Abstract
In September 2006, the seven-valent pneumococcal conjugate vaccine (PCV7; Prevenar) was introduced into the childhood vaccination schedule in the United Kingdom. We monitored the population of invasive pneumococci in Scotland in the 5 years preceding the introduction of PCV7 by using serogrouping, multilocus sequence typing (MLST), and eBURST analysis. Here, we present a unique analysis of a complete national data set of invasive pneumococci over this time. We observed an increase in invasive pneumococcal disease (IPD) caused by serotypes 1, 4, and 6 and a decrease in serogroup 14-, 19-, and 23-associated disease. Analysis of sequence type (ST) data shows a significant increase in ST306, associated with serotype 1, and a decrease in ST124, associated with serotype 14. There have also been increases in the amounts of IPD caused by ST227 (serotype 1) and ST53 (serotype 8), although these increases were not found to reach significance (P = 0.08 and 0.06, respectively). In the course of the study period preceding the introduction of PCV7, we observed considerable and significant changes in serogroup and clonal distribution over time.
Streptococcus pneumoniae (the pneumococcus) is regarded as an opportunistic pathogen, as it may exist asymptomatically as part of the normal flora of the nasopharynx but is also considered an important global pathogen, causing otitis media, pneumonia, septicemia, and meningitis. Pneumococcal pneumonia is a major cause of childhood mortality in the developing world and of adulthood mortality worldwide. Pneumococci can be divided into more than 90 serotypes on the basis of the immunohistochemistry of their polysaccharide capsule. The heptavalent pneumococcal conjugate vaccine (PCV7) is now in use in many countries and contains seven of these serotypes (serotypes 4, 6B, 9V, 14, 18C, 19F, and 23F). Although serotype is important in determining invasiveness (3), several studies have demonstrated that genotype also plays a significant role (19, 44, 45, 48); virulence factors of the pneumococcus have now been found to vary in presence and sequence according to serotype (28, 30) and genotype (38, 47). Multilocus sequence typing (MLST) is a sequence-based typing method that allows division of bacterial species by genotyping of housekeeping genes (13). This method has resulted in the recognition of over 3,000 pneumococcal sequence types (STs), or clones (http://www.mlst.net/).
Fluctuations in the prevalences of serotypes and genotypes may occur naturally in pneumococcal populations in the absence of conjugate vaccine pressure (43). We and others have previously reported the clonal expansion of the serotype 1 clone ST306 in recent years (22, 30). In order to investigate changes in the pneumococcal population prior to introduction of PCV7 in the United Kingdom, we carried out temporal analysis of the invasive pneumococci for the 5 years leading up to the introduction of PCV7. In the United States, clonal expansion (the increase in the number of previously rare clones expressing nonvaccine serotypes) has also been documented (2). Nonvaccine serotypes have been reported in a number of postvaccine studies (11, 23, 27). It is not clear whether such changes are directly driven by vaccine pressure or are due in some part to the natural fluctuations that occur in this naturally transformable species.
Here, we show, for the first time, changes in the epidemiology of pneumococcal disease immediately prior to the introduction of PCV7 in a whole country. This has the advantage of providing longitudinal data rather than a population snapshot. We used the eBURST (based upon related sequence types) algorithm to analyze clonal and serotype changes in pneumococcal isolates causing invasive disease in Scotland during the period from April 2001 to April 2006. We observed that trends in incidence of invasive pneumococcal disease (IPD) due to certain serotypes and clones were already occurring prior to introduction of PCV7, namely, that IPD associated with the nonvaccine serotype 1 ST306 clone has increased significantly over the 5-year period and that IPD associated with serotype 14 has decreased. eBURST analysis of each yearly data set has shown that the majority of clones isolated from IPD patients occur transiently and that a small number of stable clones, expressing serotypes represented in PCV7, cause the majority of invasive disease. eBURST analysis has also shown that most clonal complexes (CCs) are associated with one major serotype but that there is evidence for a small number of serotype switch events even in the absence of any conjugate vaccine. It should be noted that the 23-valent polysaccharide antipneumococcal vaccine (PPV23) was recommended for all those aged 65 and over in Scotland midway through the study period (winter 2003-2004) and therefore that the use of this vaccine may have affected the phenotypic and genotypic distributions of IPD-associated pneumococci observed here (37).
MATERIALS AND METHODS
Pneumococcal isolates.
A total of 3,066 pneumococci isolated from blood and cerebrospinal fluids (CSF) from patients of all ages between April 2001 and April 2006 were included in the study. A total of 2,838 of these isolates were available for full characterization by serogrouping/serotyping and MLST. Pneumococci were isolated in Scottish diagnostic microbiology laboratories and sent to the Scottish Meningococcus and Pneumococcus Reference Laboratory (SMPRL) as part of the enhanced pneumococcal surveillance program in Scotland. This collection is considered a complete national data set, as more than 90% of pneumococci isolated from IPD patients in Scotland are sent to the SMPRL (31). Isolates were stored at −80°C on Protect beads (Technical Service Consultants, United Kingdom) and were grown on Columbia blood agar plates (E & O Laboratories, United Kingdom) in 5% (vol/vol) CO2 at 37°C.
Scottish population data.
Population figures for each year of the study were taken from the General Register Office for Scotland (http://www.gro-scotland.gov.uk/).
Serogrouping/serotyping.
Isolates were characterized at the SMPRL by serogrouping or serotyping, which was performed by coagglutination using reagents from the Statens Serum Institut, Denmark, as previously described (49). Prior to 2003, serogrouping was routinely performed, that is, the use of typing sera to obtain serotypes (such as differentiation of serotypes 6A and 6B within serogroup 6) was not part of the routine service. In 2003, serotyping of all invasive isolates became the routine procedure at the SMPRL. For this reason, isolates have been analyzed by serotype only for study years 3, 4, and 5, that is, for the period from 1 April 2003 to 31 March 2006.
MLST.
MLST was performed on all 2,838 viable isolates by using the method of Enright and Spratt (13), but with an automated protocol (51). Briefly, internal fragments of seven housekeeping genes, aroE, gdh, gki, recP, spi, xpt, and ddl, were amplified using PCR and the nucleotide sequences on both strands determined using a 96-well format liquid-handling robot and an automated DNA-sequencing system. Alleles and STs for MLST were assigned with reference to the S. pneumoniae MLST database (http://spneumoniae.mlst.net/). Novel profiles arising from the description of a new allele or new combination of alleles were submitted to the curator of the MLST database and assigned new STs.
Analysis of clonal relationships.
Data were divided into five yearly data sets, each spanning from 1 April of one year to 31 March of the following year, and clonal groups were identified in each data set by using the eBURST program (14). Comparative eBURST analysis was used to identify STs that appeared for the first time in each year and to track the presence of pneumococcal clones throughout the study period.
Analysis of diversity within clonal complexes.
The diversity of clonal complexes was assessed using Simpson's index of diversity (46). This index provides information about community composition, rather than simply species richness, by taking the relative abundances of different species into account and is calculated using the equation 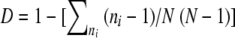 , where D represents Simpson's diversity index, ni represents the number of individuals of ST i, and N represents the number of individuals in the clonal group.
, where D represents Simpson's diversity index, ni represents the number of individuals of ST i, and N represents the number of individuals in the clonal group.
Statistical analysis.
Statistical analysis of changes in incidence was determined using the Z test for differences in proportions.
RESULTS
The study period was from 1 April 2001 until 31 March 2006. Data were broken down into yearly data sets, each spanning from 1 April of one year to 31 March of the following year. A total of 3,066 pneumococcal isolates were included in the study, of which 571 were from the 2001-2002 year, 608 from the 2002-2003 year, 665 from the 2003-2004 year, 566 from the 2004-2005 year, and 656 from the 2005-2006 year. ST was determined for 2,838 (92.6%) of the isolates. The remaining 7.4% of isolates were not available for MLST analysis, due to lack of viability after storage. These 2,838 isolates comprised 36 serogroups (45 serotypes) and 368 STs. The mean number of IPD cases per year was 567.6, corresponding to 11.24 cases of IPD per 100,000 individuals in the population annually. The most common serogroups/serotypes and clones (STs) are shown in Table 1.
TABLE 1.
Most-common pneumococcal serotypes and STs associated with IPD in Scotland from 2001 to 2006
| Serogroup or ST | Rank | Dominant ST(s) | Dominant serogroup(s) | No. of IPD cases (% of IPD incidence) | No. of cases/105 persons |
|---|---|---|---|---|---|
| Serogroups | |||||
| 14 | 1 | 9, 124 | 498 (17.5) | 9.85 | |
| 1 | 2 | 306, 227 | 261 (9.2) | 5.16 | |
| 9 | 2 | 162, 405 | 261 (9.2) | 5.16 | |
| 19 | 4 | 162, 199 | 211 (7.4) | 4.17 | |
| 6 | 5 | 191, 176, 138 | 207 (7.3) | 4.09 | |
| 4 | 6 | 246, 205, 206 | 192 (6.8) | 3.80 | |
| 23 | 7 | 311, 36 | 172 (6.1) | 3.40 | |
| 8 | 8 | 53 | 169 (6.0) | 3.34 | |
| 3 | 9 | 180 | 161 (5.7) | 3.18 | |
| 18 | 10 | 113 | 118 (4.2) | 2.33 | |
| 7 | 11 | 191 | 112 (3.9) | 2.21 | |
| 12 | 12 | 218 | 94 (3.3) | 1.86 | |
| 22 | 13 | 433 | 90 (3.2) | 1.78 | |
| 11 | 14 | 62 | 53 (1.9) | 1.05 | |
| 20 | 15 | 235 | 49 (1.7) | 0.97 | |
| STs | |||||
| 9 | 1 | 14 | 304 (10.7) | 6.01 | |
| 162 | 2 | 9, 19 | 193 (6.8) | 3.82 | |
| 306 | 3 | 1 | 151 (5.3) | 2.99 | |
| 53 | 4 | 8 | 146 (5.2) | 2.89 | |
| 180 | 5 | 3 | 129 (4.6) | 2.55 | |
| 124 | 6 | 14 | 122 (4.3) | 2.41 | |
| 191 | 7 | 7 | 106 (3.7) | 2.10 | |
| 199 | 8 | 19 | 87 (3.1) | 1.72 | |
| 218 | 9 | 12 | 83 (2.9) | 1.64 | |
| 227 | 10 | 1 | 82 (2.9) | 1.62 | |
| 311 | 11 | 23 | 79 (2.8) | 1.56 | |
| 433 | 12 | 22 | 65 (2.3) | 1.29 | |
| 246 | 12 | 4 | 65 (2.3) | 1.29 | |
| 205 | 13 | 4 | 55 (1.9) | 1.09 | |
| 176 | 14 | 6 | 52 (1.8) | 1.03 | |
| 206 | 14 | 4 | 52 (1.8) | 1.03 |
Serogroups.
Table 2 shows the changes in incidence of IPD caused by 14 common serogroups of pneumococci, expressed as percentages of annual incidence. The largest change observed was a 12.5% (P < 0.0001) increase in the incidence of IPD attributable to serotype 1 pneumococci. Serotype 14-associated IPD fell by 6.36% (P < 0.005), from 2.39/100,000 to 1.9/100,000 persons, over the study period. Serotype 14 pneumococci caused the majority of IPD in all years except year 5, when the majority of IPD was associated with pneumococci of serotype 1.
TABLE 2.
Changes in proportion of total IPD cases attributable to common serogroups in the 2001-2002 year and the 2005-2006 year
| Serogroup | 2001-2002 |
2005-2006 |
% Changea | ||||
|---|---|---|---|---|---|---|---|
| No. (%) of cases | No. of cases/105 persons | % of IPD incidence | No. of cases | No. of cases/105 persons | % of IPD incidence | ||
| 4 | 35 | 0.69 | 6.12 | 50 | 0.98 | 7.62 | 1.50 |
| 6 | 41 | 0.81 | 7.17 | 51 | 1 | 7.77 | 0.60 |
| 9 | 48 | 0.95 | 8.39 | 51 | 1 | 7.77 | −0.62 |
| 14 | 121 | 2.39 | 21.15 | 97 | 1.9 | 14.79 | −6.36* |
| 18 | 19 | 0.38 | 3.32 | 23 | 0.45 | 3.51 | 0.19 |
| 19 | 52 | 1.03 | 9.09 | 41 | 0.8 | 6.25 | −2.84 |
| 23 | 47 | 0.93 | 8.22 | 38 | 0.75 | 5.79 | −2.43 |
| 1 | 20 | 0.39 | 3.50 | 105 | 2.06 | 16.01 | 12.51** |
| 7 | 24 | 0.47 | 4.20 | 26 | 0.51 | 3.96 | −0.24 |
| 3 | 30 | 0.59 | 5.24 | 37 | 0.73 | 5.64 | 0.40 |
| 8 | 33 | 0.65 | 5.77 | 35 | 0.69 | 5.34 | −0.43 |
| 11 | 14 | 0.28 | 2.45 | 10 | 0.2 | 1.52 | −0.93 |
| 12 | 20 | 0.39 | 3.50 | 25 | 0.49 | 3.81 | 0.31 |
| 22 | 21 | 0.41 | 3.67 | 12 | 0.24 | 1.83 | −1.84 |
**, P < 0.0001; *, P < 0.005.
Changes in distribution of STs over the study period (2001 to 2006).
The largest change observed over the 5-year period was for ST306 (135.4% increase in the proportion of ST306-associated cases relative to the total number of IPD cases) (Table 3), corresponding to the increase in serotype 1 IPD. The only decrease was for ST124, corresponding to the decrease of serotype 14 IPD.
TABLE 3.
Changes in proportion of total IPD cases attributable to different STs in the 2001-2002 year and the 2005-2006 year
| ST | No. of IPD cases (% of IPD incidence) |
% Changea | |
|---|---|---|---|
| 2001-2002 (yr 1) | 2005-2006 (yr 5) | ||
| 306 | 2 (0.04) | 71 (10.82) | 135.43** |
| 162 | 27 (0.53) | 44 (6.70) | 33.06 |
| 53 | 13 (0.26) | 29 (4.42) | 31.26 |
| 227 | 14 (0.28) | 29 (4.42) | 29.28 |
| 180 | 18 (0.36) | 28 (4.27) | 19.42 |
| 218 | 14 (0.28) | 23 (3.51) | 17.51 |
| 191 | 17 (0.34) | 25 (3.81) | 15.51 |
| 199 | 14 (0.28) | 19 (2.90) | 9.65 |
| 9 | 65 (1.28) | 63 (9.60) | 4.68 |
| 124 | 26 (0.51) | 24 (3.66) | −4.23 |
| Total | 572 | 656 | |
**, P < 0.0001.
Vaccine serotypes over time.
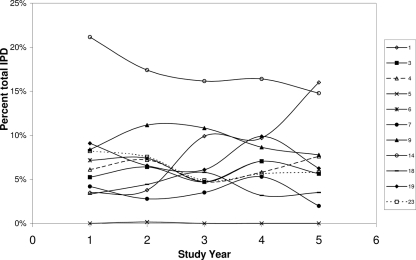
Serotype information was available only for strains isolated during and after 2003. Prior to this, only serogroup information was obtained. In order to assess changes in the proportions of IPD associated with pneumococci of serotypes included in PCV7 and in future licensed (PCV10 [Synflorix]; GlaxoSmithKline) and investigational (PCV13; Wyeth) PCVs, two separate analyses were performed. As serotype (rather than serogroup) data were available only for isolates collected from 2003 onwards, vaccine coverage calculations based on vaccine serotypes were performed for the 2003-2004 year, the 2004-2005 year, and the 2005-2006 year, and changes in the proportions of invasive-disease-associated isolates with serotypes included were assessed. The results of this analysis show that PCV7 serotypes were responsible for 47% of IPD cases in the 2003-2004 year, falling to 44% in the 2005-2006 year. When future conjugate vaccine formulations are included in the analysis, the proportions of PCV10 isolates causing IPD are found to rise from 60% in the 2003-2004 year to 63% in the 2005-2006 year and, for PCV13, from 70% to 75% over the same 3-year period. However, none of these changes are statistically significant. Figure 1 shows the changes in distribution of serotypes included in PCV7, PCV10, and PCV13 and common non-PCV7 vaccine serogroups over the course of the study. The numbers of IPD cases that could have been prevented by the use of each vaccine formulation were also calculated using vaccine serotypes (Table 4). The use of PCV10 could have prevented 298 more IPD cases than the use of PCV7, and PCV13 prevented 226 more cases than PCV10 and 524 more than PCV7 over the 3-year period from 1 April 2003 to 31 March 2006; this is in most part due to the increases in serotype 1 over this period. It should be noted that the coverage and case prevention estimates presented are likely to be underestimates, as no cross-protection for vaccine-related serotypes has been assumed. It is known that serotype 6A receives cross-protection from PCV7 (34) but that a recently reported serotype, serotype 6C, does not (39, 41). Serotype 6C capsular genes have been shown to arise from serotype 6A and would be typed as 6A with the use of phenotypic methods (42); therefore, it is likely that a proportion of the 6A isolates described here are actually serotype 6C. Serotype 19A does not receive cross-protection from PCV7 (34) and accounted for 43.8% of serogroup 19 isolates in the 3-year analysis period.
FIG. 1.
Contribution of conjugate vaccine serogroups (PCV7, PCV10, and PCV13) to IPD over 5 study years, 2001 to 2006.
TABLE 4.
Numbers of IPD cases that may have been prevented by PCV7, PCV10, and PCV13 from April 2003 to March 2006
| Yr | % Coverage |
PCV7 vs PCV10 |
PCV10 vs PCV13 |
PCV7 vs PCV13 |
|||||
|---|---|---|---|---|---|---|---|---|---|
| PCV7 | PCV10 | PCV13 | % Difference | No. of extra cases prevented | % Difference | No. of extra cases prevented | % Difference | No. of extra cases prevented | |
| 2003-2004 | 46.95 | 60.37 | 69.97 | 13.41 | 88 | 9.6 | 63 | 23.0 | 151 |
| 2004-2005 | 47.09 | 61.90 | 76.37 | 14.81 | 84 | 14.46 | 82 | 29.28 | 166 |
| 2005-2006 | 43.45 | 62.65 | 75.00 | 19.21 | 126 | 12.35 | 81 | 31.55 | 207 |
| Total | 298 | 226 | 524 | ||||||
Clonal groups.
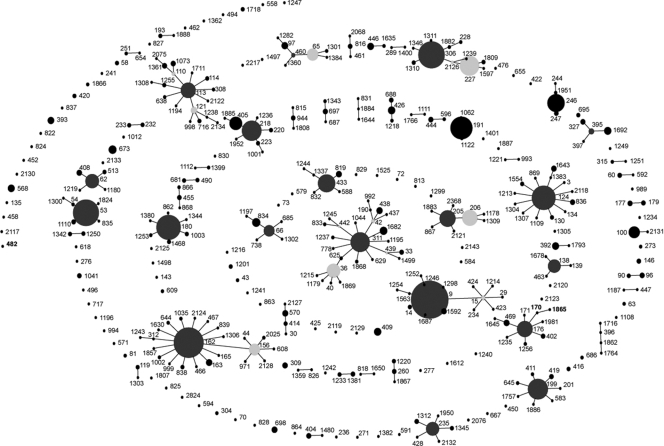
eBURST analysis of the whole data set reveals that isolates can be split into 52 clonal groups on the basis of their STs (Fig. 2). We defined a clonal group as having six out of seven identical loci. Figure 3 shows the changes in the compositions of these clonal groups over the 5-year study period, demonstrating the emergence of clonal complexes 9, 53, 124, 162, 180, and 306.
FIG. 2.
Population snapshot of pneumococcal clones isolated from IPD patients during 5 study years constructed using eBURST. Filled circles represent individual STs, and the size of each circle indicates the number of isolates belonging to that ST. Lines join STs of the same CC, i.e., those that have 6 out of 7 housekeeping gene alleles in common. Dark-gray filled circles indicate the predicted founder of a CC, and light-gray filled circles indicate subfounders.
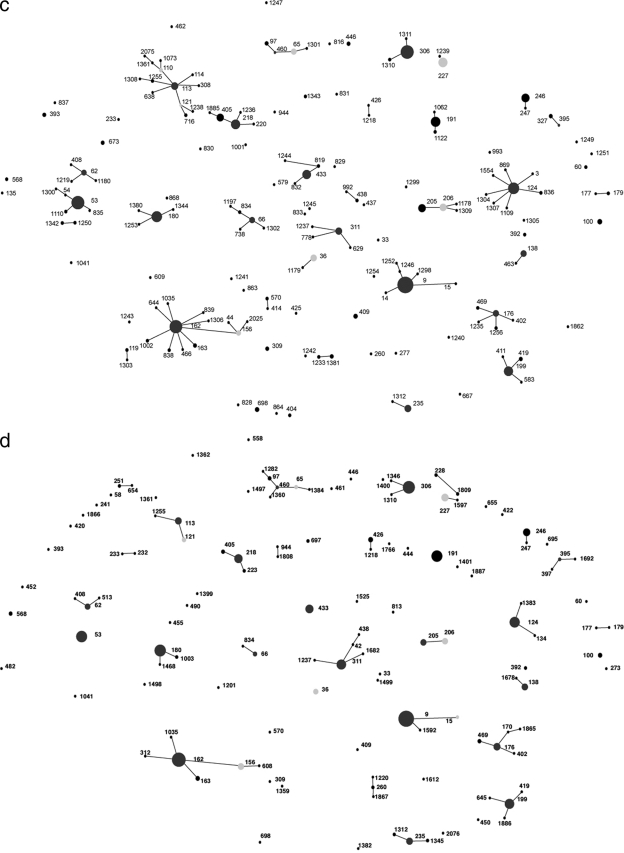
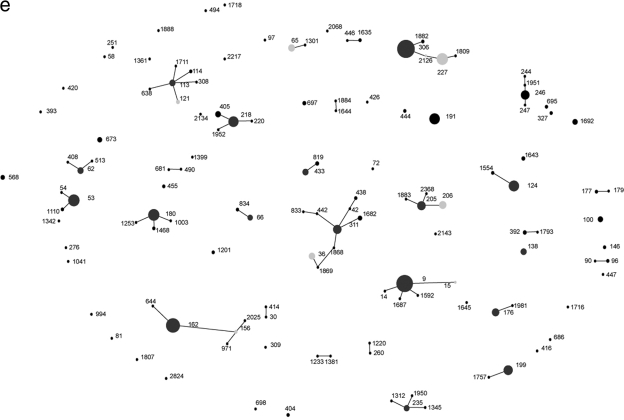
FIG. 3.
Pneumococcal clones isolated from IPD patients during the 2001-2002 year (a), the 2002-2003 year (b), the 2003-2004 year (c), the 2004-2005 year (d), and the 2005-2006 year (e). Filled circles represent individual STs observed in each study year, and the size of each circle indicates the number of isolates belonging to that ST. Positions of STs remain constant throughout Fig. 2 and 3a to 3e to track changes within the population of IPD-causing pneumococci. Lines join STs of the same CC, i.e., those that have 6 out of 7 housekeeping gene alleles in common. Dark-gray filled circles indicate the predicted founder of a CC, and light-gray filled circles indicate subfounders.
Stable and “new” STs.
Thirty-six pneumococcal clones (STs) are represented for every year of the study. We have termed these stable clones (Table 5). In order to assess changes in the invasive pneumococcal population, we also tracked the numbers of STs associated with IPD for the first time in each year and their relationship to existing clones. The majority of STs that are observed for the first time are of the same serogroup as their single-locus variants (SLVs) (data not shown). Table 6 shows those new STs that differ in capsular type from their related SLVs. Many of the “new” clones are not stable over the study period but instead can be considered “transient” clones.
TABLE 5.
Stable clones present in every year of the studya
| Clonal complex | ST | No. of STs |
|||||
|---|---|---|---|---|---|---|---|
| 2001-2002 | 2002-2003 | 2003-2004 | 2004-2005 | 2005-2006 | Total | ||
| 1 | 311 | 13 | 19 | 10 | 21 | 16 | 79 |
| 1 | 36 | 8 | 8 | 9 | 6 | 9 | 40 |
| 1 | 438 | 1 | 2 | 2 | 1 | 2 | 8 |
| 2 | 162 | 27 | 41 | 37 | 44 | 44 | 193 |
| 2 | 156 | 7 | 4 | 5 | 9 | 2 | 27 |
| 3 | 113 | 3 | 16 | 11 | 10 | 8 | 48 |
| 4 | 9 | 65 | 59 | 59 | 57 | 63 | 303 |
| 5 | 124 | 26 | 20 | 28 | 24 | 24 | 122 |
| 6 | 306 | 2 | 8 | 39 | 34 | 71 | 154 |
| 6 | 227 | 14 | 8 | 19 | 12 | 29 | 82 |
| 7 | 218 | 14 | 11 | 19 | 16 | 23 | 83 |
| 7 | 405 | 4 | 9 | 12 | 3 | 7 | 35 |
| 8 | 176 | 5 | 16 | 8 | 11 | 12 | 52 |
| 9 | 205 | 7 | 11 | 13 | 8 | 16 | 55 |
| 9 | 206 | 11 | 10 | 11 | 8 | 12 | 52 |
| 10 | 199 | 14 | 14 | 19 | 21 | 19 | 87 |
| 11 | 180 | 18 | 28 | 25 | 30 | 28 | 129 |
| 12 | 65 | 7 | 28 | 7 | 3 | 10 | 55 |
| 12 | 97 | 1 | 1 | 3 | 3 | 1 | 9 |
| 13 | 66 | 3 | 5 | 7 | 4 | 6 | 25 |
| 13 | 834 | 7 | 2 | 2 | 1 | 2 | 14 |
| 14 | 235 | 2 | 8 | 10 | 10 | 7 | 37 |
| 15 | 433 | 10 | 15 | 17 | 15 | 8 | 65 |
| 16 | 53 | 14 | 37 | 38 | 28 | 29 | 146 |
| 17 | 62 | 5 | 14 | 6 | 7 | 8 | 40 |
| 18 | 395 | 0 | 3 | 1 | 2 | 2 | 8 |
| 19 | 246 | 6 | 13 | 15 | 12 | 17 | 63 |
| 20 | 570 | 1 | 3 | 2 | 1 | 0 | 7 |
| 22 | 138 | 5 | 5 | 8 | 9 | 8 | 35 |
| 23 | 426 | 2 | 2 | 1 | 5 | 1 | 11 |
| 26 | 191 | 17 | 17 | 21 | 28 | 25 | 108 |
| 31 | 446 | 3 | 1 | 3 | 1 | 1 | 9 |
| 47 | 100 | 3 | 9 | 5 | 4 | 6 | 27 |
| 48 | 392 | 1 | 2 | 2 | 2 | 3 | 10 |
| Singleton | 673 | 3 | 2 | 3 | 2 | 3 | 13 |
| Singleton | 393 | 2 | 2 | 3 | 1 | 1 | 9 |
| Singleton | 409 | 1 | 5 | 4 | 1 | 0 | 11 |
Bold type denotes a predicted founder ST.
TABLE 6.
Clones that are observed for the first time and express capsules different from those of their related SLVs
| Yr and new ST | Serogroup | SLV (serogroup) and comment | Gene(s) with differing SLV allele |
|---|---|---|---|
| 2002-2003 | |||
| ST685a | 24 | ST66 (9N) | gdh |
| ST1563 | 6 | ST9 (14); all three ST1563 clones in database are type 14 | aroE |
| 2003-2004 | |||
| ST1062 | 15B | ST191 (7F) | ddl |
| ST1254 | 14 | ST1563 (6), not observed in the 2003-2004 yr | spi |
| ST1300 | 3 | ST54 (8) | xpt |
| ST1304a | 11A | ST124 (14) | spi |
| ST1380 | 8 | ST180 (3), ST862 (3), not observed in the 2003-2004 yr; ST1344 (3), new in the 2003-2004 yr | xpt |
| ST738 | 14 | ST66 (9N) | ddl |
| 2004-2005 (ST251) | 6A | ST654 (19F), also new in the 2004-2005 yr | recP |
| 2005-2006 (ST1884) | 34 | ST1644 (35F), also new in the 2005-2006 yr | spi |
This clone expressed the nonvaccine serotype, while the SLV was of the vaccine or vaccine-related serotype.
Diversity ratio.
Simpson's index of diversity (46) was calculated for the clonal complexes containing the largest numbers of isolates. The closer the value is to 1, the more diverse the clonal complex is. By this method, group 3 (CC113) is the most diverse complex at a genomic level even though it contains isolates of only one serogroup. The least diverse group was group 26 (CC191), which contained just three STs, despite the ranking of ST191 as the seventh-most-common cause of IPD. Such findings are in agreement with a recent study by Dagerhamn et al. (10), which found less intraclonal variation in accessory genes among STs representative of serotypes with high invasive-disease potential than among STs associated with carriage. Diversity ratios are shown in Table 7.
TABLE 7.
Simpson diversity indices for common clonal complexes
| Group | No. of isolates | No. of STs | Diversity index |
|---|---|---|---|
| CC9 | 328 | 15 | 0.14 |
| CC162 | 254 | 23 | 0.41 |
| CC306 | 250 | 12 | 0.53 |
| CC311 | 169 | 25 | 0.72 |
| CC53 | 155 | 6 | 0.11 |
| CC124 | 148 | 14 | 0.32 |
| CC180 | 140 | 7 | 0.15 |
| CC218 | 128 | 8 | 0.51 |
| CC205 | 113 | 8 | 0.29 |
| CC191 | 109 | 3 | 0.07 |
| CC199 | 99 | 8 | 0.23 |
| CC113 | 91 | 18 | 0.71 |
| CC176 | 69 | 8 | 0.46 |
DISCUSSION
In the course of the 5-year study period, during which a conjugate pneumococcal vaccine was not yet included in the childhood vaccination schedule, we observed considerable and significant changes in serogroup and clonal distributions over time among isolates from patients of all ages with invasive pneumococcal disease. Although 368 different STs were observed during the study, only 9.8% (n = 36) of these clones are “stable clones” and were associated with IPD in every year of the study. These 36 clones accounted for more than 78% of the isolates (2,241/2,838 isolates). Of the remaining 332 STs, 220 were isolated from IPD patients in only one study year. We observed that the most-common clones were stable, occurring in large numbers each year. Single- and double-locus variants of these clones were observed, some of which persisted, whereas others appeared only transiently. During the 5 years of our study, a decrease in the incidence of IPD caused by serotype 14 was observed, although this serotype remained the most common cause of IPD in Scotland over the whole period. There was also a slight rise in the number of cases of IPD in which serogroup 19 was present.
Many new STs are observed each year and seem to form a pattern of natural background fluctuation. Often, new STs do not persist within the population, suggesting that they are outcompeted by other clones. An exception to this has been the 135.4% increase in IPD caused by the serotype 1 ST306 clone over the 5 years of the study. The rise of IPD caused by this serotype has previously been noted (30). Although serotype 1-associated IPD has increased in Scotland, the outcome associated with such infections appears to be less severe than those associated with other serotypes (26). IPD associated with ST162, -53, -227, -180, -218, -191, -199, and -9 has also increased over the study period (Table 3). None of these increases were significant, although the increase in ST53 (31.25%; P = 0.0561) was approaching significance. As expected, new STs observed are most commonly of the same serogroup as closely related clones, i.e., members of the same clonal group. However, there are instances where a clone is observed for the first time in our data set and expresses a serotype different from that of its SLV(s) (Table 6). In the majority of these cases, vaccine types are replaced by vaccine types and nonvaccine types by nonvaccine types. The exception is with the appearance of an ST1304 clone of serotype 11A. ST1304 differs at the spi allele from ST124, which commonly occurs as serotype 14. This is an example of a change capable of facilitating vaccine evasion occurring within a clonal group. Certain clones appear to be well adapted to invasive disease and persist over time and space, while certain other clones, such as ST306, are able to undergo clonal expansion over just a few years. The ability of pneumococci to undergo such expansion is worrying in the face of serotype replacement in the era of conjugate vaccines. The patterns of change reported here reflect only invasive pneumococcal disease; changes in the serotypes and genotypes of circulating pneumococci carried in the nasopharynx may not be represented by these data.
We assessed the coverage of PCV7 and of the 10- and 13-valent conjugate vaccines. Coverage of 10- and 13-valent PCVs increased over the study period, and this increase was due to the increasing contribution of ST306 serotype 1 pneumococci to IPD in Scotland over the 5 years of the study. PCV7 was introduced into the childhood vaccination schedule in the whole of the United Kingdom, including Scotland, in September 2006 and has successfully reduced IPD in vaccinated children, and as yet, no serotype replacement has been observed (24) (http://www.hpa.org.uk; updated 25 November 2009). However, now, more than 2 years after the vaccine introduction, serotype replacement of vaccine types with nonvaccine types, such as serotype 1, could become a real threat, especially as serotype 1 pneumococci are associated with outbreaks (9, 17, 18, 35) and complicated pneumonia and empyema (12, 16, 32). Recently, this clone has been isolated from pediatric carriage patients (40); previously, serotype 1 pneumococci have been rare in carriage studies and are considered to have a high level of invasive potential (3, 44). Given that PCV7 reduces carriage of vaccine types, future vaccines, including serotype 1 polysaccharide, may be appropriate in areas with circulating serotype 1 pneumococci. The data presented here are evidence that PCV10 and -13 would provide significantly more protection from IPD than PCV7.
Since 1989, PPV23 has been licensed for use in the United Kingdom. This vaccine elicits a T-cell-independent response and so is not effective in young children. Prior to 2003, PPV23 was recommended for use in individuals of 2 years of age or older with an increased risk of pneumococcal disease (1). In winter 2003-2004, PPV23 was introduced for all those aged 65 years and over in Scotland and promoted in parallel with an influenza vaccination program for the same age group. The uptakes of PPV23 were estimated to be 68.1% in males and 65.5% in females during winter 2003-2004. This intervention is estimated to have resulted in a 33% reduction of IPD in the target age group (37). However, PPV23 has no demonstrable effect on nasopharyngeal carriage of pneumococci (33). We found no difference in the proportions of IPD cases associated with PPV23 serotypes between the 2005-2006 year (93.7%) and the 2001-2002 year (93.0%). It should be noted that the introduction of PPV23 in Scotland differed from the phased 3-year vaccine introduction for 10-year age bands (beginning with those aged 85 years and over) that was adopted in England and Wales and was completed in 2005/2006. The possible impact of PPV23 on the serotype and genotype distributions of invasive pneumococci should be considered when the data presented here are interpreted, although there is increasing evidence regarding the lack of efficacy of PPV23 in prevention of adult pneumonia (25) and there are conflicting data regarding its efficacy against IPD (25, 36).
Fluctuations in serotype distribution in the absence of conjugate vaccine have been described for other countries (15, 21, 43); such variations should be taken into account when the effects of PCVs are evaluated and when further licensed or investigational conjugate vaccines are considered. Global travel, including that for business, tourism, and migration, is increasing, and the European population is traveling more freely; therefore, it is possible that the use of PCV7 in other countries could also be a factor affecting the strain distribution observed for Scottish IPD. The ability of antibiotic-resistant pneumococci to undergo clonal expansion under selective pressure is well documented, and although an analysis of antimicrobial resistance has not been included here, we have previously shown that resistance is low among Scottish IPD-based isolates and confined to particular clones, for example, the erythromycin-resistant serotype 14 ST9 clone (50). For this reason, it is reasonable to presume that expansion under selective pressure is not a primary driving force for the fluctuations that we observed. This study has focused only on invasive pneumococcal isolates, but in order to gain a more complete picture of circulating pneumococcal population, it is important to carry out similar longitudinal molecular analyses of carried pneumococci. Global use of conjugate vaccines will cause profound changes in the pneumococcal population over the coming years, and monitoring of the population by using molecular methods will be vital if we are to respond to changes in the population of this organism.
Acknowledgments
We acknowledge the use of the pneumococcal MLST database which is located at Imperial College London and is funded by the Wellcome Trust. We thank the staff of the Scottish Meningococcus and Pneumococcus Reference Laboratory for access to and serotyping of invasive pneumococcal isolates and also thank the submitting Scottish diagnostic microbiology laboratories for invasive pneumococcal isolates. We also thank Graeme Cowan, now of the University of Edinburgh, for graphics expertise on eBURST diagrams.
This work was funded by an investigator-driven unrestricted-research grant from Wyeth Vaccines.
J.M.J. has received consulting fees from GlaxoSmithKline. A.J.S. has previously received unrestricted research funding from Wyeth Vaccines. S.C.C. currently receives unrestricted research funding from Wyeth Vaccines. T.J.M. has previously received unrestricted research funding from Wyeth Vaccines and GlaxoSmithKline and is currently Principal Investigator for a BBSRC/GSK CASE Ph.D. studentship.
Footnotes
Published ahead of print on 18 November 2009.
REFERENCES
- 1.Anonymous. 2006. Immunisation against infectious disease 2006: the green book. Department of Health, London, United Kingdom. http://www.dh.gov.uk/en/Publichealth/Healthprotection/Immunisation/Greenbook/DH_4097254.
- 2.Beall, B., M. C. McEllistrem, R. E. Gertz, Jr., S. Wedel, D. J. Boxrud, A. L. Gonzalez, M. J. Medina, R. Pai, T. A. Thompson, L. H. Harrison, L. McGee, and C. G. Whitney. 2006. Pre- and postvaccination clonal compositions of invasive pneumococcal serotypes for isolates collected in the United States in 1999, 2001, and 2002. J. Clin. Microbiol. 44:999-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brueggemann, A. B., D. T. Griffiths, E. Meats, T. Peto, D. W. Crook, and B. G. Spratt. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424-1432. [DOI] [PubMed] [Google Scholar]
- 4.Reference deleted.
- 5.Reference deleted.
- 6.Reference deleted.
- 7.Reference deleted.
- 8.Reference deleted.
- 9.Dagan, R., S. Gradstein, I. Belmaker, N. Porat, Y. Siton, G. Weber, J. Janco, and P. Yagupsky. 2000. An outbreak of Streptococcus pneumoniae serotype 1 in a closed community in southern Israel. Clin. Infect. Dis. 30:319-321. [DOI] [PubMed] [Google Scholar]
- 10.Dagerhamn, J., C. Blomberg, S. Browall, K. Sjostrom, E. Morfeldt, and B. Henriques-Normark. 2008. Determination of accessory gene patterns predicts the same relatedness among strains of Streptococcus pneumoniae as sequencing of housekeeping genes does and represents a novel approach in molecular epidemiology. J. Clin. Microbiol. 46:863-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dias, R., and M. Canica. 2007. Invasive pneumococcal disease in Portugal prior to and after the introduction of pneumococcal heptavalent conjugate vaccine. FEMS Immunol. Med. Microbiol. 51:35-42. [DOI] [PubMed] [Google Scholar]
- 12.Eastham, K. M., R. Freeman, A. M. Kearns, G. Eltringham, J. Clark, J. Leeming, and D. A. Spencer. 2004. Clinical features, aetiology and outcome of empyema in children in the north east of England. Thorax 59:522-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enright, M., and B. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 14.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flamaing, J., J. Verhaegen, J. Vandeven, N. Verbiest, and W. E. Peetermans. 2008. Pneumococcal bacteraemia in Belgium (1994-2004): the pre-conjugate vaccine era. J. Antimicrob. Chemother. 61:143-149. [DOI] [PubMed] [Google Scholar]
- 16.Fletcher, M., J. Leeming, K. Cartwright, and A. Finn. 2006. Childhood empyema: limited potential impact of 7-valent pneumococcal conjugate vaccine. Pediatr. Infect. Dis. J. 25:559-560. [DOI] [PubMed] [Google Scholar]
- 17.Gratten, M., F. Morey, J. Dixon, K. Manning, P. Torzillo, R. Matters, J. Erlich, J. Hanna, V. Asche, and I. Riley. 1993. An outbreak of serotype 1 Streptococcus pneumoniae infection in central Australia. Med. J. Aust. 158:340-342. [DOI] [PubMed] [Google Scholar]
- 18.Gupta, A., F. M. Khaw, E. L. Stokle, R. C. George, R. Pebody, R. E. Stansfield, C. L. Sheppard, M. Slack, R. Gorton, and D. A. Spencer. 2008. Outbreak of Streptococcus pneumoniae serotype 1 pneumonia in a United Kingdom school. BMJ 337:a2964. [DOI] [PubMed] [Google Scholar]
- 19.Hanage, W. P., T. H. Kaijalainen, R. K. Syrjanen, K. Auranen, M. Leinonen, P. H. Makela, and B. G. Spratt. 2005. Invasiveness of serotypes and clones of Streptococcus pneumoniae among children in Finland. Infect. Immun. 73:431-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reference deleted.
- 21.Henriques-Normark, B., C. Blomberg, J. Dagerhamn, P. Battig, and S. Normark. 2008. The rise and fall of bacterial clones: Streptococcus pneumoniae. Nat. Rev. Microbiol. 6:827-837. [DOI] [PubMed] [Google Scholar]
- 22.Henriques Normark, B., M. Kalin, A. Ortqvist, T. Akerlund, B. O. Liljequist, J. Hedlund, S. B. Svenson, J. Zhou, B. G. Spratt, S. Normark, and G. Kallenius. 2001. Dynamics of penicillin-susceptible clones in invasive pneumococcal disease. J. Infect. Dis. 184:861-869. [DOI] [PubMed] [Google Scholar]
- 23.Hicks, L. A., L. H. Harrison, B. Flannery, J. L. Hadler, W. Schaffner, A. S. Craig, D. Jackson, A. Thomas, B. Beall, R. Lynfield, A. Reingold, M. M. Farley, and C. G. Whitney. 2007. Incidence of pneumococcal disease due to non-pneumococcal conjugate vaccine (PCV7) serotypes in the United States during the era of widespread PCV7 vaccination, 1998-2004. J. Infect. Dis. 196:1346-1354. [DOI] [PubMed] [Google Scholar]
- 24.Reference deleted.
- 25.Huss, A., P. Scott, A. E. Stuck, C. Trotter, and M. Egger. 2009. Efficacy of pneumococcal vaccination in adults: a meta-analysis. CMAJ 180:48-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inverarity, D., K. Lamb, M. A. Diggle, C. Robertson, D. Greenhalgh, T. J. Mitchell, A. J. Smith, J. M. C. Jefferies, J. McMenamin, S. C. Clarke, and G. F. Edwards. 2008. Death or survival form invasive pneumococcal disease in Scotland: associations with serogroups and multilocus sequence types, p. 163, abstr. P2-136. Abstr. Sixth International Symposium on Pneumococci and Pneumococcal Diseases, Reykjavik, Iceland. [DOI] [PMC free article] [PubMed]
- 27.Jacobs, M. R., C. E. Good, S. Bajaksouzian, and A. R. Windau. 2008. Emergence of Streptococcus pneumoniae serotypes 19A, 6C, and 22F and serogroup 15 in Cleveland, Ohio, in relation to introduction of the protein-conjugated pneumococcal vaccine. Clin. Infect. Dis. 47:1388-1395. [DOI] [PubMed] [Google Scholar]
- 28.Jefferies, J. M., C. H. Johnston, L. A. Kirkham, G. J. Cowan, K. S. Ross, A. Smith, S. C. Clarke, A. B. Brueggemann, R. C. George, B. Pichon, G. Pluschke, V. Pfluger, and T. J. Mitchell. 2007. Presence of nonhemolytic pneumolysin in serotypes of Streptococcus pneumoniae associated with disease outbreaks. J. Infect. Dis. 196:936-944. [DOI] [PubMed] [Google Scholar]
- 29.Reference deleted.
- 30.Kirkham, L. A., J. M. Jefferies, A. R. Kerr, Y. Jing, S. C. Clarke, A. Smith, and T. J. Mitchell. 2006. Identification of invasive serotype 1 pneumococcal isolates that express nonhemolytic pneumolysin. J. Clin. Microbiol. 44:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kyaw, M. H., P. Christie, S. C. Clarke, J. D. Mooney, S. Ahmed, I. G. Jones, and H. Campbell. 2003. Invasive pneumococcal disease in Scotland, 1999-2001: use of record linkage to explore associations between patients and disease in relation to future vaccination policy. Clin. Infect. Dis. 37:1283-1291. [DOI] [PubMed] [Google Scholar]
- 32.Langley, J. M., J. D. Kellner, N. Solomon, J. L. Robinson, N. Le Saux, J. McDonald, R. Ulloa-Gutierrez, B. Tan, U. Allen, S. Dobson, and H. Joudrey. 2008. Empyema associated with community-acquired pneumonia: a Pediatric Investigator's Collaborative Study on Infections in Canada (PICNIC) study. BMC Infect. Dis. 8:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ledwith, M. 2001. Pneumococcal conjugate vaccine. Curr. Opin. Pediatr. 13:70-74. [DOI] [PubMed] [Google Scholar]
- 34.Lee, H., M. H. Nahm, R. Burton, and K.-H. Kim. 2009. Immune response in infants to the heptavalent pneumococcal conjugate vaccine against vaccine-related serotypes 6A and 19A. Clin. Vaccine Immunol. 16:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leimkugel, J., A. Adams Forgor, S. Gagneux, V. Pfluger, C. Flierl, E. Awine, M. Naegeli, J. P. Dangy, T. Smith, A. Hodgson, and G. Pluschke. 2005. An outbreak of serotype 1 Streptococcus pneumoniae meningitis in northern Ghana with features that are characteristic of Neisseria meningitidis meningitis epidemics. J. Infect. Dis. 192:192-199. [DOI] [PubMed] [Google Scholar]
- 36.Moberley, S. A., J. Holden, D. P. Tatham, and R. M. Andrews. 2008. Vaccines for preventing pneumococcal infection in adults. Cochrane Database Syst. Rev. 2008(1):CD000422. [DOI] [PubMed] [Google Scholar]
- 37.Mooney, J. D., A. Weir, J. McMenamin, L. D. Ritchie, T. V. Macfarlane, C. R. Simpson, S. Ahmed, C. Robertson, and S. C. Clarke. 2008. The impact and effectiveness of pneumococcal vaccination in Scotland for those aged 65 and over during winter 2003/2004. BMC Infect. Dis. 8:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moschioni, M., C. Donati, A. Muzzi, V. Masignani, S. Censini, W. P. Hanage, C. J. Bishop, J. N. Reis, S. Normark, B. Henriques-Normark, A. Covacci, R. Rappuoli, and M. A. Barocchi. 2008. Streptococcus pneumoniae contains 3 rlrA pilus variants that are clonally related. J. Infect. Dis. 197:888-896. [DOI] [PubMed] [Google Scholar]
- 39.Nahm, M. H., J. Lin, J. A. Finkelstein, and S. I. Pelton. 2009. Increase in the prevalence of the newly discovered pneumococcal serotype 6C in the nasopharynx after introduction of pneumococcal conjugate vaccine. J. Infect. Dis. 199:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nunes, S., R. Sa-Leao, L. C. Pereira, and H. Lencastre. 2008. Emergence of a serotype 1 Streptococcus pneumoniae lineage colonising healthy children in Portugal in the seven-valent conjugate vaccination era. Clin. Microbiol. Infect. 14:82-84. [DOI] [PubMed] [Google Scholar]
- 41.Park, I. H., M. R. Moore, J. J. Treanor, S. I. Pelton, T. Pilishvili, B. Beall, M. A. Shelly, B. E. Mahon, and M. H. Nahm. 2008. Differential effects of pneumococcal vaccines against serotypes 6A and 6C. J. Infect. Dis. 198:1818-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park, I. H., D. G. Pritchard, R. Cartee, A. Brandao, M. C. Brandileone, and M. H. Nahm. 2007. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J. Clin. Microbiol. 45:1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruckinger, S., R. von Kries, R. R. Reinert, M. van der Linden, and A. Siedler. 2008. Childhood invasive pneumococcal disease in Germany between 1997 and 2003: variability in incidence and serotype distribution in absence of general pneumococcal conjugate vaccination. Vaccine 26:3984-3986. [DOI] [PubMed] [Google Scholar]
- 44.Sandgren, A., B. Albiger, C. J. Orihuela, E. Tuomanen, S. Normark, and B. Henriques-Normark. 2005. Virulence in mice of pneumococcal clonal types with known invasive disease potential in humans. J. Infect. Dis. 192:791-800. [DOI] [PubMed] [Google Scholar]
- 45.Sandgren, A., K. Sjostrom, B. Olsson-Liljequist, B. Christensson, A. Samuelsson, G. Kronvall, and B. Henriques Normark. 2004. Effect of clonal and serotype-specific properties on the invasive capacity of Streptococcus pneumoniae. J. Infect. Dis. 189:785-796. [DOI] [PubMed] [Google Scholar]
- 46.Simpson, E. H. 1949. Measurement of diversity. Nature 163:688. [Google Scholar]
- 47.Sjostrom, K., C. Blomberg, J. Fernebro, J. Dagerhamn, E. Morfeldt, M. A. Barocchi, S. Browall, M. Moschioni, M. Andersson, F. Henriques, B. Albiger, R. Rappuoli, S. Normark, and B. Henriques-Normark. 2007. Clonal success of piliated penicillin nonsusceptible pneumococci. Proc. Natl. Acad. Sci. U. S. A. 104:12907-12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sjostrom, K., C. Spindler, A. Ortqvist, M. Kalin, A. Sandgren, S. Kuhlmann-Berenzon, and B. Henriques-Normark. 2006. Clonal and capsular types decide whether pneumococci will act as a primary or opportunistic pathogen. Clin. Infect. Dis. 42:451-459. [DOI] [PubMed] [Google Scholar]
- 49.Smart, L. E. 1986. An alternative approach to typing of Streptococcus pneumoniae strains by coagglutination. Acta Pathol. Microbiol. Immunol. Scand. B 94:409-413. [DOI] [PubMed] [Google Scholar]
- 50.Smith, A. J., J. Jefferies, S. C. Clarke, C. Dowson, G. F. Edwards, and T. J. Mitchell. 2006. Distribution of epidemic antibiotic-resistant pneumococcal clones in Scottish pneumococcal isolates analysed by multilocus sequence typing. Microbiology 152:361-365. [DOI] [PubMed] [Google Scholar]
- 51.Sullivan, C. B., J. M. Jefferies, M. A. Diggle, and S. C. Clarke. 2006. Automation of MLST using third-generation liquid-handling technology. Mol. Biotechnol. 32:219-226. [DOI] [PubMed] [Google Scholar]