Abstract
A novel chromogenic medium, Spectra MRSA (Remel, Lenexa, KS), was designed to detect methicillin-resistant Staphylococcus aureus (MRSA) rapidly and more efficiently than traditional media (i.e., tryptic soy agar with 5% sheep blood [SBA] and mannitol salt agar [MSA]). A multicenter study (including four clinical trial sites and the Medical College of Wisconsin [MCW] Milwaukee, WI) compared the performance characteristics of Spectra MRSA to those of the traditional media for the detection of MRSA. For this study, 767 nasal swab specimens from the multicenter study (traditional medium used, SBA) and 667 nasal swab specimens from MCW (traditional medium used, MSA) were plated on each test medium and examined after 24 and 48 h of incubation. At 24 h, the sensitivity and the specificity of each medium were as follows: in the multicenter study, 95.4% and 99.7%, respectively, for Spectra MRSA and 93.6% and 100%, respectively, for SBA; at MCW, 95.2% and 99.5%, respectively, for Spectra MRSA and 88.7% and 94.0%, respectively, for MSA. The positive predictive values of each medium at 24 h were as follows: in the multicenter study, 98.1% for Spectra MRSA and 100% for SBA; at MCW, 95.2% for Spectra MRSA and 60.4% for MSA. In our evaluation, we found that Spectra MRSA was able to rapidly identify and differentiate methicillin-resistant S. aureus from methicillin-susceptible S. aureus on the basis of the utilization of chromogens that result in denim blue colonies, thus eliminating the need for biochemical analysis and antimicrobial susceptibility testing. Extending the incubation beyond 24 h did not significantly improve the recovery of MRSA and resulted in decreased specificity.
A significant effort has been put forth to determine effective infection control practices that may be used to limit the spread of methicillin-resistant Staphylococcus aureus (MRSA) and minimize its impact on patient care and hospital budgets in response to the increasing rates of occurrence of MRSA in health care settings. Although most hospitals adhere to a policy of contact isolation and attempt to limit inappropriate antimicrobial usage, there is strong evidence that active surveillance cultures (ASCs) for patients at risk for MRSA colonization can increase the chance of identifying occult MRSA reservoirs and further limit the nosocomial spread of the organism (16). Recent reports by Salgado and Farr (17) and Lucet et al. (13) have shown that the organisms in positive cultures of clinical specimens routinely submitted from patients represent only a small fraction of the reservoir of antibiotic-resistant pathogens, and the largest source for nosocomial spread was attributed to asymptomatic, colonized patients who went unrecognized and unisolated in the absence of ASCs. Additionally, consensus suggests that a restricted formulary alone is unlikely to prevent the emergence and persistence of MRSA and that the use of contact precautions is important to prevent the spread of MRSA from colonized patients (5).
While the anterior nares are the most common site of S. aureus colonization and the most frequently screened and recommended site for specimen collection due to the satisfactory sensitivities of tests with that type of specimen and the ease with the specimen may be obtained (15, 18, 20), other recent studies have shown that sampling of other body sites (the oropharynx, perianal region, and groin) may enhance the sensitivity of screening for MRSA and predict the likelihood of S. aureus infection (3, 7, 11, 14, 22). Most importantly, it is agreed that the culture of specimens from more body sites would yield higher rates of recovery, but the increased resources required for the screening of specimens from multiple sites outweigh the incremental increase in yield that would be attained (5). In order to reduce the economic burden related to longer patient stays and higher costs associated with therapy and infection control, rapid and accurate tests for screening for MRSA are needed to guide intervention and decrease delays in the implementation of contact precautions. A lengthy turnaround time would further augment the risk for nosocomial transmission; thus, any effective screening protocol should be deemed to have a turnaround time within 24 h.
The methods currently available for screening for MRSA include standard culture, methods that use chromogenic media, and molecular-based testing. Although PCR methods have been highly acclaimed in the recent literature for their sensitivity and speed, the cost per test and the up-front laboratory cost of implementation are very high and may generate significant pressure to demonstrate cost savings in an unrealistic time frame (5). Culture methods that employ a selective chromogenic medium have been described to be less costly, more reliable, and faster for screening for MRSA than traditional culture (5, 8). The purpose of the multicenter study described here was to compare the performance of a novel chromogenic medium, Spectra MRSA, with that of traditional culture and validate the use of this product for screening for MRSA from nasal swab specimens.
MATERIALS AND METHODS
Patient selection criteria for study inclusion.
Anterior nasal swab specimens for culture were collected from the following patient populations at each participating trial site: new hospital admissions, prior hospital admissions, admissions to the intensive care unit, patients with a history of MRSA colonization, and patients transferred from a health care setting where MRSA is known to be present. One specimen from each asymptomatic patient screened for MRSA was included in the study. This study was approved by the institutional review board of each participating institution.
Collection of nasal swab specimens and medium inoculation protocol.
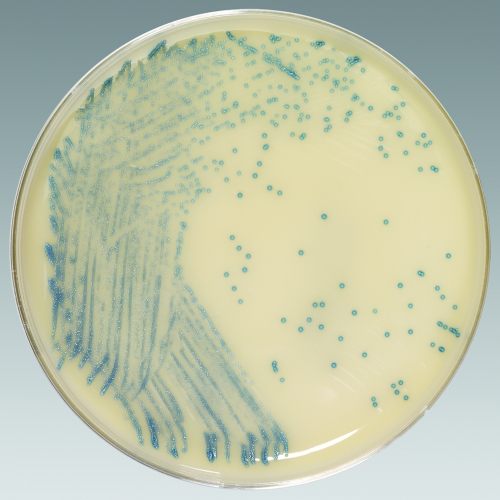
Nasal swab specimens from 767 patients at four geographically distinct trial sites and 667 patients at the Medical College of Wisconsin (MCW; Milwaukee, WI) were screened for the presence of MRSA. All participating institutions used single or double swab specimens from the anterior nares for culture. The swabs were transported in liquid Stuart or Amies gel medium and were held at 4°C for up to 24 h before they were plated. Different “gold standard” media were used at the participating clinical trial sites for MRSA detection. Four clinical trial sites (sites A to D) utilized tryptic soy agar with 5% sheep blood (SBA) as the gold standard medium; and MCW used mannitol salt agar (MSA; Remel, Lenexa, KS), a selective growth medium for Staphylococcus aureus, as the gold standard medium. Spectra MRSA (Remel) was evaluated at all participating trial sites as an experimental agar medium that may be used to screen for MRSA by chromogenically distinguishing MRSA colonies as denim blue (Fig. 1). The plates were directly inoculated by rolling the collection swab on the first quadrant of the medium and streaking for isolation by the quadrant technique. At all participating trial sites, the gold standard medium was always inoculated prior to inoculating Spectra MRSA.
FIG. 1.
Spectra MRSA medium plated with methicillin-resistant Staphylococcus aureus. The ability of Spectra MRSA to identify methicillin-resistant S. aureus based on the color of the colony, in which colonies of methicillin-resistant S. aureus appear denim blue, is shown. For the purposes of the study, nasal swab specimens were plated directly onto Spectra MRSA and a gold standard medium (SBA or MSA), and the plates were observed for growth at 24 and 48 h.
Identification of MRSA on gold standard media and Spectra MRSA.
Following incubation at 35°C in ambient air for 24 and 48 h, the colonies on Spectra MRSA were screened for a denim blue appearance, indicating MRSA. Suspected S. aureus colonies on SBA were catalase-positive, gram-positive cocci that demonstrated beta-hemolysis (1). Components of MSA, such as mannitol and a high NaCl concentration (7.5%), select for the growth of pathogenic staphylococci (i.e., S. aureus), which produce characteristic yellow-pigmented colonies (1). Following observation of the characteristic denim blue colonies on Spectra MRSA and the characteristic growth on the gold standard medium (as outlined above), all suspected colonies indicating MRSA (on Spectra MRSA) and S. aureus (on a gold standard medium) were subcultured to SBA. Following incubation at 35°C for 18 to 24 h, confirmatory identification and antimicrobial susceptibility testing were performed.
Confirmatory identification and antimicrobial susceptibility testing.
Confirmatory identification of S. aureus from all media evaluated was performed by the latex agglutination test for the detection of clumping factor and protein A (Staphaurex; Remel). Two susceptibility testing methods were performed to confirm the oxacillin resistance of all latex agglutination test-positive isolates from each medium. Latex agglutination test-negative isolates confirmed to be S. aureus following full genus and species identification were also subjected to susceptibility testing. The first susceptibility testing method included the detection of penicillin-binding protein 2a with the Oxoid PBP 2′ latex agglutination test kit (Remel); testing was performed according to the manufacturer's instructions. In addition, the oxacillin Etest (bioMérieux, Durham, NC) was performed to determine the MIC of each colony. A bacterial suspension equivalent to a 0.5 McFarland standard was prepared and thoroughly streaked onto Mueller-Hinton agar with 2% NaCl (MHA; Remel) with a sterilized Dacron swab. An oxacillin test strip was placed on the inoculated MHA, and the plate was incubated at 35°C in ambient air for 24 h. The MIC was interpreted after 24 h of incubation. The breakpoints for MRSA were interpreted as defined by the Clinical and Laboratory Standards Institute by using the MICs for oxacillin. MICs were interpreted as follows: ≤2 μg/ml, oxacillin sensitive; ≥4 μg/ml, oxacillin resistant; and values between 2 μg/ml and 4 μg/ml, intermediate oxacillin resistance (4).
Data interpretation and statistical analysis.
A true-positive (TP) result was defined as a denim blue colony on Spectra MRSA that demonstrated characteristic features on the gold standard media (as described above) and that was identified as MRSA by the Oxoid PBP 2′ test and the oxacillin Etest (≥4 μg/ml). A false positive (FP) was defined as an isolate that exhibited typical coloration on the respective medium but was not identified as representing vancomycin-resistant enterococci by Vitek 2 or antimicrobial susceptibility testing. The result for an isolate that was confirmed to be MRSA on one medium but that did not grow on another medium was considered false negative (FN). A true-negative (TN) result was defined as the lack of typical performance characteristics on any medium. Growth characteristics combined with antimicrobial susceptibility testing results were used to calculate the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of each medium evaluated (SBA, MSA, and Spectra MRSA) at 24 and 48 h. In addition, MCW calculated the analytical specificity of Spectra MRSA. Thirteen different organisms (totaling 27 isolates) were streaked onto Spectra MRSA plates by the quadrant technique, and the plates were incubated for 48 h. Observation for denim blue colonies was performed at 24 and 48 h.
RESULTS
Spectra MRSA performance characteristics.
Four geographically distinct clinical trial sites (sites A to D) yielded a total of 767 patients who were screened for nasal colonization with MRSA by using Spectra MRSA and SBA media. Following confirmation of the identity of the organism as S. aureus and oxacillin resistance, the results from sites A to D were combined to determine the sensitivities, specificities, PPVs, and NPVs at 24 and 48 h. The values for sensitivity of Spectra MRSA for the detection of MRSA at 24 and 48 h were 95.4% and 98.2%, respectively (Table 1). The sensitivities of SBA, the gold standard medium for the detection of MRSA at sites A to D, were 93.6% at 24 h and 96.3% at 48 h (Table 1). In addition, Spectra MRSA detected four MRSA colonies that were not recovered by the use of SBA. As with the sensitivity analysis, the specificity of Spectra MRSA compared to that of SBA was calculated at 24 and 48 h. The specificity analysis showed specificities of Spectra MRSA of 99.7% at 24 h and 99.2% at 48 h (Table 1). The specificity of SBA at 24 h was 100% and remained 100% after 48 h incubation (Table 1). The PPVs and NPVs of both media are shown in Table 1. Analyses of Spectra MRSA and SBA were also performed independently for clinical sites A to D to determine the sensitivities, specificities, PPVs, and NPVs. The results for the individual site are shown in Table 2.
TABLE 1.
Sensitivities, specificities, PPVs, and NPVs of Spectra MRSA and SBA for detection of MRSA from 767 nasal swab cultures at 24 and 48 h with combined data from the clinical trial sitesa
| Incubation period (h) | Medium | No. of samples with the following result: |
Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| TPa | FP | TN | FN | ||||||
| 24 | Spectra MRSA | 104 | 2 | 656 | 5 | 95.4 | 99.7 | 98.1 | 99.2 |
| SBA | 102 | 0 | 658 | 7 | 93.6 | 100.0 | 100.0 | 98.9 | |
| 48 | Spectra MRSA | 107 | 1 | 648 | 2 | 98.2 | 99.2 | 91.5 | 99.7 |
| SBA | 105 | 1 | 657 | 4 | 96.3 | 100.0 | 99.1 | 99.4 | |
A TP result is defined as a denim blue colony on Spectra MRSA that demonstrated characteristic features on the gold standard media (as described in Materials and Methods) and that was identified as MRSA by the Oxoid PBP 2′ test and the oxacillin Etest (≥4 μg/ml).
TABLE 2.
Sensitivities, specificities, PPVs, and NPVs of Spectra MRSA and SBA for detection of MRSA at 24 and 48 h for the individual clinical trial sites
| Clinical trial site and incubation period (h) | Medium | No. of samples with the following result: |
Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| TPa | FP | TN | FN | ||||||
| Site A | |||||||||
| 24 | Spectra MRSA | 29 | 0 | 91 | 0 | 100.0 | 100.0 | 100.0 | 100.0 |
| SBA | 29 | 0 | 91 | 0 | 100.0 | 100.0 | 100.0 | 100.0 | |
| 48 | Spectra MRSA | 29 | 0 | 91 | 0 | 100.0 | 100.0 | 100.0 | 100.0 |
| SBA | 29 | 0 | 91 | 0 | 100.0 | 100.0 | 100.0 | 100.0 | |
| Site B | |||||||||
| 24 | Spectra MRSA | 29 | 2 | 171 | 1 | 96.7 | 98.8 | 93.5 | 99.4 |
| SBA | 28 | 0 | 173 | 2 | 93.3 | 100.0 | 100.0 | 98.9 | |
| 48 | Spectra MRSA | 30 | 4 | 169 | 0 | 100.0 | 8.2 | 88.2 | 100.0 |
| SBA | 28 | 0 | 173 | 2 | 93.3 | 100.0 | 100.0 | 98.9 | |
| Site C | |||||||||
| 24 | Spectra MRSA | 15 | 0 | 155 | 1 | 93.8 | 100.0 | 100.0 | 99.4 |
| SBA | 16 | 0 | 155 | 0 | 100.0 | 100.0 | 100.0 | 100.0 | |
| 48 | Spectra MRSA | 16 | 1 | 154 | 0 | 100.0 | 99.4 | 94.1 | 100.0 |
| SBA | 16 | 0 | 155 | 0 | 100.0 | 100.0 | 100.0 | 100.0 | |
| Site D | |||||||||
| 24 | Spectra MRSA | 31 | 0 | 239 | 3 | 91.2 | 100.0 | 100.0 | 98.8 |
| SBA | 29 | 0 | 239 | 5 | 85.3 | 100.0 | 100.0 | 97.6 | |
| 48 | Spectra MRSA | 32 | 5 | 234 | 2 | 94.1 | 97.9 | 86.5 | 99.2 |
| SBA | 32 | 1 | 238 | 2 | 94.1 | 99.6 | 97.0 | 99.2 | |
A TP result is defined as a denim blue colony on Spectra MRSA that demonstrated characteristic features on the gold standard media (as described in Materials and Methods) and that was identified as MRSA by the Oxoid PBP 2′ test and the oxacillin Etest (≥4 μg/ml).
MCW yielded a total of 667 patients who were screened for nasal colonization with MRSA by using Spectra MRSA and MSA. Statistical analysis for each medium was performed at 24 h and 48 h, including the sensitivity, specificity, PPV, and NPV. Following confirmation of the identity of the organism as S. aureus and oxacillin resistance, the sensitivities of Spectra MRSA for the detection of MRSA at 24 h and 48 h were 95.2% and 96.0%, respectively (Table 3). The sensitivities of MSA, the gold standard medium used for the detection of MRSA, were 88.7% at 24 h and 87.9% at 48 h (Table 3). In addition, Spectra MRSA detected five MRSA colonies that were not recovered on MSA. The specificities of Spectra MRSA were 99.5% at 24 h and 95.2% at 48 h (Table 3). The specificities of MSA at 24 h and 48 h were 94.0% and 90.5%, respectively. The PPVs and NPVs of both media are shown in Table 3.
TABLE 3.
Sensitivities, specificities, PPVs, and NPVs of Spectra MRSA and MSA for detection of MRSA from 667 nasal swab cultures at 24 and 48 h conducted by MCW
| Incubation period (h) | Medium | No. of samples with the following result: |
Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |||
|---|---|---|---|---|---|---|---|---|---|
| TPa | FP | TN | FN | ||||||
| 24 | Spectra MRSA | 59 | 3 | 602 | 3 | 95.2 | 99.5 | 95.2 | 99.5 |
| MSA | 55 | 36 | 569 | 7 | 88.7 | 94.0 | 60.4 | 98.8 | |
| 48 | Spectra MRSA | 95 | 27 | 541 | 4 | 96.0 | 95.2 | 77.9 | 99.3 |
| MSA | 87 | 54 | 514 | 12 | 87.9 | 90.5 | 61.7 | 97.7 | |
A TP result is defined as a denim blue colony on Spectra MRSA that demonstrated characteristic features on the gold standard media (as described in Materials and Methods) and that was identified as MRSA by the Oxoid PBP 2′ test and the oxacillin Etest (≥4 μg/ml).
Furthermore, MCW determined the analytical specificity of Spectra MRSA. The following organisms were plated and incubated for 48 h: coagulase-negative staphylococci (10 isolates), methicillin-susceptible Staphylococcus aureus (3 isolates), Klebsiella spp. (2 isolates), viridans group streptococci (2 isolates), Streptococcus pneumonia (2 isolates), a Proteus sp., Haemophilus influenzae, a Corynebacterium sp., Moraxella spp., Escherichia coli, a Pseudomonas sp., Streptococcus pyogenes, and a Enterococcus sp. The growth of denim blue colonies was not observed on any of the plates inoculated with these organisms at 24 h and 48 h.
DISCUSSION
The incidence of methicillin-resistant Staphylococcus aureus has increased to epidemic proportions over the last two decades in both the hospital setting and the community setting (6). Consequently, a major financial burden resulting from prolonged hospital stays is placed upon the health care system (21). In addition, invasive MRSA infections result in significantly increased rates of morbidity and mortality (21). A recent report by Klevens et al. (12) estimated that in 2007, more than 94,000 life-threatening infections and nearly 19,000 deaths in the United States were attributed to MRSA, and most of these were connected with health care settings. Preventative measures, including contact isolation, proper antimicrobial usage, and hand-washing, to curb the nosocomial spread of MRSA throughout the health care setting have been evaluated (19). However, despite the implementation of education and preventative measures, the spread of MRSA via infected and asymptomatically colonized patients continues to be a problem in the health care environment. Many recent studies have shown that the use of effective control measures, including the systematic screening of persons exposed to MRSA, can confine or even eliminate the nosocomial spread of MRSA (9). Therefore, the rapid and accurate identification of individuals harboring MRSA is paramount for effective patient care and prevention of its further spread.
The traditional methods employed to identify MRSA include standard bacteriological culture with selective and/or differential medium (i.e., SBA or MSA), followed by morphological assessment and determination of biochemical characteristics and antimicrobial susceptibilities. However, identification and susceptibility testing usually require 2 to 4 days to achieve definitive results. Other methods used for MRSA detection, including conventional PCR technology, amplify the DNA obtained from clinical isolates by targeting the MRSA-specific mecA gene and an S. aureus species-specific marker (i.e., the nuc gene). Conventional PCR requires extensive, meticulous labor that is prone to the potential for contamination due to the need for post-PCR processing (6). Clearly, traditional bacteriological and conventional PCR methods are time-consuming and labor-intensive, thus failing to provide the information necessary to make timely decisions on the isolation procedures and effective antimicrobial chemotherapy that should be used (10).
FDA-approved real-time PCR assays (RT-PCR) (i.e., BD GeneOhm MRSA PCR; GeneXpert) have proven to be excellent methods for the rapid detection of MRSA. With its rapid turnaround time and exceptional testing accuracy, RT-PCR has proven to be ideal in a clinical setting of moderate MRSA endemicity where large numbers of screens need to be processed on a daily basis. However, the implementation of RT-PCR in small, community-based hospitals may be unfeasible due to limited work space (i.e., separate rooms for pre-PCR, PCR, and post-PCR work to prevent amplicon contamination), up-front costs, and the cost per test. The expenses and workload of a single PCR exceed the demands of testing one clinical specimen for the presence of MRSA (10). Bischof et al. (2) compared the following chromogenic media to BD GeneOhm MRSA (BD Diagnostics, San Diego, CA) for the detection of MRSA from nasal swab specimens: CHROMagar MRSA (BBL; Becton Dickinson, Franklin Lakes, NJ), MRSASelect (Bio-Rad, Hercules, CA), Spectra MRSA, and MSA-FX. A clear analytical advantage between the use of RT-PCR and chromogenic media was not demonstrated, and the investigators thus concluded that laboratory-specific factors (i.e., costs, turnaround time, and technical expertise) would play a significant role in deciding between screening methods.
Clinical trial sites A to D compared the performance of Spectra MRSA to that of the gold standard medium, SBA, for the detection MRSA from nasal swab specimens. The sensitivities of Spectra MRSA at 24 and 48 h were 95.4% and 98.2%, respectively, representing results better than those achieved with SBA, which had sensitivities of 93.6% at 24 h and 96.3% at 48 h. A total of five false-negative results were reported on Spectra MRSA from sites B to D at 24 h. Site D produced three false-negative results, and these were attributed to the inoculation of multiple plates with specimens containing low densities of MRSA and the fact that Spectra MRSA plates were inoculated last. The remaining two false-positive (FP) results (sites B and C, each of which produced one FP result) were attributed to a failure to correctly identify denim blue colonies, an inherent limitation of any subjective test. Underlying this issue is the importance of technician training to correctly identify the presence of denim blue colonies. Comparison of the specificity of Spectra MRSA and SBA indicated that the differences were negligible, with the specificities being 99.7% at 24 h and 99.2% at 48 h for Spectra MRSA and 100% at both 24 h and 48 h for SBA. However, methicillin-sensitive S. aureus and coagulase-negative Staphylococcus sp. colonies were commonly observed on SBA medium at 24 h, thus necessitating the need for identification and resistance confirmation.
The Medical College of Wisconsin compared the performance of Spectra MRSA to that of the gold standard medium MSA for the detection of MRSA from nasal swab specimens. The sensitivities of Spectra MRSA at 24 and 48 h were 95.4% and 96.0%, respectively, producing results far superior to those achieved with MSA, which resulted in an 88.7% sensitivity at 24 h and an 87.9% sensitivity at 48 h. A total of three false-negative results were reported on Spectra MRSA at 24 h, and these were attributed to low specimen inocula. The specificity of Spectra MRSA was better than that of MSA, with Spectra MRSA having specificities of 95.5% at 24 h and 95.2% at 48 h and with MSA having specificities of 94.0% at 24 h and 90.5% at 48 h. Similar to SBA medium, colonies of methicillin-sensitive S. aureus and coagulase-negative Staphylococcus spp. were commonly observed on MSA medium at 24 h, thus requiring further identification and resistance testing.
Spectra MRSA was developed as a selective and differential chromogenic medium for screening for MRSA on the basis of the utilization of chromogens that result in denim blue colonies. The data presented in this article demonstrate that Spectra MRSA is able to rapidly identify and differentiate methicillin-resistant S. aureus from methicillin-susceptible S. aureus without the additional steps of identification and resistance confirmation, thus allowing technologists to perform other laboratory duties. In addition, the low cost of Spectra MRSA would provide a practical and affordable means for health care facilities of all sizes to implement an effective screening method to detect and reduce the spread of MRSA.
Acknowledgments
Thermo Fisher Scientific, Remel Products, provided material support to conduct this trial.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Bannerman, T. L., and S. J. Peacock. 2007. Staphylococcus, Micrococcus, and other catalase-positive cocci, p. 390-411. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, vol. 1, 9th ed. ASM Press, Washington, DC. [Google Scholar]
- 2.Bischof, L. J., L. Lapsley, K. Fontecchio, D. Jacosalem, C. Young, R. Hanker, and D. W. Newton. 2009. Comparison of chromogenic media to BD GeneOhm methicillin-resistant Staphylococcus aureus (MRSA) PCR for detection of MRSA in nasal swabs. J. Clin. Microbiol. 47:2281-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyce, J. M., N. L. Havill, and B. Maria. 2005. Frequency and possible infection control implications of gastrointestinal colonization with methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5992-5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing; 19th informational supplement, vol. 29, no. 3. M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Diekema, D. J., and M. B. Edmond. 2007. Look before you leap: active surveillance for multidrug-resistant organisms. Clin. Infect. Dis. 44:1101-1107. [DOI] [PubMed] [Google Scholar]
- 6.Elsayed, S., B. L. Chow, N. L. Hamilton, D. B. Gregson, J. D. D. Pitout, and D. L. Church. 2003. Development and validation of a molecular beacon probe-based real-time polymerase chain reaction assay for rapid detection of methicillin resistance in Staphylococcus aureus. Arch. Pathol. Lab. Med. 127:845-849. [DOI] [PubMed] [Google Scholar]
- 7.Eveillard, M., A. de Lassence, E. Lancien, G. Barnaud, J. D. Ricard, and M. L. Joly-Guillou. 2006. Evaluation of a strategy of screening multiple anatomical sites for methicillin-resistant Staphylococcus aureus at admission to a teaching hospital. Infect. Control Hosp. Epidemiol. 27:181-184. [DOI] [PubMed] [Google Scholar]
- 8.Flayhart, D., J. F. Hindler, D. A. Bruckner, G. Hall, R. K. Shrestha, S. A. Vogel, S. S. Richter, W. Howard, R. Walther, and K. C. Carroll. 2005. Multicenter evaluation of BBL CHROMagar MRSA medium for direct detection of methicillin-resistant Staphylococcus aureus from surveillance cultures of the anterior nares. J. Clin. Microbiol. 43:5536-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huletsky, A., R. Giroux, V. Rossbach, M. Gagnon, M. Vaillancourt, M. Bernier, F. Gagnon, K. Truchon, M. Baston, F. J. Picard, A. van Belkum, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2004. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J. Clin. Microbiol. 42:1875-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonas, D., M. Speck, F. D. Daschner, and H. Grundmann. 2002. Rapid PCR-based identification of methicillin-resistant Staphylococcus aureus from screening swabs. J. Clin. Microbiol. 40:1821-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karchmer, T. B. 2005. Prevention of health care-associated methicillin-resistant Staphylococcus aureus infections: adapting to a changing epidemiology. Clin. Infect. Dis. 41:167-169. [DOI] [PubMed] [Google Scholar]
- 12.Klevens, M. R., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, G. E. Fosheim, L. K. McDougal, R. B. Carey, and S. K. Fridkin. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763-1771. [DOI] [PubMed] [Google Scholar]
- 13.Lucet, J. C., K. Grenet, L. Armand-Lefevre, M. Harnal, E. Bouvet, B. Regnier, and A. Andremont. 2005. High prevalence of carriage of methicillin-resistant Staphylococcus aureus at hospital admission in elderly patients: implications for infection control strategies. Infect. Control Hosp. Epidemiol. 26:121-126. [DOI] [PubMed] [Google Scholar]
- 14.Muto, C. A. 2006. Methicillin-resistant Staphylococcus aureus control: we didn't start the fire, but it's time to put it out. Infect. Control Hosp. Epidemiol. 27:111-115. [DOI] [PubMed] [Google Scholar]
- 15.Papia, G., M. Louie, A. Tralla, C. Johnson, V. Collins, and A. E. Simor. 1999. Screening high-risk patients for methicillin-resistant Staphylococcus aureus on admission to the hospital: is it cost effective? Infect. Control Hosp. Epidemiol. 20:473-477. [DOI] [PubMed] [Google Scholar]
- 16.Ridenour, G. A., E. S. Wong, M. A. Call, and M. W. Climo. 2006. Duration of colonization with methicillin-resistant Staphylococcus aureus among patients in the intensive care unit: implications for intervention. Infect. Control Hosp. Epidemiol. 27:271-278. [DOI] [PubMed] [Google Scholar]
- 17.Salgado, C. D., and B. M. Farr. 2006. What proportion of hospital patients colonized with methicillin-resistant Staphylococcus aureus are identified by clinical microbiological cultures? Infect. Control Hosp. Epidemiol. 27:116-121. [DOI] [PubMed] [Google Scholar]
- 18.Sanford, M. D., et al. 1994. Efficient detection and long-term persistence of the carriage of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 19:1123-1128. [DOI] [PubMed] [Google Scholar]
- 19.Siegel, J. D., E. Rhinehart, M. Jackson, L. Chiarello, and the Healthcare Infection Control Practices Advisory Committee. 2007. Guideline for isolation precautions: preventing transmission of infectious agents in healthcare settings. http://www.cdc.gov/ncidod/dhqp/pdf/isolation2007.pdf. Accessed 30 May 2009. [DOI] [PMC free article] [PubMed]
- 20.Singh, K., P. J. Gavin, T. Vescio, R. B. Thompson, Jr., R. B. Deddish, A. Fisher, G. A. Noskin, and L. R. Peterson. 2003. Microbiologic surveillance using nasal cultures alone is sufficient for detection of methicillin-resistant Staphylococcus aureus isolates in neonates. J. Clin. Microbiol. 41:2755-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Hal, S. J., D. Stark, B. Lockwood, D. Marriott, and J. Harkness. 2007. Methicillin-resistant Staphylococcus aureus (MRSA) detection: comparison of two molecular methods (IDI-MRSA PCR assay and GenoType MRSA direct PCR assay) with three selective MRSA agars (MRSA ID, MRSASelect, and CHROMagar MRSA) for use with infection-control swabs. J. Clin. Microbiol. 45:2486-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vriens, M. R., A. C. Fluit, A. Troelstra, J. Verhoef, and C. van der Werken. 2002. Staphylococcus aureus rectal carriage and its association with infections in patents in a surgical intensive care unit and a liver transplant unit. Infect. Control Hosp. Epidemiol. 23:495-501. [DOI] [PubMed] [Google Scholar]