Abstract
Burkholderia cenocepacia can cause serious infections and epidemics in patients with cystic fibrosis (CF). A CF population in the Czech Republic experienced an epidemic outbreak caused by a B. cenocepacia ST-32 strain. The clonality of the isolates was evident by multilocus sequence typing; however, fingerprinting profiles obtained by pulsed-field gel electrophoresis (PFGE) showed substantial band variability. We investigated whether the PFGE pattern diversity resulted from genomic rearrangements mediated by insertion sequences (IS); in addition, we determined whether stressful growth conditions altered the transposition activity of these IS. DNA probes for IS commonly found in B. cenocepacia were designed using the B. cenocepacia J2315 genome. Southern hybridization analysis of ST-32 isolates demonstrated diversity in both the copy number and the insertion site for a homologue of ISBcen20. Movement of the ISBcen20 homologue was detected when the ST-32 isolate CZ1238 was exposed to oxidative stress (growth in the presence of H2O2). PFGE analysis of CZ1238 derivatives exposed to oxidative stress demonstrated genomic rearrangements. Interestingly, when the closely related B. cenocepacia strain J2315 was exposed to oxidative stress, no movement of ISBcen20 was detected. Since frameshift mutations are present within the transposases of all copies of this IS in J2315, our data suggest that the transposase is inactive. In summary, we have demonstrated for the first time that IS movement can be mediated by oxidative stress and can lead to genomic rearrangements in the CF pathogen B. cenocepacia. These IS movements may alter the PFGE fingerprints of isolates that are clonal by other typing methods.
Burkholderia cenocepacia is a member of the Burkholderia cepacia complex (Bcc), a group of opportunistic pathogens that cause devastating infections in persons with cystic fibrosis (CF). Infection of a CF patient with these bacteria is usually chronic, becomes resistant to antibiotic treatment, and may be transmissible to other CF patients (15). The majority of known transmissible strains belong to the species B. cenocepacia, which is considered one of the most severe pathogens of all Bcc organisms. The epidemic lineage historically called ET-12, randomly amplified polymorphic DNA (RAPD) type 02, or the cable pilus strain (reviewed in reference 12) has been causing infections in the United Kingdom and Canada for the past 20 years (14, 20). Other epidemic clones of B. cenocepacia identified in Canada have been designated RAPD types 01, 04, and 06 (14, 19). Strains that were genetically very closely related to the Canadian RAPD 01 strain were detected a few years later in almost 30% of CF patients from the Czech Republic (an epidemic clone designated CZ1) (6).
Nearly all these transmissible CF strains were first defined by the use of pattern-matching fingerprinting techniques such as pulsed-field gel electrophoresis (PFGE) and/or RAPD (reviewed in reference 12). Despite the fact that computer software can enable the clustering of closely related fingerprints, the criteria for data analysis may not be easy to apply and may lead to results that are hard to interpret. For example, considerable variation was observed in the PFGE profiles of 77 isolates from the Czech epidemiological survey (differing by as many as 8 bands); however, RAPD analysis of the same set of isolates demonstrated that they all belonged to the single epidemic strain CZ1 (6).
The drawbacks of gel-based typing methods have recently been overcome by an alternative approach, multilocus sequence typing (MLST), that compares the nucleotide sequences of seven housekeeping genes of the Bcc (2). MLST analysis of the Bcc has a number of useful attributes: (i) since MLST is a DNA sequence-based method, all information on strain sequence types (STs) can be stored in a public database that allows lab-to-lab comparison of results; (ii) MLST provides strain genotype information as well as identifying isolates to the species level, a feature that has facilitated the recent characterization of several new Bcc species (21, 22); (iii) MLST has excellent resolving power, which has enabled the matching of clonal strains residing in the environment and in patients with clinical infections (1), as well as the tracing of the global distribution of major Bcc clones, such as Burkholderia contaminans ST-102 (13). Since MLST examines components of the core genome of Bcc bacteria, it is unlikely to be affected by genomic rearrangements within these bacteria.
We hypothesized that the intrastrain PFGE band heterogeneity observed for the Czech epidemic strain (6) may have resulted from genomic rearrangements mediated by the movement of insertion sequences (IS), which are highly abundant in Bcc genomes (11). Genome sequence analysis of B. cenocepacia strain J2315 (ST-28) revealed that it possessed 22 different types of IS elements, 4 of which were present at high copy numbers (more than 5 copies per genome): IS407 (13 copies), ISBcen8 (12 copies), ISBcen9 (9 copies), and ISBcen20 (9 copies) (7). Enhanced transposition activity of mobile elements in bacteria in response to various stress conditions, such as high temperature (17) and starvation (8), has been observed. Another stress stimulus that may play a role in potentiating IS movement in bacteria during CF infection is oxidative stress mediated by toxic reactive oxygen species (ROS). Evidence for this assumption was presented in a recent transcriptomic study of the oral bacterium Porphyromonas gingivalis, which, when exposed to H2O2, demonstrated concerted overexpression of one IS family (4). The aims of our study were (i) to characterize the distribution of selected IS elements in the Czech epidemic strain CZ1 (designated ST-32 by MLST [1]), (ii) to evaluate their possible link to the heterogeneous fingerprints of CZ1 generated by PFGE, and (iii) to examine whether oxidative stress conditions promoted the movement of IS elements within the genomes of strain CZ1 and the sequenced B. cenocepacia strain J2315 (ST-28).
MATERIALS AND METHODS
Bacterial strains.
In total, 31 isolates belonging to 4 different B. cenocepacia STs defined by RAPD/PFGE genotyping (6) or MLST (2) were included in the study: ST-32 (the ST of the Czech epidemic strain) (14 isolates), ST-28 (6 isolates), ST-234 (7 isolates), and ST-210 (4 isolates). These isolates were drawn from the Cardiff University collection, Cardiff, Wales, United Kingdom (12).
Exposure to H2O2.
Isolate CZ1238 (BCC0961; representative of strain CZ1) and strain J2315 were used to study the effect of oxidative stress on the plasticity of the B. cenocepacia genome. A 1.5-ml aliquot of a culture of each strain (grown in Luria-Bertani [LB] broth at 37°C with shaking at 150 rpm to the mid-log growth phase; ∼5 × 108 cells) was harvested by centrifugation (1,500 × g for 5 min). The bacterial pellet was resuspended in 3 ml of basal salts medium (18) containing 22 mM glucose, 0.05% Casamino Acids, and hydrogen peroxide at subinhibitory concentrations for each respective strain (0.01% for CZ1238 and 0.1% for J2315). Incubation in the presence of H2O2 was continued until the cultures reached an optical density at 600 nm (OD600) of 0.6 (∼10 to 16 h of growth), after which the cultures were serially diluted and plated out onto tryptic soya agar plates to obtain single colonies. For B. cenocepacia CZ1238, 7 single colonies were individually cultivated for storage and further study. For B. cenocepacia J2315, 96 single colonies were picked into a 96-well plate containing 150 μl of LB medium, grown overnight at 37°C, and then stored at −80°C after the addition of dimethyl sulfoxide.
Serial passage.
To assess the stability of ISBcen20 during growth passage at 37°C, B. cenocepacia CZ1238 was treated as follows. After overnight growth in LB broth, CZ1238 was plated onto an LB agar plate and left to grow for 24 h. One colony was then picked and transferred to LB broth to initiate another overnight growth. This cycle of growth and plating was repeated 8 more times to obtain a derivative of isolate CZ1238 that had passaged through 9 sequential cultivation cycles; 7 single colonies of this derivative were individually cultivated for storage and further study.
Heat shock.
To examine the effect of an elevated temperature on IS stability in B. cenocepacia CZ1238, an overnight broth culture of CZ1238 was agitated in a 42°C water bath for 30 min before being serially diluted and plated for individual colonies on tryptic soya agar plates. Seven single colonies were then picked and cultivated for storage and further study.
Multiplex PCR.
To detect all nine copies of ISBcen20 present in the genome of B. cenocepacia J2315, a universal forward primer and nine reverse primers, each specific to one of nine IS insertion points, were designed (Table 1). The distance of each reverse primer from the common forward primer differed from each other by at least 50 bp; therefore, the resulting amplicons were analyzed in a multiplex PCR format as follows. Detection of all nine IS copies was split into three multiplex PCRs, each identifying three of the ISBcen20 insertion points (i.e., PCR M1 detected insertion points 1, 4, and 7; PCR M2 detected insertion points 2, 5, and 8; and PCR M3 detected insertion points 3, 6, and 9 [see Table 1 for combinations]). A PCR for any primer combination was carried out in a volume of 20 μl with 1 μl of DNA extracted with 5% Chelex (200/400 mesh; Bio-Rad, Hemel Hempstead, United Kingdom) and with reagents of the Taq PCR core kit (Qiagen, Germany) consisting of 1× PCR buffer, 1× Solution Q, 3.7 mM MgCl2, 0.2 mM (each) deoxynucleoside triphosphate (dNTP), and 0.75 U of Taq polymerase. Primer concentrations are specified in Table 1; the concentration of the universal forward primer was equal to the concentration of the corresponding reverse primer in every respective primer pair. The PCR program was run on a Dyad DNA engine thermal cycler (Bio-Rad Laboratories, Hemel Hempstead, United Kingdom) with initial denaturation for 5 min at 94°C and a subsequent run of 30 cycles, each comprising 30 s at 94°C, 30 s at 60°C, and 45 s at 72°C. The PCR products were separated on a 2% agarose gel with 1× Tris-borate-EDTA at 3.5 V/cm.
TABLE 1.
Primers used for multiplex PCR to detect all nine copies of ISBcen20 present in the genome of B. cenocepacia J2315
| Insertion point (gene no. of ISBcen20) | Sequence (5′ to 3′) | Product size (bp) | Primer concn (μM) | Multiplex PCR no. |
|---|---|---|---|---|
| Reverse primers | ||||
| 1 (BCAL0573) | AGGAAATCGCCGCGATTC | 322 | 1 | M1 |
| 2 (BCAL1125) | TCTCGAACTGGAAGTGCC | 387 | 0.75 | M2 |
| 3 (BCAL1133) | TCGACGAAAGGGTGACAC | 491 | 0.4 | M3 |
| 4 (BCAL2480b) | TCGAACCGGAACTTCTCC | 557 | 1 | M1 |
| 5 (BCAL2496) | CAGACTGGGTTGATCGAG | 639 | 1.5 | M2 |
| 6 (BCAL3126b) | TCTCCTTGTCGAGTCGAC | 725 | 1.25 | M3 |
| 7 (BCAL3484) | TTCGATCCTGCGTTCGAC | 821 | 1 | M1 |
| 8 (BCAM1906) | TGCAGAAGCAAGGGCATC | 896 | 1.5 | M2 |
| 9 (BCAM1925) | TGTTCCCGTATATCGGGC | 944 | 1.75 | M3 |
| Forward primer, 1 to 9 | TCAGTTGTTCCGATGCCTG | |||
RFLP and Southern hybridization.
All 31 clinical isolates described above were subjected to restriction fragment length polymorphism (RFLP) and Southern hybridization as follows. High-molecular-weight genomic DNA was extracted as described previously (14), and approximately 1.5 μg of DNA was digested for 3 to 4 h with 80 U of EcoRI (Promega). The digested DNA was separated on a 1% agarose gel at 0.8 V/cm for 15 h and was then transferred to a positively charged nylon membrane.
Digoxigenin (DIG)-labeled hybridization probes specific to insertion sequences ISBcen20, ISBcen8, ISBcen9, and IS407 were amplified from B. cenocepacia J2315 using the primers shown in Table 2. To facilitate labeling, the following PCR mixture was prepared in a 25-μl reaction mixture: 1× PCR DIG labeling mix (Roche Applied Science), 1× PCR buffer (Promega), 1.5 μM MgCl2, 0.5 μM (each) primers, 0.2 mM (each) dNTP, and 1 U of Taq polymerase (Promega). The PCR thermal conditions were identical to those used for multiplex PCR. Probe-target hybridization and chemiluminescence-based detection were performed according to the manufacturer's instructions (Roche Applied Science). Each probe was added to the hybridization solution at a concentration of approximately 20 ng/ml, and the membranes were left to hybridize at 50°C overnight. The degree of pattern similarity between individual isolates was analyzed by the unweighted-pair group method using arithmetic means in GelCompar II software (version 5.1; Applied Maths, Belgium).
TABLE 2.
Primers used in IS-specific PCR
| IS detected | Sequence (5′ to 3′) | Product size (bp) |
|---|---|---|
| ISBcen20 | TGATCGAGCCGTTATTGC | 736 |
| GCAGCAACCGAGTTTGAG | ||
| ISBcen8 | GACGTCGCAAAAGCTCAC | 842 |
| GCGACCTTGGACAGTTTG | ||
| ISBcen9 | CGCAATGCCAAAAACACC | 953 |
| TTGATGCGCTCGTGATTG | ||
| IS407 | GGTTCAGTGATGCGTCGT | 1,016 |
| GGAAAGCGGCAGGAGTAT |
Southern hybridization analysis with the ISBcen20 probe was also performed on DNA extracted from cultures of each of the CZ1238 derivatives obtained from heat shock, serial passage, and exposure to H2O2.
Macrorestriction typing.
A fingerprinting method based on DNA digestion followed by PFGE was performed exactly as described previously (6). Briefly, the genomic DNA embedded in 2% low-gelling-temperature agarose (Sigma) was digested with 3 U of SpeI (Promega) at 37°C overnight and was then separated on a 1.2% pulsed-field certified agarose gel (Bio-Rad) for 24 h under the following conditions: 6 V/cm; switch time, 1 to 40 s for the initial 10 h and 30 to 90 s for the remaining 14 h. The resulting banding patterns were compared by using GelCompar II.
RESULTS
The Czech epidemic strain is part of a globally distributed B. cenocepacia lineage.
The PFGE fingerprints of isolates belonging to the Czech epidemic strain had previously been shown to be highly variable (6). MLST analysis of 10 CZ1 isolates demonstrated that they all belonged to a single sequence type, ST-32, corroborating previous RAPD analysis that had indicated they were clonal (6) and demonstrating that PFGE fingerprinting had detected rapid evolution in these isolates as a result of genomic rearrangements. Further examination of our strain collection demonstrated the presence of 35 additional ST-32 isolates. These comprised B. cenocepacia isolates recovered from individuals with CF and from humans with infections not linked to CF. One environmental ST-32 isolate from Mexico had been described in a previous study (1). Overall, the ST-32 isolates in our collection were globally distributed, indicating that this ST is another highly virulent B. cenocepacia strain IIIA lineage like the ET-12 lineage (12).
Presence of selected IS elements in B. cenocepacia.
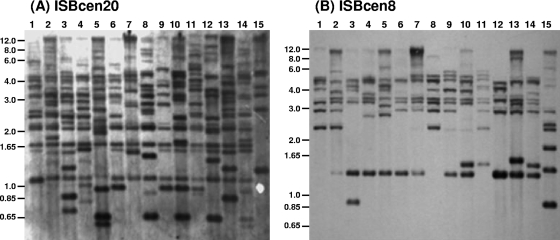
Fourteen Czech ST-32 isolates were examined by RFLP followed by Southern hybridization for the presence of homologues of the IS elements in the B. cenocepacia J2315 genome. Four IS elements were specifically looked for, since they represented those with the highest copy numbers, which were most likely to mediate genomic rearrangements (7): ISBcen20 (9 copies), ISBcen8 (12 copies), ISBcen9 (9 copies), and IS407 (13 copies). No homologues of ISBcen9 or IS407 were detected in any of the ST-32 isolates examined (data not shown). The remaining IS elements tested were found in multiple copies in the ST-32 isolates, ranging from 12 to 19 copies for ISBcen20 (Fig. 1A) and 6 to 12 copies for ISBcen8 (Fig. 1B). When the IS RFLP hybridization profiles were compared computationally, much greater heterogeneity was detected for ISBcen20, with only 6 of the 14 isolates showing more than 50% fingerprint similarity (data not shown). Application of the same cutoff to the ISBcen8 fingerprints demonstrated that a majority of the Czech ST-32 isolates (11 out of 14) fell into one cluster. These data demonstrated that the ISBcen20 homologue in the ST-32 isolates appeared far more active, with rapid evolution in the insertion site and copy number in a strain that was clonal in the core genome, as indicated by the single MLST type.
FIG. 1.
Southern hybridization of ST-32 isolates with ISBcen20 (A) and ISBcen8 (B) probes. Samples are as follows: lanes 1, BCC0817; lanes 2, BCC0818; lanes 3, BCC0961; lanes 4, BCC0851; lanes 5, BCC0896; lanes 6, BCC0885; lanes 7, BCC1110; lanes 8, BCC1114; lanes 9, BCC1111; lanes 10, BCC1112; lanes 11, BCC1113; lanes 12, BCC1121; lanes 13, BCC1126; lanes 14, BCC1128; lanes 15, reference strain J2315 (ST-28). The approximate positions of the molecular size markers (in kilobases) from the agarose gel electrophoresis of the DNA are indicated to the left of each gel.
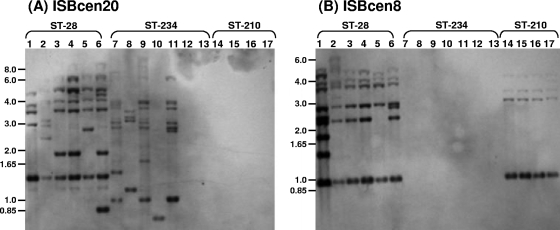
The IS RFLP hybridization profiles of three other epidemic B. cenocepacia IIIA strains (14, 19) were examined: strains belonging to ST-28 (RAPD type 02; ET-12 lineage), ST-234 (RAPD type 04), and ST-210 (RAPD type 06). The B. cenocepacia ST-28 and ST-234 strains contained multiple copies of ISBcen20 and ISBcen8 homologues; the banding patterns for ISBcen20 were more variable, results correlating with those for the Czech epidemic strain (Fig. 2). Further examination of additional ST-234 isolates demonstrated that two out of seven lacked ISBcen20 homologues, suggesting that positive and negative IS variants can exist for this lineage. Only ISBcen8 was detected in the four B. cenocepacia ST-210 isolates tested; ISBcen20 was absent from all four. Since the ISBcen20 homologue had exhibited higher diversity in its copy number and distribution in ST-32 isolates than ISBcen8, we chose to focus our further work on the former element, assuming that this was most likely an active mobile element contributing to the rapid genomic rearrangements observed by PFGE (6).
FIG. 2.
Southern hybridization of isolates of three STs (given above the lane numbers) with ISBcen20 (A) and ISBcen8 (B) probes. Samples are as follows: lanes 1, J2315; lanes 2, BCC0313; lanes 3, BCC0162; lanes 4, BCC0163; lanes 5, BCC0164; lanes 6, BCC0077; lanes 7, BCC0575; lanes 8, BCC0107; lanes 9, BCC1168; lanes 10, BCC0078; lanes 11, BCC0534; lanes 12, BCC0222; lanes 13, BCC0223; lanes 14, BCC0536; lanes 15, BCC0541; lanes 16, BCC0514; lanes 17, BCC0098. The approximate positions of the molecular size markers (in kilobases) from the agarose gel electrophoresis of the DNA are indicated to the left of each gel.
Movement of ISBcen20 in ST-32.
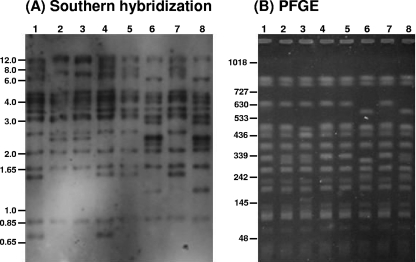
To assess the degree of ISBcen20 movement under physiological and stress conditions, isolate CZ1238, a representative of B. cenocepacia ST-32, was subjected either to serial passage on nutrient medium, to a heat shock, or to growth in a subinhibitory concentration of H2O2 (see Materials and Methods). No alterations in the intragenomic distribution of ISBcen20 were detected in a set of seven subcultures passaged on agar or in seven colonies exposed to a heat shock (data not shown). However, oxidative stress induced by exposure to 0.01% H2O2 potentiated ISBcen20 activity: only one of the seven derivatives tested had the same IS profile as wild-type CZ1238 (Fig. 3A). Overall, in addition to the wild-type IS profile, three new IS profiles were detected among the seven CZ1238 derivatives examined (Fig. 3A). The IS movement triggered by exposure to H2O2 was also reflected in changes to the macrorestriction profile obtained by PFGE analysis: four different profiles differing by as many as five bands were detected among the seven CZ1238 derivatives (Fig. 3B). Although four of the seven colonies studied shared the same PFGE profile, none of the PFGE patterns corresponded exactly to that of the parental strain (Fig. 3B).
FIG. 3.
Southern hybridization with an ISBcen20 probe (A) and PFGE (B) of seven individual colonies (lanes 1 to 7, respectively) of ST-32 strain CZ1238 exposed to 0.01% H2O2. Lanes 8, unexposed CZ1238. (A) Profiles in lanes 1 and 4, lanes 2 and 3, lanes 5 and 7, and lanes 6 and 8 match each other completely. The approximate positions of the molecular size markers (in kilobases) from the agarose gel electrophoresis of the DNA for the Southern hybridization are shown on the left. (B) Profiles in lanes 1, 4, 5, and 7 match each other completely. The positions and sizes (in kilobases) of the markers run for PFGE are shown on the left.
Movement of ISBcen20 in B. cenocepacia ST-28.
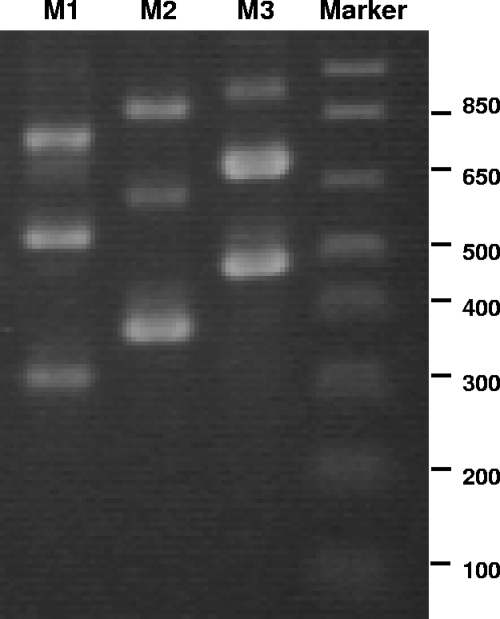
The results for the Czech epidemic strain CZ1238 prompted the study of IS behavior in the genome-sequenced epidemic B. cenocepacia strain J2315 (7). The availability of genome sequence information made it possible to follow transposition events for every single ISBcen20 copy in the genome. To detect all nine ISBcen20 copies and to determine the frequency of their transposition via a cut-and-paste mechanism, we designed a set of three multiplex PCRs that amplified a product of a slightly different size for each IS insertion point (Table 1; Fig. 4). This approach was used to screen 96 colonies of J2315 derived from a culture exposed to 0.1% H2O2. In contrast to those from strain CZ1, all the J2315 colonies tested possessed each of the nine copies of ISBcen20 at the original insertion points derived from genome sequence analysis. To check if other possible transposition mechanisms (undetectable by PCR) and any genomic rearrangements had occurred under our experimental conditions, seven random colonies of J2315 were further analyzed by PFGE and ISBcen20 hybridization profiling. Neither method displayed any visible changes in the genomes of the colonies derived, indicating an absence of oxidative-stress-primed ISBcen20 activity in B. cenocepacia strain J2315 (data not shown).
FIG. 4.
PCR detection of the nine ISBcen20 copies present in B. cenocepacia strain J2315. Lanes M1 to M3 represent multiplex PCRs, each identifying three IS copies with different insertion points within the genome. Marker, 1-kb molecular size marker. Fragments of relevant sizes (in base pairs) are indicated on the right.
DISCUSSION
The molecular typing anomalies described previously for B. cenocepacia strain CZ1 (ST-32) raised the question of the basis for the extensive genomic rearrangements within a single strain (6). We have demonstrated that the reason may lie in the diversity and activity of the large number of IS elements present in the Burkholderia genome. Based on the data available from the sequenced genome of B. cenocepacia strain J2315 (ST-28), which is closely related to strain CZ1, we selected the four IS elements that have the highest copy numbers in J2315 and tested the CZ1 genome for the presence of homologues. ISBcen20 and ISBcen8 were found in all tested CF isolates of the CZ1 clone, with ISBcen20 in particular demonstrating substantial variability in its copy number and genomic distribution. Marked variation in the RFLP-Southern hybridization profiles of ST-32 isolates probed with ISBcen20 was also observed for other B. cenocepacia strains known to have caused multiple epidemic outbreaks (i.e., ST-28 [ET-12] and ST-234 [RAPD 04]) (14 strains). It was noteworthy that two of the seven ST-234 isolates tested did not possess an ISBcen20 homologue; whether this represents a loss or a gain of the IS element remains to be determined, but it does demonstrate the capacity for very rapid evolution in terms of the foreign DNA content in clonal strains of B. cenocepacia.
The significant changes in the copy number and insertion sites of ISBcen20 homologues observed in CF isolates of the same strain suggested that in vivo transposition activity of this element may occur. We hypothesized that the movement may be stimulated by oxidative stress, which is an inherent feature of the respiratory tract in cystic fibrosis patients; the subsequent demonstration of ISBcen20 movement mediated by oxidative stress in the Czech strain is highly novel in the context of both IS elements and CF microbial pathogens. A chronically inflamed CF lung is rich in ROS, which are produced by the host immune system in response to infection. CF pathogens have developed various ROS-protective mechanisms to enable successful colonization of the lung. For example, B. cenocepacia possesses antioxidant enzymes, such as superoxide dismutase, catalase, and catalase-peroxidase (9, 10), and recent microarray analysis has demonstrated induction of a hydroperoxide resistance protein in response to ROS (5). Although these enzymes allow bacteria to survive in the airways, they are unlikely to neutralize all ROS, which may therefore still affect some processes occurring in bacteria at molecular levels.
DNA analysis of individual CZ1 colonies exposed to oxidative stress clearly demonstrated that ISBcen20 has moved and very likely contributed to changes in the PFGE profiles of these derivatives. In contrast, ISBcen20 distribution, as well as PFGE fingerprints, remained unchanged in B. cenocepacia J2315, even though this strain was also capable of good growth under the same stress conditions as CZ1. All nine copies of ISBcen20 within the J2315 genome contain a frameshift mutation within the IS element's transposase (7). The bioinformatic annotation of J2315 suggests that this transposase may be expressed if a −1 ribosomal translational slippage (16) occurs at the mutation point within its coding sequence (5′-AAAAAC-3′) (7). The lack of H2O2-induced movement of ISBcen20 in J2315 suggests that in fact this frameshift mutation in the transposase is a lethal mutation that cannot be overcome and that the ISBcen20 element is a pseudogene in this strain. These data also suggest that IS movement in response to oxidative stress is strain specific in B. cenocepacia, and the degree of genomic stability may differ considerably between closely genetically related strains.
Heat shock has been shown to increase the frequencies of transposition of 8 different IS elements in an environmental isolate of Burkholderia multivorans (17); interestingly, oxidative stress did not stimulate IS activity in this B. multivorans strain. The lack of ISBcen20 movement in B. cenocepacia after heat shock suggests either that this element is unresponsive to this form of stress or that temperature is not a sufficiently stimulating factor for epidemic B. cenocepacia CF strains, which are well adapted to growth in human lungs. However, the demonstration that an IS element in B. cenocepacia can respond to oxidative stress, an inherent feature of the CF lung environment, is highly significant for other CF pathogens. It will be interesting to explore whether other B. cepacia complex species and even Pseudomonas aeruginosa (both of which have multiple IS elements) also possess this rapid form of evolution, which, if it alters virulence, could be responsible for the wide variation in clinical outcomes associated with microbial infection in CF.
PFGE has been employed as a gold-standard bacterial-typing technique for many bacterial pathogens. However, its usefulness for B. cenocepacia has not been unequivocal, especially when large populations of organisms are to be analyzed (3). Very high discriminatory power may be misleading in terms of correct determination of epidemiologic relatedness among isolates and may not cluster together those that actually belong to the same clone. This was the pitfall of our previous epidemiological analysis, where we had great difficulty interpreting the PFGE data obtained for Czech clinical isolates (6). The present study has documented that PFGE results for the ST-32 strain were affected by oxidative stress conditions and that PFGE pattern variability was very likely affected by the transposition activity of at least one insertion sequence within this B. cenocepacia strain. The fact that this activity could be detected after a few hours of growth in the presence of oxidative stress indicates the capacity of certain B. cenocepacia strains for rapid IS-mediated evolution.
It is noteworthy that our data demonstrated only an indirect link between ISBcen20 activity and the PFGE patterns obtained after exposure to oxygen radicals (Fig. 3). Although we observed substantial ISBcen20 movement following incubation in 0.01% H2O2, PFGE data derived from the same condition could not be considered to be altered specifically due to ISBcen20. In addition, pairs of CZ1238 colonies that showed different profiles by Southern hybridization (Fig. 3A) did not match those with changed patterns by PFGE (Fig. 3B). It is likely that multiple mobile elements were potentiated by oxidative stress and that their activity eventually led to changes in PFGE. ISBcen20 may be just one of the elements that can mediate changes in PFGE patterns through mechanisms of DNA rearrangement/recombination; it is unlikely that the straightforward self-insertion of a small IS can be detected directly by PFGE. Also, since ISBcen20 itself lacks a target sequence for the SpeI restriction enzyme used for PFGE analysis, the alterations detected by PFGE (Fig. 3) were most probably the result of large-scale genomic recombinations that occurred after the oxidative-stress-primed IS movements.
We conclude that ISBcen20 is an active mobile element in the epidemic B. cenocepacia ST-32 CF strain and that its activity is inducible by reactive oxygen species. This mechanism represents a novel form of rapid evolution that B. cenocepacia may undertake during infection, and it will be very interesting to determine whether other CF pathogens that are rich in IS elements respond in the same way. The transposition activity of ISBcen20 very likely contributes to genomic rearrangements of the strain that are detected by PFGE analysis.
Acknowledgments
This work was funded by grants from the Wellcome Trust (075586) and the Ministry of Education of the Czech Republic (MSM0021620812).
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Baldwin, A., E. Mahenthiralingam, P. Drevinek, P. Vandamme, J. R. Govan, D. J. Waine, J. J. LiPuma, L. Chiarini, C. Dalmastri, D. A. Henry, D. P. Speert, D. Honeybourne, M. C. Maiden, and C. G. Dowson. 2007. Environmental Burkholderia cepacia complex isolates in human infections. Emerg. Infect. Dis. 13:458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin, A., E. Mahenthiralingam, K. M. Thickett, D. Honeybourne, M. C. Maiden, J. R. Govan, D. P. Speert, J. J. Lipuma, P. Vandamme, and C. G. Dowson. 2005. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J. Clin. Microbiol. 43:4665-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coenye, T., T. Spilker, A. Martin, and J. J. LiPuma. 2002. Comparative assessment of genotyping methods for epidemiologic study of Burkholderia cepacia genomovar III. J. Clin. Microbiol. 40:3300-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diaz, P. I., N. Slakeski, E. C. Reynolds, R. Morona, A. H. Rogers, and P. E. Kolenbrander. 2006. Role of oxyR in the oral anaerobe Porphyromonas gingivalis. J. Bacteriol. 188:2454-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drevinek, P., M. T. Holden, Z. Ge, A. M. Jones, I. Ketchell, R. T. Gill, and E. Mahenthiralingam. 2008. Gene expression changes linked to antimicrobial resistance, oxidative stress, iron depletion and retained motility are observed when Burkholderia cenocepacia grows in cystic fibrosis sputum. BMC Infect. Dis. 8:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drevinek, P., S. Vosahlikova, O. Cinek, V. Vavrova, J. Bartosova, P. Pohunek, and E. Mahenthiralingam. 2005. Widespread clone of Burkholderia cenocepacia in cystic fibrosis patients in the Czech Republic. J. Med. Microbiol. 54:655-659. [DOI] [PubMed] [Google Scholar]
- 7.Holden, M. T., H. M. Seth-Smith, L. C. Crossman, M. Sebaihia, S. D. Bentley, A. M. Cerdeno-Tarraga, N. R. Thomson, N. Bason, M. A. Quail, S. Sharp, I. Cherevach, C. Churcher, I. Goodhead, H. Hauser, N. Holroyd, K. Mungall, P. Scott, D. Walker, B. White, H. Rose, P. Iversen, D. Mil-Homens, E. P. Rocha, A. M. Fialho, A. Baldwin, C. Dowson, B. G. Barrell, J. R. Govan, P. Vandamme, C. A. Hart, E. Mahenthiralingam, and J. Parkhill. 2009. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 191:261-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ilves, H., R. Horak, and M. Kivisaar. 2001. Involvement of sigma(S) in starvation-induced transposition of Pseudomonas putida transposon Tn4652. J. Bacteriol. 183:5445-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keith, K. E., and M. A. Valvano. 2007. Characterization of SodC, a periplasmic superoxide dismutase from Burkholderia cenocepacia. Infect. Immun. 75:2451-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lefebre, M., and M. Valvano. 2001. In vitro resistance of Burkholderia cepacia complex isolates to reactive oxygen species in relation to catalase and superoxide dismutase production. Microbiology 147:97-109. [DOI] [PubMed] [Google Scholar]
- 11.Lessie, T. G., W. Hendrickson, B. D. Manning, and R. Devereux. 1996. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol. Lett. 144:117-128. [DOI] [PubMed] [Google Scholar]
- 12.Mahenthiralingam, E., A. Baldwin, and C. G. Dowson. 2008. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J. Appl. Microbiol. 104:1539-1551. [DOI] [PubMed] [Google Scholar]
- 13.Mahenthiralingam, E., A. Baldwin, P. Drevinek, E. Vanlaere, P. Vandamme, J. J. Lipuma, and C. G. Dowson. 2006. Multilocus sequence typing breathes life into a microbial metagenome. PLoS One 1:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahenthiralingam, E., M. E. Campbell, D. A. Henry, and D. P. Speert. 1996. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by randomly amplified polymorphic DNA fingerprinting. J. Clin. Microbiol. 34:2914-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahenthiralingam, E., T. A. Urban, and J. B. Goldberg. 2005. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3:144-156. [DOI] [PubMed] [Google Scholar]
- 16.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohtsubo, Y., H. Genka, H. Komatsu, Y. Nagata, and M. Tsuda. 2005. High-temperature-induced transposition of insertion elements in Burkholderia multivorans ATCC 17616. Appl. Environ. Microbiol. 71:1822-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Sullivan, L. A., A. J. Weightman, T. H. Jones, A. M. Marchbank, J. M. Tiedje, and E. Mahenthiralingam. 2007. Identifying the genetic basis of ecologically and biotechnologically useful functions of the bacterium Burkholderia vietnamiensis. Environ. Microbiol. 9:1017-1034. [DOI] [PubMed] [Google Scholar]
- 19.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun, L., R. Z. Jiang, S. Steinbach, A. Holmes, C. Campanelli, J. Forstner, U. Sajjan, Y. Tan, M. Riley, and R. Goldstein. 1995. The emergence of a highly transmissible lineage of cbl+ Pseudomonas (Burkholderia) cepacia causing CF centre epidemics in North America and Britain. Nat. Med. 1:661-666. [DOI] [PubMed] [Google Scholar]
- 21.Vanlaere, E., A. Baldwin, D. Gevers, D. Henry, E. De Brandt, J. J. LiPuma, E. Mahenthiralingam, D. P. Speert, C. Dowson, and P. Vandamme. 2009. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int. J. Syst. Evol. Microbiol. 59:102-111. [DOI] [PubMed] [Google Scholar]
- 22.Vanlaere, E., J. J. Lipuma, A. Baldwin, D. Henry, E. De Brandt, E. Mahenthiralingam, D. Speert, C. Dowson, and P. Vandamme. 2008. Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int. J. Syst. Evol. Microbiol. 58:1580-1590. [DOI] [PubMed] [Google Scholar]