Abstract
A real-time PCR assay with the ability to rapidly identify all pathogenic bacteria would have widespread medical utility. Current real-time PCR technologies cannot accomplish this task due to severe limitations in multiplexing ability. To this end, we developed a new assay system which supports very high degrees of multiplexing. We developed a new class of mismatch-tolerant “sloppy” molecular beacons, modified them to provide an extended hybridization range, and developed a multiprobe, multimelting temperature (Tm) signature approach to bacterial species identification. Sloppy molecular beacons were exceptionally versatile, and they were able to generate specific Tm values for DNA sequences that differed by as little as one nucleotide to as many as 23 polymorphisms. Combining the Tm values generated by several probe-target hybrids resulted in Tm signatures that served as highly accurate sequence identifiers. Using this method, PCR assays with as few as six sloppy molecular beacons targeting bacterial 16S rRNA gene segments could reproducibly classify 119 different sequence types of pathogenic and commensal bacteria, representing 64 genera, into 111 Tm signature types. Blinded studies using the assay to identify the bacteria present in 270 patient-derived clinical cultures including 106 patient blood cultures showed a 95 to 97% concordance with conventional methods. Importantly, no bacteria were misidentified; rather, the few species that could not be identified were classified as “indeterminate,” resulting in an assay specificity of 100%. This approach enables highly multiplexed target detection using a simple PCR format that can transform infectious disease diagnostics and improve patient outcomes.
Human bloodstream infections (BSI) must be treated rapidly and effectively in order to avoid significant morbidity and mortality (17). A rising incidence of drug-resistant infections has complicated antibiotic selection (33), emphasizing the importance of rapidly determining the identity and drug susceptibility profile of each infecting bacterial species. Unfortunately, conventional microbiological identification and drug susceptibility determination methods are often too time-consuming to allow quick treatment decisions. Many bacterial species have specific antibiotic indications while most have local antibiotic resistance patterns that can be predicted by periodic examination of antibiotic resistance profiles (10, 19). Consequently, species identification may be used to guide antibiotic therapy pending final antibiotic susceptibility tests. A rapid bacterial species identification method would dramatically speed up the diagnosis of serious diseases, enabling rapid and definitive treatment, and concomitantly decrease the use of broad-spectrum antibiotics.
Ideally, a rapid assay for BSI should be functionally equivalent to the current diagnostic standard, blood culture (15), in being able to identify all clinically significant pathogens as well as commensals. To this end, molecular assays have targeted bacterial 16S rRNA genes or 16S-23S rRNA gene spacer regions (11, 24, 27, 29). These DNA segments contain hypervariable sequences that can be used to identify virtually all known bacterial species. Hypervariable sequences are flanked by highly conserved DNA sequences that permit universal amplification of the diagnostic targets, utilizing a limited primer set. DNA sequencing methods, including some used in clinical diagnostics (2, 16), have confirmed the utility of this approach although the clinical matrix has usually been culture-positive material rather than uncultured patient blood. The roadblock to its wider clinical use has been an inability to develop user-friendly real-time PCR methods that can distinguish among the same diverse set of sequences as differentiated by DNA sequencing. DNA probes, such as those used in real-time PCR assays, are able to bind to only a limited range of DNA sequences. Thus, currently available PCR-based detection methods are usually limited to differentiating among bacterial species within a single genus (8, 11, 20, 26). Bacterial species identification can be moderately increased by multiplexing several probes to perform different species detection assays simultaneously (18, 30); however, the fundamental limitation caused by a narrow probe binding range remains the same. Moreover, it is difficult to distinguish more than six different fluorophores in a single real-time PCR assay using current real-time PCR instruments due to limitations in the discriminatory capacity of real-time PCR instruments. This places a practical limitation on the number of real-time PCR probes that can be used in a bacterial species identification system designed to be carried out in a single assay well.
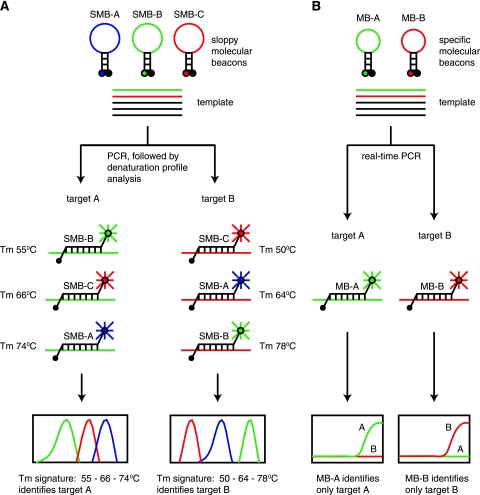
Our approach for identifying bacterial species utilizes modified molecular beacon probes termed “sloppy molecular beacons” (SMBs). These probes, by virtue of their greater length (40 to 60 nucleotides) and the choice of their probe sequence, are able to form probe-target hybrids with PCR amplicons generated from a wide range of bacterial species. When the resulting hybrids are melted apart, the temperature at which they dissociate (Tm) is characteristic of the particular bacterial DNA sequence that is present in the sample. Although a single Tm value is not sufficiently precise to unambiguously identify each species that is present, a unique set of these specially designed probes, each with a unique sequence, provides a set of Tm values (Tm signature) that distinctively identifies the species that is present (Fig. 1A). This approach, which was initially utilized in the speciation of mycobacteria (9), differs fundamentally from conventional real-time PCR assays that use a single DNA probe to detect a single (or closely related) target (Fig. 1B). Here, we demonstrate how the variable hybridization signals generated by six specially modified SMB probes can be combined with a pattern-based analytic approach to reproducibly identify and distinguish several hundred different sequences, thereby classifying a wide range of bacteria into clinically relevant genera and species. This novel approach enabled us to develop a universal bacterial identification assay and to validate this assay on cultured clinical isolates and positive patient blood cultures.
FIG. 1.
Two paradigms of target identification. (A) Sloppy molecular beacon approach. Three sloppy molecular beacons with different semispecific probe regions are placed in a PCR with an unknown target. Each probe is distinguished by a unique fluorophore. Asymmetrical PCR is performed, and the Tm of each probe target-hybrid is measured. Each probe generates a Tm that is specific for a limited number of targets such that probe A, probe B, and probe C generate Tm values of 55°C, 66°C, and 74°C, respectively, with target A and Tm values of 64°C, 78°C, and 50°C, respectively, with target B. All three Tm values are combined into a single species-specific signature that identifies the unknown target. Hundreds of different targets can theoretically be identified and distinguished with the same three probes in this manner. (B) Conventional approach. Two molecular beacons with probe regions that are specific for a single target sequence are placed in a PCR with an unknown target. Each molecular beacon is distinguished by a unique fluorophore or reaction well. PCR is performed, and the presence or absence of each molecular beacon signal is assessed. The unknown target is identified by determining which of the molecular beacons in a reaction produces a positive signal. The number of different target sequences that can be identified is limited to the number of different molecular beacons that are used in the assay.
MATERIALS AND METHODS
Bacterial isolates.
Initial studies were performed on a reference set of bacterial isolates and a limited number of artificial DNA templates representing 64 genera and 133 species obtained from various sources (see Table S1 in the supplemental material). The identity of each isolate was confirmed by sequencing a 700-bp region of the 16S rRNA gene, which included the hypervariable regions V3 through V6 (3). To perform confirmatory studies, we also created a test set of clinical isolates by randomly selecting 164 clinical isolates cultured from 158 patient's clinical cultures of blood, sputum, and urine from the New Jersey Medical School University Hospital's clinical microbiology laboratory. An additional 81 positive and 25 negative blood cultures, selected at random over an 8-month period, were obtained from the same laboratory. The investigator performing confirmatory studies on test set or blood culture-derived isolates was blinded to all prior species assignments. All clinical samples were stripped of personal identifiers prior to study entry, and the use of these samples for the current study was approved by the New Jersey Medical School Institutional Review Board. Additional details on clinical specimen and isolate collection and characterization are provided in Methods in the supplemental material.
DNA isolation.
Bacterial DNA was isolated from reference and test set isolates by boiling a loop of pure bacterial culture in Insta Gene Matrix solution (Bio-Rad Laboratories, Hercules, CA) for 20 min. DNA used for limit-of-detection analysis was subjected to RNase digestion followed by phenol-chloroform purification and ethanol precipitation. DNA was isolated from blood culture bottles by mixing 1 ml of the blood culture with an equal volume of 8% NaOH and incubating the samples for 15 min with occasional mixing. The solution was then transferred to a Cepheid GeneXpert open sample processing cartridge (Cepheid, Sunnyvale, CA) and then automatically processed for DNA by a GeneXpert DX system (Cepheid).
Primers and sloppy molecular beacons.
We had previously identified the 16S rRNA gene V3 and V6 hypervariable segments that provided a high degree of sequence variation as most appropriate for bacterial species identification using DNA probes (3). We designed universal primer sets to amplify each region from all clinically relevant bacterial species using linear-after-the-exponential (LATE)-PCR (23) (detailed in Methods in the supplemental material). SMBs were designed using the in silico DNA folding program mfold (http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/dna-form1.cgi), and the probe-target hybrid folding program DINAMelt (http://dinamelt.bioinfo.rpi.edu/twostate.php) was used to predict the possible hybrid structures and Tms. Unlike conventional molecular beacons, we designed the SMBs so that their probe lengths ranged from 40 to 60 nucleotides, and stem lengths varied between 5 and 7 nucleotides. All the probes were purchased from Sigma Genosys (St. Louis, MO). Two probes, SMB45 and V6P3, were used to test melting temperature variation as a function of probe-target mismatches. Six probes were designed for species identification by making them partially complementary to the V3 (two probes) and V6 (four probes) hypervariable regions of the 16S rRNA gene. Details of the molecular beacon sequences are listed in Methods in the supplemental material.
PCR conditions and post-PCR Tm analysis.
PCR assays of the reference and clinical isolates used 2 to 10 ng of DNA or 108 to 106 copies of synthetic template, except where noted, for limit-of-detection analyses. Individual SMBs were tested in individual PCR wells. The finalized assay was performed in six wells. For the assay, all six wells contained DNA from a given sample and a PCR master mix. One of the six sloppy molecular beacons used in the assay was then added to each well. Each PCR contained 1× PCR buffer, a 200 mM concentration of each deoxyribonucleoside triphosphate, 2.5 mM (for V3) or 4 mM (for V6) MgCl2, 0.5 μM excess primer (V3R or V6F), 0.0167 μM limiting primer (V3F or V6R, respectively), 0.06 U/μl of Taq Polymerase Stoffel fragment (Applied Biosystems, Foster City, CA), 1 ng/μl of any one of the six probes, and bacterial DNA or synthetic target in a final volume of 20 μl in each well of a 384-well plate. PCR was performed using an ABI Prism 7900HT Sequence Detection System (Applied Biosystems) with the following cycling parameters: 94°C for 2 min followed by 45 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 15 s. A post-PCR Tm analysis was then performed by heating the resulting amplicons to 94°C for 5 min, followed by gradual cooling to 35°C. After a 10-min hold at 35°C, the temperature was gradually raised by increments of 1° to 90°C, and the reaction mixture was held at each temperature for 1 min, with fluorescence monitored at each temperature for the last 30 s. The Tm data were plotted as the first derivative of fluorescence versus temperature after first smoothing the data by calculating a four-temperature-point rolling average and normalizing each curve to the fluorescence at 85°C. Tm values were identified by selecting the peak of each curve, and a six-point Tm signature was generated for each DNA sample.
Tm signature matching.
Bacterial species in the test set of clinical isolates or in the blood culture study were identified by finding the closest matches between the Tm signature of each unknown bacteria to the Tm signatures of the known bacterial species in a reference set look-up table. A program was created to express each signature as a single point in six-dimensional space (where each axis is used to plot one of the six Tm values). The distance between that point and the point determined for each of the known species was then calculated to produce a series of distance indexes, or D values. The known species that was at the closest distance to the unknown species (i.e., which had the smallest D value) was then identified as a correct match. The calculated distance was usually in the range of 0 to 3 for matching species. An unknown species generating a D value of ≥5 was considered “indeterminate.” The program used to calculate D values from experimentally derived Tms is available in Excel format (see Data in the supplemental material).
RESULTS
Sloppy molecular beacons hybridize differentially to a wide range of DNA sequences.
We explored modifications in conventional molecular beacon design to enable them to hybridize to a wide range of sequences and generate sequence-specific Tm values. We were able to design probes with stable stem-and-loop conformations, avoiding aberrant secondary structures by introducing a small number of nucleotide substitutions into the probe sequence. The substitutions were chosen to correspond to one or more of the bacterial sequences in our reference set so that at the same time they also increased the range of species to which the probe would hybridize.
We created two sloppy molecular beacons, a 45-nucleotide-long SMB45 with a 5-bp stem and a 60-nucleotide-long V6P3 with a 6-bp stem (see Methods in the supplemental material) which were highly complementary to a segment within the V6 hypervariable region of the bacterial 16S rRNA gene. We then examined their hybridization characteristics utilizing sets of artificial PCR amplicons. The amplicons either had core sequences that were fully complementary to the probes or contained from 1 to 17 nucleotide mismatches. Mismatches were introduced to span the entire target at roughly regular intervals. In addition, to explore the effect of the position of the mismatch within the target sequence, mismatches were omitted from one end, creating an “anchor” sequence over which the SMB probe and targets were perfectly complementary (see Tables S2, S3, and S4 in the supplemental material).
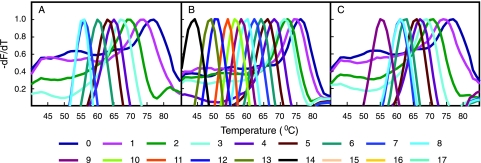
Each sloppy molecular beacon hybridized and generated distinguishable Tm values in the presence of targets that differed by as many as 8 nucleotides for SMB45 with a 45-nucleotide probe (Fig. 2A) and 14 nucleotides for V6P3 with a 60-nucleotide probe region (Fig. 2B); the increased probe lengths of the molecular beacons resulted in a larger number of mismatches being tolerated and still resulted in measurable probe-target hybrid Tm values. Tm values for each probe-target hybrid decreased in a regular fashion with an increasing number of mismatches between the probe and the target, thereby generating distinct Tm values for different probe-target hybrids with various numbers of mismatches for each probe (Fig. 2A and B). We also observed that the presence of a 15-nucleotide-long anchor sequence stabilized the binding of the probe to its targets, increasing Tm values for each mismatch by 1 to 5°C, which increased the number of detectable mismatches by SMB45 to nine (Fig. 2C). Experiments with different anchor lengths and mismatches incorporated into the anchors themselves enabled us to determine that 15- to 18-nucleotide-long anchors were optimal for a balance between the versatility of probe binding and discrimination of the different mismatched targets by the SMBs (data not shown). These results indicate that both sloppy molecular beacon probes with semiconserved anchor sequences and relatively large nonanchored probes generate a wide range of Tm values in the presence of divergent target sequences.
FIG. 2.
Effect of probe-target mismatches on sloppy molecular beacon hybridization. The first derivatives of normalized melting plots are shown for two different sloppy molecular beacons in the presence of different oligonucleotide targets. The Tm value for each hybridization is indicated by the peak of each curve. (A) SMB45 in the presence of targets that do not contain an anchor sequence. (B) V6P3 in the presence of targets that do not contain an anchor sequence. (C) SMB45 in the presence of targets that contain a 15-nucleotide-long anchor sequence at their 3′ ends. Numbers indicate mismatches between the target and probe sequences.
In examining the target sequences in the V6 hypervariable regions of the 133 species included in the test set of our study, we noticed that the high degree of sequence diversity present in this region would require that we use hybridization probes that were able to stably bind to very divergent sequences. Based on our above observations with artificial amplicons, we reasoned that both probes with very long sequences and probes with partial sequence extensions to the relatively conserved regions of the target sequence (i.e., anchored probes) are necessary to generate measurable Tm values from a reasonable number of target sequences. In addition to the long nonanchored V6P3 SMB, which virtually encompassed the entire V6 hypervariable region, we designed the relatively short V6P1 SMB including a 40-nucleotide-long probe with partially conserved sequences that served as a 15-nucleotide anchor. Each of these SMBs produced measurable Tm values in the presence of 57% and 95%, respectively, of the reference set bacteria (see Table S5 in the supplemental material). Both V6P1 and V6P3 tolerated up to 23 mismatches, producing Tm values of 44°C (Burkholderia mallei) and 46°C (Providencia alcalifaciens), respectively, which is attributable to their anchor sequence and long probe length, respectively. In spite of the longer probe lengths, each probe also retained the ability to distinguish between targets that differed by a small number of nucleotides. As expected from our studies with artificial amplicons, there was generally an inverse relationship between the number of probe-target mismatches and Tm value. However, the positional dependence of the mismatches, involvement of the probe stem in forming the probe-target hybrids, and formation of G-T pairings in the bacterial DNA samples introduced an additional level of Tm variability. Thus, in some cases, two targets with identical numbers of mismatches produced different Tm values, and several targets with large number of mismatches produced higher Tm values than targets with comparatively lower numbers of mismatches (as shown in Table 1 for V6P3). This enhanced variability improved the ability of each SMB probe to differentiate among bacterial targets.
TABLE 1.
Illustrative probe target hybrid Tm values and number of mismatches of probe V6P3 with selected bacterial sequences
| Bacteria | Tm (°C) | No. of mismatches to the probe (loop) | G-T pairing | No. of stem nucleotides binding to target sequence |
|---|---|---|---|---|
| Providencia alcalifaciens | 46 | 23 | None | 3 |
| Yersinia pestis | 47 | 21 | None | 3 |
| Shigella flexneri | 47 | 22 | None | 3 |
| Pseudomonas aeruginosa | 48 | 22 | 2 | 6 |
| Acinetobacter calcoaceticus | 49 | 23 | 1 | 6 |
| Citrobacter freundii | 50 | 22 | 1 | 3 |
| Serratia marcescens | 51 | 21 | 2 | 3 |
| Haemophilus influenzae | 52 | 20 | None | 3 |
| Proteus vulgaris | 54 | 20 | 2 | 3 |
| Pasteurella multocida | 55 | 16 | 4 | 3 |
| Listeria grayi | 55 | 14 | 4 | 2 |
| Staphylococcus warneri | 55 | 13 | 4 | 2 |
| Staphylococcus hominis | 56 | 13 | 4 | 2 |
| Staphylococcus lugdunensis | 58 | 12 | 4 | 2 |
| Streptococcus pneumoniae | 60 | 12 | 4 | 2 |
| Streptococcus agalactiae | 61 | 12 | 1 | 2 |
| Staphylococcus haemolyticus | 62 | 10 | 3 | 2 |
| Aerococcus viridans | 63 | 11 | 2 | 2 |
| Bacillus subtilis | 67 | 8 | 3 | 2 |
| Bacillus cereus | 71 | 6 | 2 | 2 |
| Bacillus anthracis | 72 | 5 | 2 | 2 |
Tm signature, DNA sequence types, and bacterial classification.
Concatenating and aligning the target regions of the V3 and V6 hypervariable stretches probed in our assay revealed that the 133 reference set bacteria were represented by 119 distinguishable V3-plus-V6 sequence types. We hypothesized that a small set of SMB probes could be selected so that each target sequence type would generate a unique Tm signature when the Tms produced by each individual SMB were combined. We designed a set of six different SMB probes seeking the maximum amount of Tm information from these 119 sequence types. Thus, in addition to V6P1 and V6P3, we designed four probes, V3P1 and V3P2, targeting the V3 region of 16S rRNA gene, and V6P2 and V6P4, targeting the V6 region of 16S rRNA gene. Each SMB probe, except V6P3, contained an anchor sequence within the probe, and each probe, except V6P1 (which was perfectly complementary to its target in Staphylococcus aureus), contained deliberately introduced mutations so that the probes were not perfectly complementary to specific bacterial sequences.
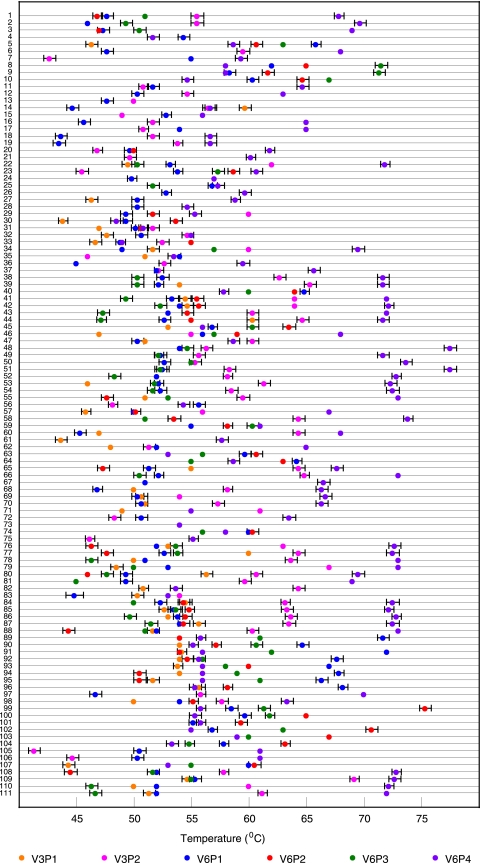
We tested the six-probe assay against DNA (or, in a few cases, artificial PCR target amplicons) from each of the 133 bacterial species in the reference set. This procedure was repeated six different times with different master mixes over a period of 3 months in order to introduce as much experimental variability as possible. The results were used to create a Tm signature look-up table, which listed the mean Tm value that each SMB generated in the presence of each target (see Table S5 in the supplemental material). The assay was then repeated blindly using coded DNA aliquots, and the identity of the samples was determined by calculating the distance index (D) between each unknown Tm signature with signatures in the look-up table. The reference species with the smallest D value identified the blinded sample (Table 2). The assay reproducibly generated 111 Tm signatures out of the 119 distinct sequence types (Fig. 3). A total of 104 of the Tm signatures corresponded to unique sequence types while 7 Tm signatures could not completely resolve DNA sequences but instead specifically identified six groups of two sequence types each and one group of three sequences types. With two exceptions, each group contained bacterial species from the same genera. The exceptions were the species pairs (i) Bartonella henselae and Campylobacter jejuni and (ii) Enterobacter cloacae and Klebsiella oxytoca, which produced indistinguishable Tm signatures even though their target sequences differed marginally. Thus, the assay was highly successful in detecting DNA sequence differences within the region probed by the SMBs. As a consequence, virtually all of the bacterial species within the reference set were either individually identified or else categorized into clinically useful groups of related species (Table 2). Significantly, the Tm values generated by each SMB probe hybridizing to DNA from the same bacterial species was highly reproducible, usually varying less than ±0.5°C (Fig. 3). The species-specific signatures were stable regardless of the quantity of DNA added to the PCR, to a lower limit of detection of ∼500 genome equivalents for different bacteria spiked into blood, or if large amounts (up to 50 mg) of interfering human DNA was present (data not shown).
TABLE 2.
Bacterial species identification using Tm signature D values
| Blinded test species | Match (D value) |
|---|---|
| Acinetobacter baumanii | A. baumanii (0.65) |
| Acinetobacter calcoaceticus | A. calcoaceticus (0.67) |
| Acinetobacter lwoffii | A. lwoffii (0.60) |
| Actinimyces meyeri | A. meyeri (0.47) |
| Aerococcus viridans | A. viridans (0.60) |
| Aeromonas hydrophila | A. hydrophila (0.60) |
| Arcanobacterium pyogenes | A. pyogenes (0.47) |
| Bacillus anthracis | B. anthracis (0.43) |
| Bacillus cereus | B. cereus (0.74) |
| Bacillus subtilis | B. subtilis (0.18) |
| Bartonella henselae/Campylobacter jejuni | B. henselae/C. jejuni (0.37) |
| Bartonella quintana | B. quintana (0.47) |
| Bifidobacterium breve | B. breve (0.33) |
| Bordetella holmesii/Bordetella pertussis/Bordetella parapertussis | |
| B. holmesii/B. pertussis/B. parapertussis (0.71) | |
| Borrelia burgdorferi | B. burgdorferi (0.17) |
| Brucella abortus | B. abortus (0.47) |
| Brucella canis/Brucella melitensis/suis | B. canis/B. melitensis/suis (0.97) |
| Burkholderia cepacia | B. cepacia (0.58) |
| Burkholderia mallei | B. mallei (0.62) |
| Chlamydia psittaci | C. psittaci (2.04) |
| Chlamydia trachomatis | C. trachomatis (0.37) |
| Citrobacter freundii | C. freundii (0.62) |
| Clostridium botulinum | C. botulinum (0.76) |
| Clostridium acetobutylicum | C. acetobutylicum (0.17) |
| Clostridium difficile | C. difficile (0.74) |
| Clostridium perfringens | C. perfringens (0.37) |
| Clostridium septicum | C. septicum (0.50) |
| Clostridium tetani | C. tetani (0.47) |
| Corynebacterium amycolatum | C. amycolatum (1.53) |
| Corynebacterium diptheriae | C. diptheriae (0.78) |
| Corynebacterium jeikeium | C. jeikeium (0.78) |
| Corynebacterium pseudodiptheriticum | C. pseudodiptheriticum (0.82) |
| Corynebacterium xerosis | C. xerosis (0.90) |
| Coxiella burnetti | C. burnetti (1.12) |
| Dermabacter hominis | D. hominis (0.90) |
| Ehrlichia chaffeensis | E. chaffeensis (0.83) |
| Eikenella corrodens | E. corrodens (0.91) |
| Enterobacter aerogenes | E. aerogenes (1.07) |
| Enterobacter cloacae/Klebsiella oxytoca | E. cloacae/K. oxytoca (0.87) |
| Enterococcus spp.a | Enterococcus spp. (0.24) |
| Escherichia coli/Shigella sp. | E. coli/Shigella sp. (1.11) |
| E. coli/Shigella sp. | E. coli/Shigella sp. (1.00) |
| E. coli/Shigella sp. | E. coli/Shigella sp. (1.03) |
| E. coli/Shigella sp. | E. coli/Shigella sp. (0.98) |
| Exiguobacterium sp. | Exiguobacterium sp. (0.68) |
| Franciscella tularensis | F. tularensis (0.00) |
| Fusobacterium necrophorum | F. necrophorum (0.67) |
| Haemophilus aegyptius/Pasteurella multocida | H. aegyptius/P. multocida (1.00) |
| Haemophilus aphrophilus | H. aphrophilus (0.59) |
| Haemophilus ducreyi | H. ducreyi (1.82) |
| Haemophilus influenzae | H. influenzae (0.89) |
| Haemophilus parahemolyticus | H. parahemolyticus (0.37) |
| Haemophilus parainfluenzae | H. parainfluenzae (0.97) |
| Haemophilus paraprophilus | H. paraprophilus (0.83) |
| Hafnia alvei | H. alvei (0.60) |
| Helicobacter pylori | H. pylori (0.50) |
| Kingella kingae | K. kingae (0.85) |
| Klebsiella pneumoniae | K. pneumoniae (1.73) |
| Lactobacillus fermentans | L. fermentans (0.88) |
| Legionella pneumophilla | L. pneumophilla (0.75) |
| Leptospira interrogans/Leptospira kirschneri | L. interrogans/L. kirschneri (0.6) |
| Leptospira biflexa | L. biflexa (0.33) |
| Leuconostoc mesenteroides | L. mesenteroides (1.53) |
| Listeria grayi/Listeria monocytogenes | L. grayi/L. monocytogenes (1.49) |
| Moraxella catarrhalis | M. catarrhalis (1.67) |
| Morganella morganii | M. morganii (0.52) |
| Mycoplasma pneumoniae | M. pneumoniae (0.50) |
| Neisseria gonorrhoeae | N. gonorrhoeae (0.41) |
| Neisseria meningitidis/Neisseria lactamica | N. meningitidis/N. lactamica (1.00) |
| Neisseria mucosa | N. mucosa (0.82) |
| Nocardia brasiliensis/Nocardia farcinica/Nocardia pseudobrasiliensis | |
| N. brasiliensis/N. Farcinica/N. pseudobrasiliensis (1.00) | |
| Oligella urethralis | O. urethralis (2.03) |
| Porphyromonas gingivalis | P. gingivalis (0.00) |
| Prevotella melaninogenica | P. melaninogenica (1.20) |
| Propionibacterium acnes | P. acnes (0.85) |
| Proteus mirabilis | P. mirabilis (0.80) |
| Proteus vulgaris | P. vulgaris (0.97) |
| Providencia alcalifaciens | P. alcalifaciens (0.67) |
| Providencia rettgeri | P. rettgeri (0.50) |
| Pseudomonas aeruginosa | P. aeruginosa (0.99) |
| Psychrobacter faecalis | P. faecalis (1.37) |
| Rhodococcus equi | R. equi (0.82) |
| Rickettsia prowazekii/Rickettsia rickettsii | R. prowazekii/R. rickettsii (0.85) |
| Salmonella arizonae | S. arizonae (1.19) |
| Salmonella enteritidis | S. enteritidis (1.48) |
| Salmonella paratyphi | S. paratyphi (0.96) |
| Salmonella typhi | S. typhi (1.20) |
| Serratia marcescens | S. marcescens (1.14) |
| Staphylococcus aureus | S. aureus (0.97) |
| Staphylococcus capitis/S. epidermidis | S. capitis/S. epidermidis (1.08) |
| Staphylococcus cohnii/S. haemolyticus | S. haemolyticus/S. cohnii (0.83) |
| Staphylococcus hominis | S. hominis (0.82) |
| Staphylococcus lugdunensis | S. lugdunensis (0.83) |
| Staphylococcus sciuri | S. sciuri (0.97) |
| Staphylococcus simulans | S. simulans (1.34) |
| Staphylococcus warneri | S. warneri (1.34) |
| Stenotrophomonas maltophilia | S. maltophilia (1.34) |
| Streptobacillus moniliformis | S. moniliformis (1.42) |
| Streptococcus agalactiae | S. agalactiae (1.02) |
| Streptococcus constellatus | S. constellatus (0.76) |
| Streptococcus equi/mutans | S. equi/S. mutans (0.41) |
| Streptococcus dysgalactiae | S. dysgalactiae (0.69) |
| Streptococcus mitis/S. pneumoniae | S. mitis/S. pneumoniae (1.00) |
| Streptococcus pyogenes/S. uberis | S. pyogenes/S. uberis (0.43) |
| Treponema denticola | T. denticola (0.60) |
| Treponema palidum | T. palidum (0.47) |
| Ureaplasma urealyticum | U. urealyticum (1.03) |
| Vibrio alginolyticus/Vibrio campbellii | V. alginolyticus/V. campbellii (1.03) |
| Vibrio cholerae | V. cholerae (0.71) |
| Yersinia enterocolitica | Y. enterocolitica (0.46) |
| Yersinia pestis | Y. pestis (1.51) |
Enterococcus spp. represents E. avium, E. faecium, E. faecalis, and E. gallinarum.
FIG. 3.
Distribution of Tm signatures among 111 Tm signature types. Each line represents one of 111 Tm signature types. The colored dots along each line correspond to the mean Tm values (from six independent measurements) generated by each of the six sloppy molecular beacon probes in the presence of that sequence type. Error bars show ±1 standard deviation from the mean values. The absence of a dot indicates that the corresponding probe did not generate a Tm value for that particular bacterial DNA. The numbers indicate the Tm profile of the species (species groups) as outlined in Table S6 in the supplemental material.
Blinded studies on clinical samples.
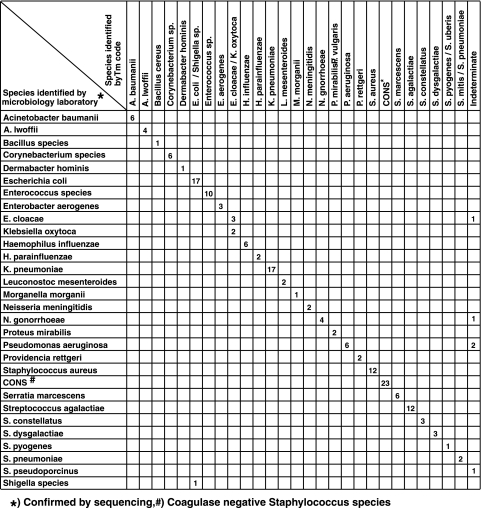
We performed a blinded evaluation of 164 clinical isolates from 158 patients in the course of their clinical diagnostic evaluations. Subsequent decoding showed that these samples represented 19 genera and 41 species, which included virtually all of the bacterial species commonly isolated in clinical laboratories (12, 13) (Fig. 4). Six separate PCR assays, each containing a different SMB probe, were performed with the DNA from each isolate. The resulting Tm signatures were used to generate D values for each isolate. In the relatively rare cases where the Tm signatures were identical in more than one species, D values were used to assign each unknown to a species “group.” To account for the clinical isolates that were not present in our species reference set, we added an indeterminate category to this study, defined as samples generating D values of >5.0. We were able to make correct assignments for 159 out of 164 isolates (a concordance of 97%; 95% confidence interval [CI], 94.3% to 99.6%). The five isolates that could not be correctly identified were assigned to the indeterminate group due to high D values; and no isolate was wrongly identified (specificity, 100%). Examining the five indeterminate samples showed that four consisted of species (two Pseudomonas aeruginosa and one each of E. cloacae and Neisseria gonorrhoeae) that should have been identified. In each case, miss-assignment to the indeterminate category was due to a manual error in deciphering the low Tm values by the blinded data interpreter, causing one or more Tm values to be erroneously entered as zero. DNA sequence analysis of the fifth isolate revealed it to be Streptococcus pseudoporcinus, which was absent from our reference set and, thus, not expected to be detected by our assay. When this one isolate was excluded from our study, the corrected assay concordance was 97.5% (95% CI, 95.2% to 99.9%).
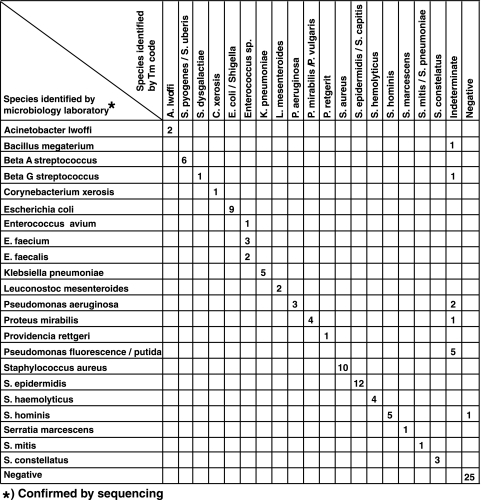
FIG. 4.
Assay performance on cultured clinical isolates. The results of the sloppy molecular beacon assay are compared to a gold standard of microbiology species identification confirmed by 16S rRNA sequencing. Due to space limitations Corynebacterium species represent C. jeikeium, C. diphtheriae, C. xerosis, and C. amycolatum, which were all identified to the species level by our assay; Enterococcus species represent E. faecalis, E. faecium, E. gallinarum, and E. avium; the coagulase-negative staphylococcal species represent S. epidermidis, S. haemolyticus, S. lugdunensis, S. capitis, S. hominis, S. sciuri, and S. cohnii. Two Shigella isolates were not identifiable to the species level and are listed as Shigella species.
As this assay will be particularly useful for rapidly identifying a bacterial species present in a blood culture sample, we performed a blinded assay on 81 positive and 25 negative clinical blood cultures. We expected to find occasional blood cultures that were positive for more than one bacterial species. Therefore, we added a “mixed” category for this study, defined as samples generating more than one Tm peak in any of the six SMB probes. All suspected mixed cultures with double Tm profiles for any probe were visually deconvolved to generate the species-specific signatures of the individual bacterial species present; otherwise, automated species assignments were made using the D value threshold. For deconvolution of the double Tm profiles, all the possible Tm signature patterns resulting from the presence of the double Tm peaks were checked for their D values against the reference set of Tm signatures. The two Tm signature patterns generating the smallest D values identified the bacteria in the mixture. Overall, we were able to correctly identify 76/86 (concordance of 88.4%; 95% CI, 81.6% to 95.1%) of the bacterial species present in the 81 positive cultures (Fig. 5). The assay made the correct species assignment for 67/76 (88%) of the blood cultures which contained a single bacterial species and for 9/10 (90%) of the bacterial species present in the five mixed blood cultures. Of the 10 indeterminate instances, four bacterial species (two P. aeruginosa and one each for Proteus mirabilis and group G Streptococcus) were called indeterminate when they should have been identified by the assay. An additional bacterial isolate (Staphylococcus hominis) was called negative. This single false-negative result occurred in a mixed culture which also contained a Streptococcus dysgalactiae species that was correctly identified. Finally, six isolates, five Pseudomonas putida and one Bacillus megaterium, were called indeterminate. However, we noted that these two species were absent from our reference and, thus, were not expected to be detected. Excluding these last six isolates from our study, the corrected assay concordance was 95.0% (95% CI, 90.2% to 99.8%). Importantly, the specificity of the assay was also high as none of the 25 negative blood cultures resulted in a bacterial species assignment, and there were no misidentifications.
FIG. 5.
Assay performance in tests of clinical blood cultures. The results of the sloppy molecular beacon assay are compared to a gold standard of microbiology species identification confirmed by 16S rRNA sequencing.
DISCUSSION
This study presents a paradigm shift in PCR-based molecular diagnostics. Our approach uses a combination of probes and Tm signature to identify target sequences. Compared to assays which use individual real-time PCR probes to identify individual DNA targets, this approach makes it possible to identify a very large number of different target sequences with a very small number of probes. Our novel probe design strategies which included the use of anchor sequences or very long probe sequences (up to 60 nucleotides in length) were critical to its success. These modifications expanded the range of sequences to which the sloppy probes could hybridize while providing sufficient probe-target stability to generate reproducible Tm values and permit single nucleotide sequence discrimination. The variable length of the anchor sequences in the different probes and presence of mismatches in the anchor sequences themselves added to the differential sequence discrimination and hybridization versatility of the probes. The “sloppiness” of the probe hybridizations did not affect the overall assay precision, which resulted in a high level of sensitivity and specificity in our validation studies on clinical isolates and blood cultures, with not a single bacterial isolate being misidentified. This is attributable to the novel Tm signature principle combined with the distance index paradigm that we used in identifying the bacterial species. Thus, each sequence was identified by a distinct and unique combination of six different Tm values. In blinded assays, any deviant or missing Tm values in the Tm signature for any sequence always resulted in unusual D values, which easily enabled us to identify the assay as indeterminate and prevented false positive misinterpretations.
Our assay also identified most instances where two different bacterial species were present in the same blood culture by deconvolving double peaks into Tm values that were characteristic for two different bacterial species. This is because judicious designing of probes caused most bacteria to generate distinctly different Tm values for the same probe, generating discrete double peaks in the case of mixed DNA samples. Mixed blood cultures are uncommon, and when they occur, mixtures are often due to the presence of a contaminating commensal skin organism plus the true pathogen (18). We have confirmed in DNA spiking studies that we can identify the presence of common commensals when they are mixed with any of the bacterial species frequently seen in blood cultures (data not shown). It is interesting that the only bacterial species that we could not detect in a blood culture mixture was also the commensal S. hominis, which we suspect to be a contaminant present at a very low level in the clinical blood culture.
Sloppy molecular beacons represent one of several new approaches that have been proposed for rapid pathogen detection and speciation. A number of commercial and in-house molecular diagnostic PCR, ligase chain reaction, nucleic acid sequence-based amplification (22, 32), and pyrosequencing assays (4) for bloodstream infections have recently been described. These approaches work well in research settings or in clinical laboratories with substantial technical capacity; however, none of these methods combine the range of bacterial species identification, robustness, and the ease of use that is possible using our approach. Newer technologies including matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectroscopy, DNA microarrays, deep sequencing, and technologies like PCR coupled with electrospray ionization mass spectrometry may provide additional diagnostic modalities (1, 6, 7, 21, 31). However, these technologies are currently too costly, complex, and/or time-consuming for clinical use; and it is not yet clear whether they will ever offer a substantial advantage over our diagnostic method that requires little more than a real-time PCR.
Our current assay has some limitations. Several pairs of bacterial species could not be distinguished. These few pairs of organisms were identified as small groups rather than as individual species. A seventh sloppy molecular beacon designed to probe an additional target or sequence-specific modification of the existing probe sequences would be expected to resolve these remaining groups into individual species. The assay is clearly not able to identify a bacterial species which is not present in the reference set, which as a matter of fact increases its specificity. This would recur rarely once large collections of clinical isolates are tested and new species-specific signatures are added to the reference set. The assay was also not able to identify a few clinical isolates that were present in our reference set. This was attributable to manual errors in Tm determination, usually in the few Tm values that were below 45°C. Improved instrumentation and automated Tm identification would resolve this only true source of error. The assay has a sensitivity and specificity that rival conventional species identification performed in clinical laboratories (5, 14, 25, 28) even with the current limitations. However, the assay is far more rapid and has the capability of being far more sensitive than conventional assays, suggesting that our universal bacterial identification system can provide a significant advance to infectious disease diagnostics.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health grants U0I-AI-056689 and U0I-AI-175490.
D.A., F.R.K., and S.A.E.M. are among a group of inventors who earn royalties for molecular beacon usage.
Footnotes
Published ahead of print on 18 November 2009.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Baldwin, C. D., G. B. Howe, R. Sampath, L. B. Blyn, H. Matthews, V. Harpin, T. A. Hall, J. J. Drader, S. A. Hofstadler, M. W. Eshoo, K. Rudnick, K. Studarus, D. Moore, S. Abbott, J. M. Janda, and C. A. Whitehouse. 2009. Usefulness of multilocus polymerase chain reaction followed by electrospray ionization mass spectrometry to identify a diverse panel of bacterial isolates. Diagn. Microbiol. Infect. Dis. 63:403-408. [DOI] [PubMed] [Google Scholar]
- 2.Bosshard, P. P., R. Zbinden, S. Abels, B. Boddinghaus, M. Altwegg, and E. C. Bottger. 2006. 16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting Gram-negative bacteria in the clinical laboratory. J. Clin. Microbiol. 44:1359-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakravorty, S., D. Helb, M. Burday, N. Connell, and D. Alland. 2007. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 69:330-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claesson, M. J., O. O'Sullivan, Q. Wang, J. Nikkila, J. R. Marchesi, H. Smidt, W. M. de Vos, R. P. Ross, and P. W. O'Toole. 2009. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.d'Azevedo, P. A., C. A. Dias, A. L. Goncalves, F. Rowe, and L. M. Teixeira. 2001. Evaluation of an automated system for the identification and antimicrobial susceptibility testing of enterococci. Diagn. Microbiol. Infect. Dis. 40:157-161. [DOI] [PubMed] [Google Scholar]
- 6.Degand, N., E. Carbonnelle, B. Dauphin, J. L. Beretti, M. Le Bourgeois, I. Sermet-Gaudelus, C. Segonds, P. Berche, X. Nassif, and A. Ferroni. 2008. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J. Clin. Microbiol. 46:3361-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards, J. R., H. Ruparel, and J. Ju. 2005. Mass-spectrometry DNA sequencing. Mutat. Res. 573:3-12. [DOI] [PubMed] [Google Scholar]
- 8.Edwards, K. J., M. E. Kaufmann, and N. A. Saunders. 2001. Rapid and accurate identification of coagulase-negative staphylococci by real-time PCR. J. Clin. Microbiol. 39:3047-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Hajj, H. H., S. A. Marras, S. Tyagi, E. Shashkina, M. Kamboj, T. E. Kiehn, M. S. Glickman, F. R. Kramer, and D. Alland. 2009. Identification of mycobacterial species utilizing sloppy molecular beacon probes. J. Clin. Microbiol. [DOI] [PMC free article] [PubMed]
- 10.Fridkin, S. K., J. R. Edwards, F. C. Tenover, R. P. Gaynes, and J. E. McGowan, Jr. 2001. Antimicrobial resistance prevalence rates in hospital antibiograms reflect prevalence rates among pathogens associated with hospital-acquired infections. Clin. Infect. Dis. 33:324-330. [DOI] [PubMed] [Google Scholar]
- 11.Fujita, S., Y. Senda, T. Iwagami, and T. Hashimoto. 2005. Rapid identification of staphylococcal strains from positive-testing blood culture bottles by internal transcribed spacer PCR followed by microchip gel electrophoresis. J. Clin. Microbiol. 43:1149-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jorgensen, J. H. 2004. Need for susceptibility testing guidelines for fastidious or less-frequently isolated bacteria. J. Clin. Microbiol. 42:493-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlowsky, J. A., M. E. Jones, D. C. Draghi, C. Thornsberry, D. F. Sahm, and G. A. Volturo. 2004. Prevalence and antimicrobial susceptibilities of bacteria isolated from blood cultures of hospitalized patients in the United States in 2002. Ann. Clin. Microbiol. Antimicrob. 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, M., S. R. Heo, S. H. Choi, H. Kwon, J. S. Park, M. W. Seong, D. H. Lee, K. U. Park, J. Song, and E. C. Kim. 2008. Comparison of the MicroScan, VITEK 2, and Crystal GP with 16S rRNA sequencing and MicroSeq 500 v2.0 analysis for coagulase-negative staphylococci. BMC Microbiol. 8:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klouche, M., and U. Schroder. 2008. Rapid methods for diagnosis of bloodstream infections. Clin. Chem. Lab. Med. 46:888-908. [DOI] [PubMed] [Google Scholar]
- 16.Kolbert, C. P., and D. H. Persing. 1999. Ribosomal DNA sequencing as a tool for identification of bacterial pathogens. Curr. Opin. Microbiol. 2:299-305. [DOI] [PubMed] [Google Scholar]
- 17.Kollef, M. H., G. Sherman, S. Ward, and V. J. Fraser. 1999. Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients. Chest 115:462-474. [DOI] [PubMed] [Google Scholar]
- 18.Lehmann, L. E., K. P. Hunfeld, T. Emrich, G. Haberhausen, H. Wissing, A. Hoeft, and F. Stuber. 2008. A multiplex real-time PCR assay for rapid detection and differentiation of 25 bacterial and fungal pathogens from whole blood samples. Med. Microbiol. Immunol. 197:313-324. [DOI] [PubMed] [Google Scholar]
- 19.Loeb, M., D. W. Bentley, S. Bradley, K. Crossley, R. Garibaldi, N. Gantz, A. McGeer, R. R. Muder, J. Mylotte, L. E. Nicolle, B. Nurse, S. Paton, A. E. Simor, P. Smith, and L. Strausbaugh. 2001. Development of minimum criteria for the initiation of antibiotics in residents of long-term-care facilities: results of a consensus conference. Infect. Control Hosp. Epidemiol. 22:120-124. [DOI] [PubMed] [Google Scholar]
- 20.Mangold, K. A., M. Regner, M. Tajuddin, A. M. Tajuddin, L. Jennings, H. Du, and K. L. Kaul. 2007. Neisseria species identification assay for the confirmation of Neisseria gonorrhoeae-positive results of the COBAS Amplicor PCR. J. Clin. Microbiol. 45:1403-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Out, A. A., I. J. van Minderhout, J. J. Goeman, Y. Ariyurek, S. Ossowski, K. Schneeberger, D. Weigel, M. van Galen, P. E. Taschner, C. M. Tops, M. H. Breuning, G. J. van Ommen, J. T. den Dunnen, P. Devilee, and F. J. Hes. 4 September 2009. Deep sequencing to reveal new variants in pooled DNA samples. Hum. Mutat. doi: 10.1002/humu.21122. [Epub ahead of print.] [DOI] [PubMed]
- 22.Pfaller, M. A. 2001. Molecular approaches to diagnosing and managing infectious diseases: practicality and costs. Emerg. Infect. Dis. 7:312-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pierce, K. E., J. A. Sanchez, J. E. Rice, and L. J. Wangh. 2005. Linear-after-the-exponential (LATE)-PCR: primer design criteria for high yields of specific single-stranded DNA and improved real-time detection. Proc. Natl. Acad. Sci. U. S. A. 102:8609-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rafi, W., A. Chandramuki, R. Mani, P. Satishchandra, and S. K. Shankar. 2008. Rapid diagnosis of acute bacterial meningitis: role of a broad range 16S rRNA polymerase chain reaction. J. Emerg. Med. doi: 10.1016/j/jemermed.2008.02.053. [DOI] [PubMed]
- 25.Rhoads, S., L. Marinelli, C. A. Imperatrice, and I. Nachamkin. 1995. Comparison of MicroScan WalkAway system and Vitek system for identification of gram-negative bacteria. J. Clin. Microbiol. 33:3044-3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sen, K. 2000. Rapid identification of Yersinia enterocolitica in blood by the 5′ nuclease PCR assay. J. Clin. Microbiol. 38:1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shang, S., G. Chen, Y. Wu, L. Du, and Z. Zhao. 2005. Rapid diagnosis of bacterial sepsis with PCR amplification and microarray hybridization in 16S rRNA gene. Pediatr. Res. 58:143-148. [DOI] [PubMed] [Google Scholar]
- 28.Snyder, J. W., G. K. Munier, and C. L. Johnson. 2008. Direct comparison of the BD phoenix system with the MicroScan WalkAway system for identification and antimicrobial susceptibility testing of Enterobacteriaceae and nonfermentative gram-negative organisms. J. Clin. Microbiol. 46:2327-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, M., B. Cao, Q. Yu, L. Liu, Q. Gao, L. Wang, and L. Feng. 2008. Analysis of the 16S-23S rRNA gene internal transcribed spacer region in Klebsiella species. J. Clin. Microbiol. 46:3555-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wellinghausen, N., B. Wirths, A. Essig, and L. Wassill. 2004. Evaluation of the Hyplex BloodScreen multiplex PCR-enzyme-linked immunosorbent assay system for direct identification of gram-positive cocci and gram-negative bacilli from positive blood cultures. J. Clin. Microbiol. 42:3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoo, S. M., J. Y. Choi, J. K. Yun, J. K. Choi, S. Y. Shin, K. Lee, J. M. Kim, and S. Y. Lee. 7 October 2009. DNA microarray-based identification of bacterial and fungal pathogens in bloodstream infections. Mol. Cell Probes. doi: 10.1016/j.mcp.2009.09.003. [Epub ahead of print.] [DOI] [PubMed]
- 32.Zhao, Y., S. Park, B. N. Kreiswirth, C. C. Ginocchio, R. Veyret, A. Laayoun, A. Troesch, and D. S. Perlin. 2009. Rapid real-time nucleic Acid sequence-based amplification-molecular beacon platform to detect fungal and bacterial bloodstream infections. J. Clin. Microbiol. 47:2067-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zinner, S. H. 2007. Antibiotic use: present and future. New Microbiol. 30:321-325. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.