Our observations indicate that a multistep algorithm consisting of GDH EIA, toxin EIA, and selective DPCR can provide a specific and cost-effective approach to the laboratory detection of toxigenic C. difficile that is more sensitive than CYT or EIA alone but not as sensitive as toxigenic culture or DPCR.
MATERIALS AND METHODS
Study description.
From January 2008 through July 2008, 699 soft or liquid stool samples from adult patients at Harborview Medical Center were submitted for detection of toxigenic C. difficile by both the tissue culture cytotoxicity assay (CYT) and a three-step EIA/PCR algorithm. Specimens were refrigerated upon receipt, and duplicates from the same day were excluded. The C. difficile EIA/PCR algorithm began with the C Diff Quik Chek (GDH-Q) EIA, with a positive GDH-Q result triggering the second-step Tox A/B Quik Chek (AB-Q) (Wampole/TechLab, Blacksburg, VA) EIA for toxins A and B. Specimens with indeterminate EIA results (GDH-Q positive, AB-Q negative) were tested by direct stool PCR (DPCR) to detect the tcdB gene. AB-Q-positive specimens were considered positive and GDH-Q-negative specimens were considered negative for toxigenic C. difficile and were not tested further by PCR in view of previous reports documenting the high specificity and sensitivity of the Quik Chek toxin and GDH EIAs, respectively (20, 23, 47).
EIAs.
The Quik Chek assays are lateral-flow, membrane-bound enzyme immunoassays that are visually interpreted. The C Diff Quik Chek (GDH-Q) detects the glutamate dehydrogenase antigen of C. difficile, whereas the Tox A/B Quik Chek (AB-Q) detects toxins A and B without differentiation of the toxins. Each EIA was contained in a separate cassette. Both assays were performed per the manufacturer's instructions.
CYT.
Specimens submitted for C. difficile cytotoxin detection by tissue culture assay were processed within 24 h by the University of Washington Clinical Virology Laboratory. After dilution and centrifugation of fecal material in Hanks' solution, the supernatant was filtered and added to human diploid fibroblast cell monolayers with and without C. difficile antitoxin (TechLab, Blacksburg, VA). Samples producing characteristic cytopathic effects only in the absence of C. difficile antitoxin were considered positive for C. difficile toxin B, with the final interpretation of results after 48 h of incubation.
DPCR extraction.
Non-liquid stools were diluted and mixed well with sufficient molecular-grade phosphate-buffered saline (Invitrogen, Carlsbad, CA) to allow aspiration with a wide-bore pipette. The liquefied specimen was thoroughly mixed by vortexing and then centrifuged for 1 min at low speed (80 × g) to sediment solid fecal material while retaining bacteria in the supernatant fluid. An occasional mucoid specimen required further mixing and centrifugation at 320 × g, with careful aspiration of supernatant. A 100-μl aliquot of supernatant fluid aspirated from just above the sediment with a genomic wide-bore tip was lysed with NucliSENS lysis buffer, and nucleic acids were extracted and eluted with the NucliSENS miniMAG system (bioMérieux, Durham, NC) per the manufacturer's instructions. The final eluted specimen volume was 60 μl.
DPCR real-time PCR.
Amplification to detect tcdB was performed on the Rotor-Gene-Q 6000 thermocycler (Qiagen, Inc., Valencia, CA). The NK104/NK105 primer set, designed by Kato et al. (27), was used to amplify a 204-bp sequence. Each 20-μl DPCR mixture contained 10 μl of 2× QuantiTect SYBR green master mix (Qiagen, Valencia, CA), 1 μl each of 10 μM forward and reverse primers, 6 μl of H2O, and 2 μl of extracted specimen. Amplification began with a 15-min step at 95°C, followed by 40 cycles of 15 s at 95°C, 25 s at 53°C, and 20 s at 72°C, which was finally followed by a 30-s step at 72°C before a stepwise (1°C/5 s) temperature increase to 90°C. Detection of the 16S rRNA-encoding gene of C. difficile was also performed in a separate reaction on each extracted GDH-Q-positive specimen as an internal control, using primer set CD3/CD6 (45) (5′-GGCGGCGTGCCTAAC-3′ and 5′-TGGCTCACCTTTGATATTC-3′) and the same amplification conditions as for tcdB detection. Distinct single-melt peaks at 76.3 ± 1°C and 82.0 ± 1°C were considered positive for tcdB and C. difficile 16S rRNA gene detection, respectively. Because several GDH-Q-positive specimens had no detectable C. difficile 16S rRNA gene and were culture negative for C. difficile, an additional amplification control of heat-extracted DNA from a nontoxigenic C. difficile isolate was added to each extracted specimen and a reagent control in a 1/15 dilution. Subsequently, amplification with universal bacterial 16S rRNA gene primers (25) was employed as an internal extraction and amplification control, with an expected melt peak of 86.5 ± 2°C and fewer than 20 cycles.
Statistical analysis.
Graphpad software (http://graphpad.com/quickcalcs) was used to determine 95% confidence intervals (CI) and significance by McNemar's test of proportions and the chi-square test with Yates' correction.
RESULTS
In data not shown (32), the detection of toxigenic C. difficile by DPCR performed as a second-step assay on Triage C difficile panel-indeterminate (GDH-positive, toxin A-negative) specimens was validated by comparing detection with the combined yield of positives by the cytotoxin assay (CYT) and a toxigenic culture method that identified toxigenic C. difficile colonies by real-time PCR for tcdB. The Triage EIA-DPCR combination detected 93.4% (71/76) of specimens positive for toxigenic C. difficile, with specificity and positive and negative predictive values (PPVs and NPVs, respectively) of 100%, 100%, and 98.9%, respectively. The Triage panel is a membrane-bound lateral-flow EIA that is no longer commercially available.
For evaluation of the three-step GDH-Q/AB-Q/DPCR algorithm, all DPCR and CYT positives were considered true positives (Table 1), based on the DPCR performance described above and the high specificity of CYT. All CYT-positive specimens were also positive by GDH-Q. CYT was negative on two of 28 specimens that were positive by both GDH-Q and AB-Q.
TABLE 1.
Performance of C Diff Quik Chek EIA, Tox A/B Quik Chek EIA, CYT assay, and direct PCR for detection of toxigenic C. difficile
| No. of samples | Result by assaya: |
Interpretation of results | |||
|---|---|---|---|---|---|
| GDH-Q | A/B-Q | CYT | DPCRb | ||
| 585 | − | ND | − | ND | − |
| 43 | + | − | − | − | − |
| 2 | + | + | − | ND | − |
| 26 | + | + | + | ND | + |
| 19 | + | − | + | + | + |
| 2 | + | − | + | − | + |
| 22 | + | − | − | + | + |
Abbreviations: GDH-Q, C Diff Quik Chek EIA; AB-Q, Tox A/B Quik Chek EIA; CYT, cytotoxin tissue culture assay; DPCR, direct real-time PCR detection of tcdB; ND, not done.
PCR results: 1.9% (4/211) of consecutive GDH-Q-negative specimens in a subsequent time period were DPCR positive for tcdB; therefore, 11 of 585 are expected to be GDH-Q negative, DPCR positive, as shown in Table 2.
In the initial evaluation of data, 585 (83.7% of 699) fecal specimens that were negative by GDH-Q and CYT assays were considered to be true negatives (Table 1). However, in December 2008, DPCR performed on 211 consecutive GDH-Q-negative specimens revealed that 1.9% (n = 4) of GDH-Q-negative specimens contained toxigenic C. difficile, which was confirmed by toxigenic culture, including GDH-Q, AB-Q, and DPCR testing of C. difficile isolates. By applying this false-negative rate to the 585 GDH-Q-negative specimens, 11 additional specimens were designated as true positives, with 574 remaining as true negatives, as shown in Table 2.
TABLE 2.
Detection of toxigenic C. difficile by cytotoxin, GDH-Q EIA, and sequential algorithms compared with gold standard (cytotoxin assay and PCR)
| Assaya | Result | Comparison to CYT and DPCR resultsb |
|||||
|---|---|---|---|---|---|---|---|
| No. of specimens |
% Sensitivity (95% CI) | % Specificity (95% CI) | % PPV (95% CI) | % NPV (95% CI) | |||
| Either positivec | Negative | ||||||
| CYT (tissue culture) | Positive | 47 | 0 | 58.8 | 100 | 100 | 94.9 |
| Negative | 33 | 619 | (47.8-68.9) | (93.0-96.4) | |||
| GDH-Q | Positive | 69 | 45 | 86.3 | 92.7 | 60.5 | 98.1 |
| Negative | 11 | 574 | (76.9-92.3) | (90.4-94.5) | (51.3-69.0) | (96.6-99.0) | |
| Two-step GDH-Q/AB-Q | Positive | 26 | 2d | 32.5 | 99.7 | 92.9 | 92.0 |
| Negative | 54 | 617 | (23.2-43.4) | (98.8-100) | (76.3-99.1) | (89.6-93.8) | |
| Three-step GDH-Q/AB-Q/DPCR | Positive | 67 | 2d | 83.8 | 99.7 | 97.1 | 97.9 |
| Negative | 13 | 617 | (74.0-90.4) | (98.8-100) | (89.4-99.8) | (96.5-98.8) | |
Abbreviations: AB-Q, Tox A/B Quik Chek EIA; GDH-Q, C Diff Quik Chek EIA; DPCR, direct real-time PCR; CI, confidence interval.
GDH-Q was performed on all specimens, with AB-Q EIA performed on GDH-Q-positives and DPCR on indeterminate (GDH-Q-positive, AB-Q-negative) specimens.
GDH-Q-negative specimens were not tested by DPCR in this study, so the rate of DPCR positivity (1.9%) in 211 GDH-Q-negative, CYT-negative specimens from a subsequent time period was applied to 585 GDH-Q negatives, resulting in 11 specimens designated as GDH-Q negative, DPCR positive.
CYT negative and DPCR not done.
After adjusting for false-negative results, GDH-Q was 86.3% (69/80) sensitive with a 60.5% positive predictive value (Table 3) but only 92.7% specificity. AB-Q performed on GDH-Q-positive specimens was only 32.5% sensitive but highly specific (99.7%). CYT achieved only 58.8% (47/80) sensitivity, whereas the three-step algorithm of GDH-Q, AB-Q, and DPCR was more sensitive, detecting 83.8% (67/80) of positive specimens, even though two GDH-Q-positive, CYT-positive specimens were not detected by DPCR. The difference in detection of positive specimens between CYT and the EIA-DPCR combination was significant (P < 0.001 by McNemar's test for paired proportions) and represented a 43% increase in the yield of positive specimens over CYT alone. An algorithm combining the GDH-Q and AB-Q EIAs with CYT testing of GDH-Q-positive, AB-Q-negative specimens would only have improved sensitivity to 63.8% (51/80). In additional data not shown, the presence of toxigenic C. difficile was confirmed in 10 out of 10 consecutive CYT-negative, DPCR-positive specimens by anaerobic culture followed by GDH-Q, AB-Q, and DPCR performed on suspensions of C. difficile isolates.
TABLE 3.
Estimated annual cost, processing time, and yield of positive results for toxigenic Clostridium difficile assays
| Assaya | No. of specimens positive for toxigenic C. difficileb | Cost (US$)c |
Daily processing time (h) (% of specimens) | ||
|---|---|---|---|---|---|
| Annual | Per specimen | Per positive specimen | |||
| AB-Q | 115d | 46,562 | 15.02 | 405 | <1 |
| Two-step GDH-Q/AB-Q | 115 | 55,942 | 18.05 | 486 | <1.5 |
| CYT | 208 | 110,546 | 35.66 | 533 | ≤48 |
| Three-step GDH-Q/AB-Q→CYT | 208 | 75,680 | 24.41 | 365 | <1.5 (87.7) |
| ≤48 (12.3) | |||||
| DPCR | 346e | 125,767 | 40.57 | 363 | ≤5 |
| Two-step GDH-Q/DPCR | 297 | 109,193 | 35.22 | 368 | <1 (83.7) |
| ≤5 (16.3) | |||||
| Three-step GDH-Q/AB-Q/DPCR | 297 | 126,121 | 40.68 | 425 | <1.5 (87.7) |
| ≤5 (12.3) | |||||
For abbreviations, see Table 1.
Based on annual volume of 3,100 specimens (506 tested in 2nd step, 381 in 3rd step), 11.4% (n = 354) prevalence, and data from Tables 1 and 2.
Estimated cost per test with labor and benefits at $50/h: AB-Q, $15.02 batched and $21.68 singly; GDH-Q, $14.51 batched; CYT, $51.75 batched; DPCR, $40.57 in batch of nine tests and $184 singly, for $127 in an average batch of 1.5 tests.
Yield assumes all GDH-Q-negative specimens to be AB-Q-negative.
Yield assumes all GDH-Q-positive, AB-Q-positive specimens to be DPCR-positive.
Repeat specimens after GDH-Q-negative results.
We investigated the value of submitting additional specimens after an initial negative GDH-Q result on patients with no history of a positive toxin result. From January through July 2008, repeat GDH-Q tests were performed for 171 patients on 194 specimens submitted one to seven days after the initial test. Duplicate specimens from the same day were rejected. All specimens submitted 1 to 3 days after the first test were either GDH-Q negative (117/119) or GDH-Q positive, AB-Q negative, and DPCR negative (2/119). Of 75 repeat specimens tested at 4 to 6 days, 13 were GDH-Q positive, with six (8.0% of 75) of 13 positive for toxigenic C. difficile by either AB-Q (n = 3) or DPCR (n = 3). The difference in yield of toxin-positive samples between the two collection periods was significant (P < 0.02) by chi-square contingency table with Yates' correction.
Cost estimates.
Table 3 displays the estimated costs of the various approaches to detect toxigenic C. difficile. Figures were derived from our 2007-2008 test volume, estimated labor and benefit costs of $50/hour (U.S. dollars), and the yield of toxigenic C. difficile-positive specimens shown in Tables 1 and 2. Labor and materials costs for quality control, test preparation, cleanup, and result interpretation were included, but equipment expenses were excluded. Note that costs may vary significantly according to region and specimen volume.
Estimates of annual laboratory expenses for toxigenic C. difficile detection ranged from $47,000 and $56,000 for AB-Q alone or a two-stage GDH-Q/AB-Q algorithm, respectively, to $126,000 for DPCR as either a stand-alone test or in the three-step EIA/DPCR protocol (Table 3). However, the two-step GDH-Q/AB-Q approach would miss 231 (66.7%) of 346 positive specimens detectable by a stand-alone DPCR test, and assuming that all AB-Q positive specimens are also GDH-Q positive, the AB-Q yield of positive specimens would be equivalent to that of the two-step GDH-Q/AB-Q. A three-step EIA/CYT protocol would miss 138 (39.9%) of 346 specimens, with 505 (16.3% of 3,100) specimens requiring 24 to 48 h of incubation before reporting, whereas the three-step EIA/DPCR algorithm would miss only 49 (14.2%) positive specimens, and processing could be completed within one day.
EIA costs per specimen were lowest at $15 for AB-Q and $18 for the two-step GDH-Q/AB-Q, respectively, but when evaluated on a cost-per-positive-test basis, the PCR-based three-step algorithm was comparable to EIA testing (Table 3). A less-expensive approach that employs DPCR is a two-step GDH-Q/DPCR algorithm, which would delay results for 4% of positive specimens that would otherwise be detected by the AB-Q in 30 to 40 min. DPCR performed on all specimens had the lowest cost per positive specimen and highest sensitivity (346 out of 354 positive specimens), but the three-step EIA/DPCR option has the advantage of shorter turnaround time and ease of test performance for the 87.7% of specimens that do not require DPCR.
DISCUSSION
The accurate diagnosis of C. difficile infection (CDI) has become increasingly important, yet the laboratory methods for diagnosis have remained problematic and controversial. The tissue culture assay for C. difficile cytotoxin (CYT) is often cited as the “gold standard” for toxigenic C. difficile detection (10, 29). However, while the CYT is highly specific, it is only moderately sensitive and has been well documented to miss cases of CDI (14, 33, 35, 44). For this reason, the Society for Healthcare Epidemiology of America (SHEA) has recommended that both stool culture and toxin testing be performed, “preferably with confirmation of organism toxicity if a direct stool toxin test is negative or not done” (21). Nevertheless, this recommendation is not followed by the vast majority of clinical laboratories, which largely rely on insensitive toxin EIA kits (6).
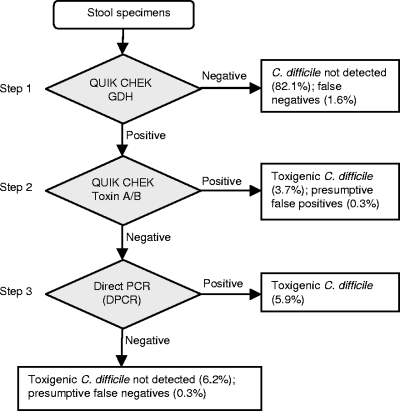
Proposed multistep diagnostic algorithms have improved efficiency but continue to depend upon insensitive combinations of EIA and CYT assays (23, 57). The goal of the present study was to determine whether DPCR could be integrated with EIAs to optimize the detection of toxigenic C. difficile while taking advantage of the convenience of the rapid commercial assays. As suggested by previous investigators (9, 51, 58), our study used a GDH screening method to rapidly report the majority of specimens. The GDH-Q, a lateral-flow EIA which can be completed in 30 to 40 min, allowed 84% of specimens to be reported as negative for C. difficile. Second-step toxin EIA testing by the AB-Q, also a 30- to 40-min assay, identified 4% of specimens as toxin positive, leaving 12% for DPCR testing (Fig. 1).
FIG. 1.
Three-step algorithm for detection of toxigenic C. difficile. This algorithm provides rapid, sensitive, specific, and cost-effective detection of toxigenic C. difficile. The Quik Chek EIAs (Wampole/TechLab, Blacksburg, VA) are C Diff Quik Chek for glutamate dehydrogenase antigen (GDH) and Tox A/B Quik Chek for toxins A and B. DPCR detects the tcdB gene.
Consistent with SHEA recommendations, our DPCR method had been validated using a gold standard of both toxigenic culture (with real-time PCR detection of tcdB in colonies) and CYT. Against this standard, we found DPCR to be a sensitive and specific method for the detection of toxigenic C. difficile in fecal specimens (32) and, therefore, did not include toxigenic culture in this study but instead compared results with the total yield of positive specimens by CYT and DPCR, as shown in Table 1.
A shortcoming of this approach was the assumption of the high sensitivity and negative predictive value of GDH-Q. Although Reyes et al. reported that the GDH-Q detected 46 of 46 specimens with toxigenic C. difficile (47), and data from Gilligan suggest a high negative predictive value (23), we found that four (1.9%) of 211 consecutive GDH-Q negative specimens in a subsequent time period contained the tcdB gene by DPCR testing, which was confirmed by toxigenic culture. Chart review of the patients with falsely negative GDH-Q results revealed that three of four received antibiotic treatment for CDI based on the positive DPCR results. If DPCR had not been performed on the GDH-Q-negative specimens, all of which were also CYT negative, the sensitivity and NPV of the three-step Quik Chek EIA/DPCR algorithm would have appeared to be 97.1% and 99.7%, whereas the more accurate adjusted values were determined to be 83.8% and 97.9%, respectively (Fig. 1 and Table 2).
It should be noted that our laboratory performs PCR and toxigenic culture on request when warranted by the clinical presentation, regardless of the GDH-Q results. For example, on one occasion subsequent to this investigation, we encountered a patient with refractory diarrhea, colitis by endoscopy, and repeated specimens negative by GDH-Q and a toxin immunoassay. After detection of toxigenic C. difficile by both DPCR and toxigenic culture, the patient underwent appropriate treatment for CDI. Occasional specimens may contain inhibitory substances or insufficient numbers of C. difficile organisms to produce a positive EIA result. In such cases, DPCR and toxigenic culture can be very helpful and should be employed irrespective of a negative EIA result when clinical suspicion of CDI is high.
The results of our EIA/DPCR algorithm are similar to the results of direct real-time PCR assays described by Sloan et al. (50) and Stamper et al. (54), who reported sensitivities of 86% and 83.6% and specificities of 97% and 98.2%, respectively, in their comparisons with toxigenic culture results. The sensitivity of our three-step approach was less than that of the method of Peterson et al., who found their real-time PCR to be 93.4% sensitive and 97.4% specific when compared to a gold standard that combined clinical information (≥3 loose stools/day) with ≥2 positive toxin assays (44). To estimate the sensitivity and specificity of performing DPCR on all specimens, 26 specimens that were positive by GDH-Q, AB-Q, and CYT-Q but not tested by DPCR (per the three-step algorithm) must be assumed to be DPCR positive. Using this assumption, the sensitivity and specificity of DPCR as a stand-alone test are estimated to be 97.5% (78/80) and 99.7% (617/619), respectively.
For the real-time PCR detection of toxigenic C. difficile, we chose to target a conserved region of the tcdB gene (24), in accordance with most PCR-based C. difficile assays (12, 24, 44, 60). The tcdB gene is a preferable target to tcdA because it allows the detection of increasingly prevalent toxin A-negative, toxin B-positive strains (4, 13, 15, 26, 27, 30, 36, 41, 48, 49), as well as strains producing both toxins A and B. Strains that carry tcdB but exhibit low levels of expression (38) or produce toxin A only (17) appear to be very uncommon. Although Sloan et al. have suggested tcdC to be a suitable target for PCR detection of toxigenic C. difficile (50), the variability of tcdC (53, 55) and its dispensability for virulence raise concerns about its long-term stability as a diagnostic target. A recent study has documented that toxin B is essential for C. difficile virulence (37), but five of the strains described by Sloan as toxigenic contained only the tcdA and tcdC genes and may actually have been nontoxigenic due to the absence of toxin B.
In our study, CYT detected only 58.8% of positive specimens, comparable to previous studies describing the insensitivity of CYT (18, 22, 39, 46). The AB-Q, performed on GDH-Q-positive specimens, was more than 99% specific but very insensitive (32.5%), consistent with previous studies of the AB-Q (23, 47) and other toxin immunoassays (44, 50). High specificity (99.7%), rapid turnaround time, and technical simplicity make the AB-Q a useful intermediate-step assay. However, the failure to detect toxin in 41 of 67 specimens found to be DPCR positive highlights the unreliability of negative toxin EIA results. Two specimens with positive AB-Q results on GDH-positive specimens were not confirmed by CYT and were unavailable for DPCR testing. Gilligan has also noted a small percentage of AB-Q-positive specimens that could not be confirmed by CYT (23). These may have been cross-reacting Clostridium sordellii isolates, as mentioned in the AB-Q package insert. We have observed positive AB-Q results with the C. sordellii ATCC 9714 strain and one patient C sordellii isolate, both of which were GDH-Q negative. Restricting AB-Q testing to GDH-Q-positive specimens thus might reduce the likelihood of false-positive results.
Submission of repeat specimens from patients with an initial GDH-Q-negative specimen and no history of a previous positive toxin result produced positive toxin results in 3.0% of specimens tested within 7 days of the negative initial test, comparable to previous reports of 1.3 to 2.1% (1, 16, 40). All repeat GDH-Q specimens submitted within one to three days of the first test were negative, whereas 8% of those specimens submitted after four to six days were found to be toxin positive. This is similar to the results of Cardona and Rand, who recommended against repeating AB-Q testing within 48 h of a negative result (16).
Clearly, GDH-Q is insufficiently specific and CYT and AB-Q are insufficiently sensitive to be relied upon as assays for toxigenic C. difficile, whether used alone or in combination with each other, although EIA testing is the least expensive method for the laboratory budget. The incorporation of DPCR into an algorithm using EIAs as screening steps increases overall costs of testing but not the cost per case detected (Table 3). The increased number of cases detected (approximately 182 specimens per year positive by three-step EIA/DPCR but negative by EIA) may compensate for higher labor and reagent costs by reducing the nosocomial spread of CDI. Infected patients are an important reservoir of C. difficile in institutional settings, and person-to-person transmission has been demonstrated in 10 to 20% of cases of hospital-associated CDI (8, 56). Emerging 027/NAP1 epidemic strains may have even greater transmissibility due to enhanced toxin and spore production (2).
Patients who develop nosocomial CDI incur increased attributable costs ranging from $3,791 to $6,959 per episode (19, 34, 42, 52) (median costs adjusted to 2009 Consumer Price Index dollars [59]) and are hospitalized from 2.9 to 5.5 additional days (31, 34, 42, 52), compared to patients without CDI. The additional annual costs of a DPCR or three-step EIA/DPCR algorithm are justified by the earlier detection of CDI, which would reduce the number of cases resulting from nosocomial transmission. In comparison to the toxin EIA (AB-Q) alone, the three-step EIA/DPCR algorithm would cost $437 per additional case of CDI detected, while a strategy of performing DPCR on every specimen would cost $343 per additional case detected, as calculated from Table 3 figures. The additional expense is even more modest when compared to the “gold standard” of tissue culture cytotoxin assay (CYT), with the three-step EIA/DPCR algorithm costing $175 per additional case detected and DPCR only $110 per additional case detected. A further justification for improving the sensitivity of C. difficile detection is the resulting decrease in length of stay due to earlier diagnosis. With the daily cost of hospitalization attributable to CDI estimated at between $1,300 and $2,000 (31, 34, 42, 52), implementing DPCR or a three-step EIA/DPCR algorithm could save more than $200,000 annually simply by making the diagnosis of C. difficile one day earlier in cases missed by EIA, as these patients would otherwise be likely to require repeat testing.
The choice of real-time PCR methods for direct detection from stools now includes the Xpert C. difficile test (Cepheid, Sunnyvale, CA), the ProGastro Cd assay (Prodesse, Waukesha, WI), and the BD GeneOhm Cdiff assay (BD Diagnostics, San Diego, CA), with published GeneOhm sensitivity values of 83.6 and 93.9% and NPVs of 97.1% to 99.2%, when compared to toxigenic culture (7, 54), although unresolved GeneOhm results were as high as 7.3% in one study (7). Our observations confirm that DPCR is a sensitive, specific, and cost-effective method for the detection of toxigenic C. difficile. A multistep algorithm consisting of a GDH EIA screen followed by a specific toxin EIA screen and direct real-time PCR provides a convenient, rapid, and specific alternative strategy, but with the trade-off of some reduction in sensitivity.
Acknowledgments
The NucliSENS miniMAG instrument and reagents were provided by bioMérieux, Inc., for the duration of the 2005 study. We are grateful to Carol I. Johnson, Cheryl A. McMillan, Thomas R. Smith, and Audrey A. Twite for their professionalism and technical support.
Footnotes
Published ahead of print on 18 November 2009.
REFERENCES
- 1.Aichinger, E., C. D. Schleck, W. S. Harmsen, L. M. Nyre, and R. Patel. 2008. Nonutility of repeat laboratory testing for detection of Clostridium difficile by use of PCR or enzyme immunoassay. J. Clin. Microbiol. 46:3795-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Åkerlund, T., I. Persoon, M. Unemo, T. Norén, B. Svenungsson, M. Wullt, and L. G. Burman. 2008. Increased sporulation rate of epidemic Clostridium difficile type 027/NAP1. J. Clin. Microbiol. 46:1530-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alcala, L., L. Sánchez-Cambronero, M. P. Catalán, M. Sánchez-Somolinos, M. T. Peláez, M. Marín, and E. Bouza. 2008. Comparison of three commercial methods for rapid detection of Clostridium difficile toxins A and B from fecal specimens. J. Clin. Microbiol. 46:3833-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfa, M. J., A. Kabani, D. Lyerly, S. Moncrief, L. M. Neville, A. Al-Barrak, G. K. Harding, B. Dyck, K. Olekson, and J. M. Embil. 2000. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile responsible for a nosocomial outbreak of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 38:2706-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alfa, M. J., B. Swan, B. VanDekerhove, P. Pang, and G. K. Harding. 2002. The diagnosis of Clostridium difficile-associated diarrhea: comparison of Triage C. difficile panel, EIA for ToxA/B and cytotoxin assays. Diagn. Microbiol. Infect. Dis. 43:257-263. [DOI] [PubMed] [Google Scholar]
- 6.Anonymous. 2008. CAP Bacteriology Survey Set D-A. Summary. College of American Pathologists, Northfield, IL.
- 7.Barbut, F., M. Braun, B. Burghoffer, V. Lalande, and C. Eckert. 2009. Rapid diagnosis of toxigenic strains of Clostridium difficile in diarrheal stools by real-time PCR. J. Clin. Microbiol. 47:1276-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbut, F., B. Gariazzo, L. Bonné, V. Lalande, B. Burghoffer, R. Luiuz, and J. Petit. 2007. Clinical features of Clostridium difficile-associated infections and molecular characterization of strains: results of a retrospective study, 2000-2004. Infect. Control Hosp. Epidemiol. 28:131-139. [DOI] [PubMed] [Google Scholar]
- 9.Barbut, F., V. Lalande, G. Daprey, P. Cohen, N. Marle, B. Burghoffer, and J. C. Petit. 2000. Usefulness of simultaneous detection of toxin A and glutamate dehydrogenase for the diagnosis of Clostridium difficile-associated diseases. Eur. J. Clin. Microbiol. Infect. Dis. 19:481-484. [DOI] [PubMed] [Google Scholar]
- 10.Bartlett, J. G. 2002. Clinical practice. Antibiotic-associated diarrhea. N. Engl. J. Med. 346:334-339. [DOI] [PubMed] [Google Scholar]
- 11.Bartlett, J. G. 2006. Narrative review: the new epidemic of Clostridium difficile-associated enteric disease. Ann. Intern. Med. 145:758-764. [DOI] [PubMed] [Google Scholar]
- 12.Belanger, S. D., M. Boissinot, N. Clairoux, F. J. Picard, and M. G. Bergeron. 2003. Rapid detection of Clostridium difficile in feces by real-time PCR. J. Clin. Microbiol. 41:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borriello, S. P., B. W. Wren, S. Hyde, S. V. Seddon, P. Sibbons, M. M. Krishna, S. Tabaqchali, S. Manek, and A. B. Price. 1992. Molecular, immunological, and biological characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect. Immun. 60:4192-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouza, E., T. Peláez, R. Alonso, P. Catalán, P. Muñoz, and M. R. Créixems. 2001. ‘Second-look’ cytotoxicity: an evaluation of culture plus cytotoxin assay of Clostridium difficile isolates in the laboratory diagnosis of CDAD. J. Hosp. Infect. 48:233-237. [DOI] [PubMed] [Google Scholar]
- 15.Brazier, J. S., S. L. Stubbs, and B. I. Duerden. 1999. Prevalence of toxin A negative/B positive Clostridium difficile strains. J. Hosp. Infect. 42:248-249. [PubMed] [Google Scholar]
- 16.Cardona, D. M., and K. H. Rand. 2008. Evaluation of repeat Clostridium difficile enzyme immunoassay testing. J. Clin. Microbiol. 46:3686-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen, S. H., Y. J. Tang, B. Hansen, and J. J. Silva. 1998. Isolation of a toxin B-deficient mutant strain of Clostridium difficile in a case of recurrent C. difficile-associated diarrhea. Clin. Infect. Dis. 26:410-412. [DOI] [PubMed] [Google Scholar]
- 18.Delmee, M., J. Van Broeck, A. Simon, M. Janssens, and V. Avesani. 2005. Laboratory diagnosis of Clostridium difficile-associated diarrhoea: a plea for culture. J. Med. Microbiol. 54:187-191. [DOI] [PubMed] [Google Scholar]
- 19.Dubberke, E. R., K. A. Reske, M. A. Olsen, L. C. McDonald, and V. J. Fraser. 2008. Short- and long-term attributable costs of Clostridium difficile-associated disease in nonsurgical inpatients. Clin. Infect. Dis. 46:497-504. [DOI] [PubMed] [Google Scholar]
- 20.Fenner, L., A. F. Widmer, G. Goy, S. Rudin, and R. Frei. 2008. Rapid and reliable diagnostic algorithm for detection of Clostridium difficile. J. Clin. Microbiol. 46:328-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerding, D. N., S. Johnson, L. R. Peterson, M. E. Mulligan, and J. J. Silva. 1995. Clostridium difficile-associated diarrhea and colitis. Infect. Control. Hosp. Epidemiol. 16:459-477. [DOI] [PubMed] [Google Scholar]
- 22.Gerding, D. N., M. M. Olson, L. R. Peterson, D. G. Teasley, R. L. Gebhard, M. L. Schwartz, and J. T. Lee, Jr. 1986. Clostridium difficile-associated diarrhea and colitis in adults. A prospective case-controlled epidemiologic study. Arch. Intern. Med. 146:95-100. [PubMed] [Google Scholar]
- 23.Gilligan, P. H. 2008. Is a two-step glutamate dehydrogenase antigen-cytotoxicity neutralization assay algorithm superior to the premier toxin A and B enzyme immunoassay for laboratory detection of Clostridium difficile? J. Clin. Microbiol. 46:1523-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guilbault, C., A. C. Labbe, L. Poirier, L. Busque, C. Beliveau, and M. Laverdiere. 2002. Development and evaluation of a PCR method for detection of the Clostridium difficile toxin B gene in stool specimens. J. Clin. Microbiol. 40:2288-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen, G., S. Swanzy, R. Gupta, B. Cookson, and A. P. Limaye. 2008. In vitro activity of fluoroquinolones against clinical isolates of Nocardia identified by partial 16S rRNA sequencing. Eur. J. Clin. Microbiol. Infect. Dis. 27:115-120. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, S., S. P. Sambol, J. S. Brazier, M. Delmée, V. Avesani, M. M. Merrigan, and D. N. Gerding. 2003. International typing study of toxin A-negative, toxin B-positive Clostridium difficile variants. J. Clin. Microbiol. 41:1543-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato, H., N. Kato, K. Watanabe, K. Watanabe, N. Iwai, H. Nakamura, T. Yamamoto, K. Suzuki, S. M. Kim, Y. Chong, and E. B. Wasito. 1998. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J. Clin. Microbiol. 36:2178-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelly, C. P., and J. T. LaMont. 2008. Clostridium difficile—more difficult than ever. N. Engl. J. Med. 359:1932-1940. [DOI] [PubMed] [Google Scholar]
- 29.Kelly, C. P., C. Pothoulakis, and J. T. LaMont. 1994. Clostridium difficile colitis. N. Engl. J. Med. 330:257-262. [DOI] [PubMed] [Google Scholar]
- 30.Kim, H., T. V. Riley, M. Kim, C. K. Kim, D. Yong, K. Lee, Y. Chong, and J. Park. 2008. Increasing prevalence of toxin A-negative, toxin B-positive isolates of Clostridium difficile in Korea: impact on laboratory diagnosis. J. Clin. Microbiol. 46:1116-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kyne, L., M. B. Hamel, R. Polavaram, and C. P. Kelly. 2002. Health care costs and mortality associated with nosocomial diarrhea due to Clostridium difficile. Clin. Infect. Dis. 34:346-353. [DOI] [PubMed] [Google Scholar]
- 32.Larson, A. M., A. M. Fung, and F. C. Fang. 2006. Direct PCR detection of toxigenic Clostridium difficile from stool specimens, abstr. C-033. Abstr. 106th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 33.Lashner, B. A., J. Todorczuk, D. F. Sahm, and S. B. Hanauer. 1986. Clostridium difficile culture-positive toxin-negative diarrhea. Am. J. Gastroenterol. 81:940-943. [PubMed] [Google Scholar]
- 34.Lawrence, S. J., L. A. Puzniak, B. N. Shadel, K. N. Gillespie, M. H. Kollef, and L. M. Mundy. 2007. Clostridium difficile in the intensive care unit: epidemiology, costs, and colonization pressure. Infect. Control Hosp. Epidemiol. 28:123-130. [DOI] [PubMed] [Google Scholar]
- 35.Lee, S. D., D. K. Turgeon, C. W. Ko, T. R. Fritsche, and C. M. Surawicz. 2003. Clinical correlation of toxin and common antigen enzyme immunoassay testing in patients with Clostridium difficile disease. Am. J. Gastroenterol. 98:1569-1572. [DOI] [PubMed] [Google Scholar]
- 36.Lyerly, D. M., L. A. Barroso, T. D. Wilkins, C. Depitre, and G. Corthier. 1992. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect. Immun. 60:4633-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyras, D., J. R. O'Connor, P. M. Howarth, S. P. Sambol, G. P. Carter, T. Phumoonna, R. Poon, V. Adams, G. Vedantam, S. Johnson, D. N. Gerding, and J. I. Rood. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathis, J. N., L. Pilkinton, and D. E. McMillin. 1999. Detection and transcription of toxin DNA in a nontoxigenic strain of Clostridium difficile. Curr. Microbiol. 38:324-328. [DOI] [PubMed] [Google Scholar]
- 39.McFarland, L. V., G. W. Elmer, W. E. Stamm, and M. E. Mulligan. 1991. Correlation of immunoblot type, enterotoxin production, and cytotoxin production with clinical manifestations of Clostridium difficile infection in a cohort of hospitalized patients. Infect. Immun. 59:2456-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohan, S. S., B. P. McDermott, S. Parchuri, and B. A. Cunha. 2006. Lack of value of repeat stool testing for Clostridium difficile toxin. Am. J. Med. 119:356.e7-356.e8. [DOI] [PubMed] [Google Scholar]
- 41.Moncrief, J. S., L. Zheng, L. M. Neville, and D. M. Lyerly. 2000. Genetic characterization of toxin A-negative, toxin B-positive Clostridium difficile isolates by PCR. J. Clin. Microbiol. 38:3072-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Brien, J. A., B. J. Lahue, J. J. Caro, and D. M. Davidson. 2007. The emerging infectious challenge of Clostridium difficile-associated disease in Massachusetts hospitals: clinical and economic consequences. Infect. Control Hosp. Epidemiol. 28:1219-1227. [DOI] [PubMed] [Google Scholar]
- 43.Peterson, L. R., and P. J. Kelly. 1993. The role of the clinical microbiology laboratory in the management of Clostridium difficile-associated diarrhea. Infect. Dis. Clin. North Am. 7:277-293. [PubMed] [Google Scholar]
- 44.Peterson, L. R., R. U. Manson, S. M. Paule, D. M. Hacek, A. Robicsek, R. B. Thomson, Jr., and K. L. Kaul. 2007. Detection of toxigenic Clostridium difficile in stool samples by real-time polymerase chain reaction for the diagnosis of C. difficile-associated diarrhea. Clin. Infect. Dis. 45:1152-1160. [DOI] [PubMed] [Google Scholar]
- 45.Rakeman, J. L., N. Olson, E. D. Stefansson, T. R. Fritsche, A. P. Limaye, and B. T. Cookson. 2005. A two-step approach to toxigenic Clostridium difficile detection increases diagnostic yield, abstr. C-335. Abstr. 105th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 46.Reller, M. E., C. A. Lema, T. M. Perl, M. Cai, T. L. Ross, K. A. Speck, and K. C. Carroll. 2007. Yield of stool culture with isolate toxin testing versus a two-step algorithm including stool toxin testing for detection of toxigenic Clostridium difficile. J. Clin. Microbiol. 45:3601-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reyes, R. C., M. A. John, D. L. Ayotte, A. Covacich, S. Milburn, and Z. Hussain. 2007. Performance of TechLab C. DIFF QUIK CHEK and TechLab. C. DIFFICILE TOX A/B II for the detection of Clostridium difficile in stool samples. Diagn. Microbiol. Infect. Dis. 59:33-37. [DOI] [PubMed] [Google Scholar]
- 48.Rupnik, M., N. Kato, M. Grabnar, and H. Kato. 2003. New types of toxin A-negative, toxin B-positive strains among Clostridium difficile isolates from Asia. J. Clin. Microbiol. 41:1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samra, Z., S. Talmor, and J. Bahar. 2002. High prevalence of toxin A-negative toxin B-positive Clostridium difficile in hospitalized patients with gastrointestinal disease. Diagn. Microbiol. Infect. Dis. 43:189-192. [DOI] [PubMed] [Google Scholar]
- 50.Sloan, L. M., B. J. Duresko, D. R. Gustafson, and J. E. Rosenblatt. 2008. Comparison of real-time PCR for detection of the tcdC gene with four toxin immunoassays and culture in diagnosis of Clostridium difficile infection. J. Clin. Microbiol. 46:1996-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snell, H., M. Ramos, S. Longo, M. John, and Z. Hussain. 2004. Performance of the TechLab C. DIFF CHEK-60 enzyme immunoassay (EIA) in combination with the C. difficile Tox A/B II EIA kit, the Triage C. difficile panel immunoassay, and a cytotoxin assay for diagnosis of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 42:4863-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song, X., J. G. Bartlett, K. Speck, A. Naegeli, K. Carroll, and T. M. Perl. 2008. Rising economic impact of Clostridium difficile-associated disease in adult hospitalized patient population. Infect. Control Hosp. Epidemiol. 29:823-828. [DOI] [PubMed] [Google Scholar]
- 53.Spigaglia, P., and P. Mastrantonio. 2002. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J. Clin. Microbiol. 40:3470-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stamper, P. D., R. Alcabasa, D. Aird, W. Babiker, J. Wehrlin, I. Ikpeama, and K. C. Carroll. 2009. Comparison of a commercial real-time PCR assay for tcdB detection to a cell culture cytotoxicity assay and toxigenic culture for direct detection of toxin-producing Clostridium difficile in clinical samples. J. Clin. Microbiol. 47:373-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stare, B. G., M. Delmee, and M. Rupnik. 2007. Variant forms of the binary toxin CDT locus and tcdC gene in Clostridium difficile strains. J. Med. Microbiol. 56:329-335. [DOI] [PubMed] [Google Scholar]
- 56.Svenungsson, B., L. G. Burman, K. Jalakas-Pörnull, A. Lagergren, J. Struwe, and T. Åkerlund. 2003. Epidemiology and molecular characterization of Clostridium difficile strains from patients with diarrhea: low disease incidence and evidence of limited cross-infection in a Swedish teaching hospital. J. Clin. Microbiol. 41:4031-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ticehurst, J. R., D. Z. Aird, L. M. Dam, A. P. Borek, J. T. Hargrove, and K. C. Carroll. 2006. Effective detection of toxigenic Clostridium difficile by a two-step algorithm including tests for antigen and cytotoxin. J. Clin. Microbiol. 44:1145-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turgeon, D. K., T. J. Novicki, J. Quick, L. Carlson, P. Miller, B. Ulness, A. Cent, R. Ashley, A. Larson, M. Coyle, A. P. Limaye, B. T. Cookson, and T. R. Fritsche. 2003. Six rapid tests for direct detection of Clostridium difficile and its toxins in fecal samples compared with the fibroblast cytotoxicity assay. J. Clin. Microbiol. 41:667-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.U.S. Department of Labor, Bureau of Labor Statistics. Accessed 21 July 2009. Consumer Price Index. http://www.bls.gov/data/inflation_calculator.htm.
- 60.van den Berg, R. J., N. Vaessen, H. P. Endtz, T. Schulin, E. R. van der Vorm, and E. J. Kuijper. 2007. Evaluation of real-time PCR and conventional diagnostic methods for the detection of Clostridium difficile-associated diarrhoea in a prospective multicentre study. J. Med. Microbiol. 56:36-42. [DOI] [PubMed] [Google Scholar]