Abstract
We report the first case of Gordonibacter pamelaeae bacteremia, identified by phenotypic tests and 16S rRNA sequencing in a patient with disseminated rectosigmoid carcinoma and responsive to amoxicillin-clavulanate. The bacterium was a nonsporulating, anaerobic, Gram-positive, nonmotile, coccobacillus that was catalase, arginine dihydrolase, and arginine acrylamidase positive. The gastrointestinal tract is probably its reservoir.
CASE REPORT
An 82-year-old Chinese man was admitted because of fever and lethargy for 1 day. He was diagnosed to have rectosigmoid carcinoma with lung metastasis which was treated conservatively. He also had hypertension and deafness. His fever was associated with decreased appetite, nausea, and vomiting. He noticed blood-stained stool with mucus for 3 days and recent weight loss. On admission, his body temperature was 37.9°C. There were no localizing signs. The total white cell count was 12.9 × 109/liter (neutrophil count was 11.7 × 109/liter), the hemoglobin level was 9.3 g/dl, and the platelet count was 366 × 109/liter. The results of liver and renal function tests were normal. Blood, stool, and urine cultures were performed, and empirical intravenous amoxicillin-clavulanate was commenced.
On day 2 postincubation, the anaerobic blood culture bottle turned positive with a nonsporulating Gram-positive bacillus. Stool culture was negative for diarrheal pathogens, and urine culture showed no growth. The fever gradually subsided after 3 days of amoxicillin-clavulanate, and the patient's general condition gradually improved. The patient was discharged after 9 days of antibiotics. There was no relapse of the bacteremia up to the time of writing, 2 years after discharge.
Microbiological data.
All clinical data were collected prospectively. Clinical specimens were collected and handled according to standard protocols (8). All suspect colonies were identified by standard conventional biochemical methods (8). All tests were performed in triplicate with freshly prepared media on separate occasions. In addition, the Vitek system (ANI) (bioMerieux Vitek), the API system (20A) (bioMerieux Vitek, Hazelwood, MO), and the ATB Expression system (ID32A) (bioMerieux Vitek) were used for the identification of the bacterial isolate. In vitro susceptibilities to penicillin, metronidazole, vancomycin, and amoxicillin-clavulanate were determined using the Etest method. The blood culture isolate was a Gram-positive, non-spore-forming coccobacillus, with occasional elongated forms. It grew on sheep blood agar as nonhemolytic, gray colonies of 0.5 mm in diameter after 48 h of incubation at 37°C in an anaerobic environment but did not grow in ambient air or 5% CO2. It was catalase positive and nonmotile, confirmed by flagellar staining. It was arginine dihydrolase and arginine acrylamidase positive (Table 1). The Vitek system (ANI) identified the bacterium as 95% Clostridium limosum, the API system (20A) identified it as 91.9% Eggerthella lenta and 7.9% Actinomyces viscosus, and the ATB Expression system (ID32A) identified it as 96.4% Eggerthella lenta. The MICs of penicillin, metronidazole, vancomycin, and amoxicillin-clavulanate for the isolate were 0.5 μg/ml, 0.125 μg/ml, 1 μg/ml, and 0.5 μg/ml, respectively.
TABLE 1.
Biochemical profiles of the blood culture isolate, Gordonibacter pamelaeae, Eggerthella hongkongensis, Eggerthella sinensis, and Eggerthella lenta by a combination of conventional biochemical tests and Vitek ANI, API 20A, and ATB ID32A systems
| Biochemical reaction, enzyme, or substrate | Result for bacteriuma: |
||||
|---|---|---|---|---|---|
| Blood culture isolate | Gordonibacter pamelaeae (n = 1) (reference 14) | Eggerthella hongkongensis (n = 4) (reference 5) | Eggerthella sinensis (n = 1) (reference 5) | Eggerthella lenta (n > 10) (references 2 and 8) | |
| Catalase | + | + | + | + | V |
| Arginine dihydrolase | + | + | + | + | + |
| Alkaline phosphatase | − | − | V | − | − |
| Phosphate choline | − | NA | − | − | − |
| Reduction of nitrate | − | − | − | − | V |
| Reduction of triphenyl tetrazolium | − | NA | − | − | − |
| α-Fucosidase | − | − | − | − | V |
| β-Fucosidase | − | NA | − | − | − |
| β-Glucosidase | − | − | + | − | − |
| α-Mannosidase | − | NA | − | − | − |
| β-Lactosidase | − | NA | − | − | − |
| β-Xylosidase | − | NA | − | − | − |
| Alanine arylamidase | − | − | V | − | V |
| Arginine arylamidase | + | − | V | V | V |
| Benzoyl-arginine arylamidase | − | NA | − | − | − |
| Glycine arylamidase | − | − | − | − | V |
| Histidine arylamidase | − | − | − | − | V |
| Leucine arylamidase | − | − | V | − | V |
| Lysine arylamidase | − | NA | V | + | V |
| Phenylalanine arylamidase | − | − | − | − | V |
| Proline arylamidase | − | − | − | − | V |
| Serine arylamidase | − | − | − | − | V |
| Tyrosine arylamidase | − | − | − | − | V |
All strains are negative for glutamic acid decarboxylase; indole production; urease; oxidation/fermentation of arabinose, glucose, mannose, raffinose, trehalose, and xylose; α-arabinosidase; α-galactosidase; β-galactosidase; β-galactosidase-6-phosphate; α-glucosidase; β-glucuronidase; N-acetyl-glucosaminidase; glutamyl glutamic acid arylamidase; leucyl glycine arylamidase; and pyroglutamic acid arylamidase. V, variable; NA, not available.
16S rRNA gene sequencing.
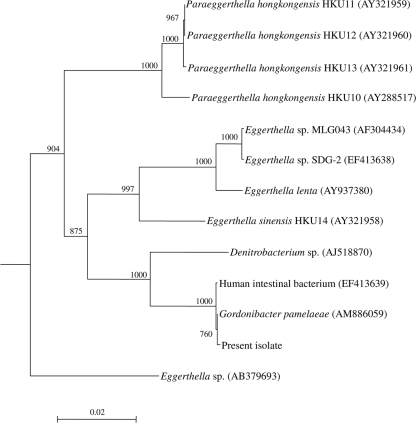
Bacterial DNA extraction, PCR amplification, and DNA sequencing of the 16S rRNA gene of the anaerobic Gram-positive bacillus were performed according to our previous publications (3, 4, 10, 11, 12, 13), using LPW8429 (5′-TTGATCCTGGCTCAGGAC-3′) and LPW58 (5′-AGGCCCGGGAACGTATTCAC-3′) (Sigma-Proligo, Singapore) as the PCR and sequencing primers. The sequences of the PCR products were compared with sequences of closely related species in GenBank by multiple sequence alignment using ClustalX 1.83 (9). Phylogenetic relationships were determined using the neighbor-joining method. Sequencing of the 16S rRNA gene of the isolate showed that there was a 1 (0.08%)-base difference between the 16S rRNA gene sequence of the isolate and that of Gordonibacter pamelaeae (GenBank accession no. AM886059), a 2 (0.16%)-base difference between the 16S rRNA gene sequence of the isolate and that of human intestinal bacterium ARC-1 (GenBank accession no. EF413639), a 77 (6.1%)-base difference between the 16S rRNA gene sequence of the isolate and that of Eggerthella sinensis (GenBank accession no. AY321958), an 84 (6.7%)-base difference between the 16S rRNA gene sequence of the isolate and that of Eggerthella hongkongensis (GenBank accession no. AY288517), and a 95 (7.5%)-base difference between the 16S rRNA gene sequence of the isolate and that of Eggerthella lenta (GenBank accession no. AY937380), indicating that the isolate was a strain of Gordonibacter pamelaeae (Fig. 1).
FIG. 1.
Phylogenetic tree showing the relationships of the patient's isolate to related species. The tree was inferred from 16S rRNA data by the neighbor-joining method and rooted using the 16S rRNA gene sequence of Bacteroides fragilis (X83943). Bootstrap values were calculated from 1,000 trees. The scale bar indicates the estimated number of substitutions per 50 bases. Names and accession numbers are given as cited in the GenBank database.
The family Coriobacteriaceae is currently composed of 13 genera, of which Eggerthella is the one most commonly associated with clinical infections. In 2004, we reported the discovery of two novel Eggerthella species, E. hongkongensis and E. sinensis, from patients with bacteremia (5). Furthermore, these two novel species were shown to account for half of the cases of Eggerthella bacteremia, with an incidence similar to that of E. lenta (5). Recently, Gordonibacter pamelaeae, a proposed novel genus and species of the family Coriobacteriaceae, was isolated from a colonic biopsy specimen of a 33-year-old male patient with Crohn's disease on azathioprine, mutaflor, and cortisone (14). G. pamelaeae was phylogenetically most closely related to the Eggerthella species. Furthermore, based on chemotaxonomic data, it was also proposed that E. hongkongensis be reclassified as Paraeggerthella hongkongensis.
We defined the first case of bacteremia associated with G. pamelaeae with the help of both phenotypic tests and 16S rRNA gene sequencing. The clinical significance of the bacterium was evident by its isolation from blood culture as pure growth, the patient's systemic inflammatory response (fever, leukocytosis, and neutrophilia) to the bacteremia, and the prompt response to amoxicillin-clavulanate treatment. E. lenta, E. hongkongensis, E. sinensis, and G. pamelaeae are all anaerobic, Gram-positive, nonsporulating, coccobacilli that grow as nonhemolytic, gray colonies of 0.5 mm in diameter after 48 h of incubation at 37°C and are catalase and arginine dihydrolase positive. Similar to E. lenta, E. hongkongensis, or E. sinensis, the strain isolated in the present study is nonmotile, which is in contrast to the reported strain of G. pamelaeae, which is motile and has a subpolar flagellum (14). 16S rRNA gene sequencing confirmed unambiguously that the present isolate is G. pamelaeae, with only one base difference between its 16S rRNA gene sequence and that of the reported G. pamelaeae (Fig. 1).
The gastrointestinal tract is likely to be the reservoir of G. pamelaeae, E. hongkongensis, and E. sinensis and the source of bacteremia in these patients. E. lenta is an important member of the normal flora in the human intestine (1, 6). In addition to cases of bacteremia, the bacterium has been isolated from appendix tissues, peritoneal fluid, intra-abdominal and peritoneal abscesses, and intestinal tumors (7). The sources of infections in these patients were probably the gastrointestinal tract. For the four cases of E. hongkongensis and one case of E. sinensis that we reported previously, the four cases of E. hongkongensis bacteremia suffered from perianal abscess, infected rectal tumor, liver abscess, and acute appendicitis, respectively, and the case of E. sinensis bacteremia had acute proctitis (5). Moreover, four of these five cases had polymicrobial bacteremia, with other bacteria of the gastrointestinal tract, including Bacteroides fragilis, Peptostreptococcus species, Streptococcus constellatus, and Escherichia coli, recovered (5). As for G. pamelaeae, the bacterium was originally discovered in the colonic biopsy specimen of a patient with Crohn's disease (14). The patient of the present report had rectosigmoid carcinoma with fresh blood and mucus in his stool. We speculate that he might have had an infected tumor with secondary G. pamelaeae bacteremia or, alternatively, that the normal integrity of the gastrointestinal tract was breached by the tumor and that he had primary G. pamelaeae bacteremia. Moreover, the 16S rRNA gene sequences of these two strains were almost identical to another clone from the human intestine (GenBank accession no. EF413639). All these showed that the gastrointestinal tract is probably the reservoir of these bacterial species of closely related genera. Further studies will determine the relative predominance of G. pamelaeae, E. lenta, E. hongkongensis, and E. sinensis as members of the human intestinal flora as well as their relative importance in causing bacteremia and other infections.
Nucleotide sequence accession number.
The 16S rRNA gene sequence of the isolate has been deposited within the GenBank sequence database under accession number GQ292558.
Acknowledgments
This work is partly supported by the University Development Fund and the Committee for Research and Conference Grant, The University of Hong Kong.
Footnotes
Published ahead of print on 18 November 2009.
REFERENCES
- 1.Eggerth, A. 1935. The gram-positive non-spore-bearing anaerobic bacilli of human faeces. J. Bacteriol. 30:277-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kageyama, A., Y. Benno, and T. Nakase. 1999. Phylogenetic evidence for the transfer of Eubacterium lentum to the genus Eggerthella as Eggerthella lenta gen. nov., comb. nov. Int. J. Syst. Bacteriol. 49:1725-1732. [DOI] [PubMed] [Google Scholar]
- 3.Lau, S. K., K. H. Ng, P. C. Woo, K. T. Yip, A. M. Fung, G. K. Woo, K. M. Chan, T. L. Que, and K. Y. Yuen. 2006. Usefulness of Microseq 500 16S rDNA bacterial identification system for identification of anaerobic gram-positive bacilli isolated from blood cultures. J. Clin. Pathol. 59:219-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau, S. K., P. C. Woo, A. M. Fung, K. M. Chan, G. K. Woo, and K. Y. Yuen. 2004. Anaerobic, non-sporulating, Gram-positive bacilli bacteraemia characterized by 16S rRNA gene sequencing. J. Med. Microbiol. 53:1247-1253. [DOI] [PubMed] [Google Scholar]
- 5.Lau, S. K., P. C. Woo, G. K. Woo, A. M. Fung, M. K. Wong, K. M. Chan, D. M. Tam, and K. Y. Yuen. 2004. Eggerthella hongkongensis sp. nov. and Eggerthella sinensis sp. nov., two novel Eggerthella species, account for half of the cases of Eggerthella bacteremia. Diagn. Microbiol. Infect. Dis. 49:255-263. [DOI] [PubMed] [Google Scholar]
- 6.Moore, W. E., and L. V. Holdeman. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosca, A., P. Summanen, S. M. Finegold, G. De Michele, and G. Miragliotta. 1998. Cellular fatty acid composition, soluble-protein profile, and antimicrobial resistance pattern of Eubacterium lentum. J. Clin. Microbiol. 36:752-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray, P. R., E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.). 2007. Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 9.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woo, P. C., A. M. Fung, S. K. Lau, and K. Y. Yuen. 2002. Identification by 16S rRNA gene sequencing of Lactobacillus salivarius bacteremic cholecystitis. J. Clin. Microbiol. 40:265-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woo, P. C., A. M. Fung, S. K. Lau, E. Hon, and K. Y. Yuen. 2002. Diagnosis of pelvic actinomycosis by 16S ribosomal RNA gene sequencing and its clinical significance. Diagn. Microbiol. Infect. Dis. 43:113-118. [DOI] [PubMed] [Google Scholar]
- 12.Woo, P. C., A. M. Fung, S. K. Lau, J. L. Teng, B. H. Wong, M. K. Wong, E. Hon, G. W. Tang, and K. Y. Yuen. 2003. Actinomyces hongkongensis sp. nov. A novel Actinomyces species isolated from a patient with pelvic actinomycosis. Syst. Appl. Microbiol. 26:518-522. [DOI] [PubMed] [Google Scholar]
- 13.Woo, P. C., S. K. Lau, A. W. Lin, S. O. Curreem, A. M. Fung, and K. Y. Yuen. 2007. Surgical site abscess caused by Lactobacillus fermentum identified by 16S ribosomal RNA gene sequencing. Diagn. Microbiol. Infect. Dis. 58:251-254. [DOI] [PubMed] [Google Scholar]
- 14.Würdemann, D., B. J. Tindall, R. Pukall, H. Lünsdorf, C. Strömpl, T. Namuth, H. Nahrstedt, M. Wos-Oxley, S. Ott, S. Schreiber, K. N. Timmis, and A. P. Oxley. 2009. Gordonibacter pamelaeae gen. nov., sp. nov., a new member of the Coriobacteriaceae isolated from a patient with Crohn's disease, and reclassification of Eggerthella hongkongensis Lau et al. 2006 as Paraeggerthella hongkongensis gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 59:1405-1415. [DOI] [PubMed] [Google Scholar]