Abstract
The analysis of A/H1N1 and A/H3N2 influenza viruses collected between 2005 and 2008 in Cambodia detected strains resistant to oseltamivir and confirmed widespread resistance to adamantanes. Phylogenetic analyses revealed intrasubtype reassortment, probable reemergence of A/H3N2 viruses in two consecutive seasons, and cocirculation of different lineages in each subtype.
Influenza A/H3N2 and A/H1N1 viruses are recurrent respiratory pathogens causing 250,000 to 500,000 deaths annually worldwide (16). Vaccination is the primary means of protection against influenza virus infections. Antiviral drugs are alternative options to prevent or treat infections and are divided into two classes: M2 protein inhibitors (adamantanes) and neuraminidase inhibitors (NAI). However, in recent years, the emergence of resistance due to several amino acid substitutions has become widespread (1, 13, 17). We analyzed the genetic relationships of 25 A/H3N2 and 21 A/H1N1 influenza strains, representative of the overall influenza virus circulation from 2005 to 2008 in different regions of Cambodia.
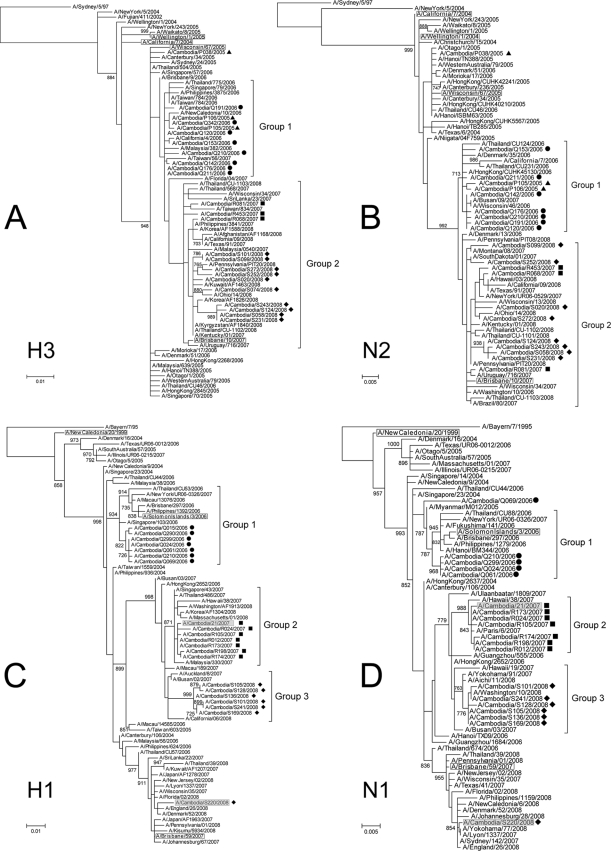
Maximum likelihood-based phylogenies (4) showed that most of the H3N2 Cambodian isolates formed two major groups (Fig. 1A and 1B), where group 1 and group 2 contained isolates from 2005 to 2006 and from 2007 to 2008, respectively. The phylogenies of the H1N1 subtype genes showed that the Cambodian isolates formed a separate group during each influenza season (Fig. 1C and 1D).
FIG. 1.
Rooted phylogenies of H3 hemagglutinin (A), N2 neuraminidase (B), H1 hemagglutinin (C), and N1 neuraminidase (D) gene sequences obtained from influenza viruses isolated between 2005 and 2008 in Cambodia. Cambodian isolates harboring a mutation H275Y on the NA protein are shaded. Cambodian isolates sequenced in 2005, 2006, 2007, and 2008 were labeled with triangles, circles, squares, and diamonds, respectively. The vaccine reference strains are boxed.
Although the A/Cambodia/Q069/2006 (H1N1) isolate was part of group 1, as shown by the phylogenetic relatedness of the HA and M (data not shown) segments, the phylogeny of the segment NA suggested a different evolutionary history, where A/Cambodia/Q069/2006 (H1N1) was located between group 1 and 2. Furthermore, the NA protein of A/Cambodia/Q069/2006 (H1N1) differed from those of the other 2006 Cambodian viruses by a significant number of amino acid changes. Reassortment is believed to play an important role in generating antigenically novel viruses. For instance, intersubtype reassortments between human and avian influenza viruses were responsible for past and current influenza pandemics (14). Intrasubtype reassortment generates diversity with the possibility of increasing the frequency of immune escape variants (3, 5, 10, 11).
The A/Cambodia/S220/2008 (H1N1) and A/Cambodia/21/2007 (H1N1) isolates contained the H275Y mutation (N1 numbering), which is associated with a high level of resistance to oseltamivir, a major NAI widely used for influenza infection treatment and chemoprophylaxis. Phylogenetically, A/Cambodia/S220/2008 (H1N1) belongs to clade 2B (6), which is characterized by many resistant variants. This suggests that this isolate was probably part of the overall emergence of that resistant variant but introduced at a very low level in Cambodia. A/Cambodia/21/2007 (H1N1) was more of a transient strain. The virus belonged to the A/Hong Kong/2652/2006-like clade (group 2, Fig. 1C), which usually did not include strains resistant to oseltamivir (6). It is worth noting that the use of NAI for influenza treatment is very uncommon in Cambodia. The A/Cambodia/S220/2008 (H1N1) strain did not grow in Madin-Darby canine kidney cells, even after several passages, but the A/Cambodia/21/2007 (H1N1) strain was tested successfully for susceptibility to neuraminidase inhibitor drugs. The mean concentrations of oseltamivir and zanamivir required to inhibit 50% of neuraminidase activity (IC50) for this virus were 1,144 nM and 0.81 nM, respectively. These results are in concordance with those obtained with other H275Y mutant viruses tested under the same conditions (6).
The S31N mutation responsible for adamantane resistance was present in the majority of the circulating A/Hong Kong/2652/2006-like H1N1 viruses, including Cambodian isolates from 2007 and 2008, with the exception of A/Cambodia/S220/2008 (H1N1), which belongs to the A/Brisbane/59/2007-like clade (where the S31N mutation is not common). A/Cambodia/P038/2005 (H1N1) and all the H3N2 sequences obtained from 2007 and 2008 Cambodian strains contained the S31N mutation.
These results suggest that NAI would be the drugs of choice for influenza treatment and chemoprophylaxis in Cambodia, as adamantanes are no longer expected to be effective.
The HA, NA, and M gene trees showed that A/Cambodia/P105/2005 (H3N2) and A/Cambodia/P106/2005 (H3N2) clustered with 2006 isolates from Cambodia but also with strains isolated in 2007 in other countries. Few polymorphic sites were observed between the neuraminidase proteins of A/Cambodia/P105/2005 (H3N2) and A/Cambodia/P106/2005 (H3N2) and those of the 2006 isolates, while the proteins coded by the M gene were identical. The 2005 and 2006 Cambodian isolates probably shared close antigenic properties, since no differences in the HA antigenic sites were found. Since these two 2005 sequences belonged to a cluster supported by a significant bootstrap, it is possible that 2006 viruses resulted from a reemergence phenomenon. The seasonality that characterizes influenza virus circulation is still poorly understood. Endogenous seeding has been previously reported (2, 5, 7, 15), although its apparent random emergence is not well understood. The frequency and the underlying mechanisms of endogenous seeding are important factors for predicting the next candidate vaccine composition. Furthermore, Southeast Asian countries are believed to play an important role in the spread of influenza due to the atypical year-round infections that could ignite the epidemic every year (12). It is then of paramount importance to unravel the circulation and the seasonality patterns of human influenza by expanding its surveillance, particularly in Southeast Asia, during influenza seasons and also between seasons, during which most antigenic drift is believed to occur (9, 10).
The phylogenies of the three genes suggested that multiple introductions of both subtypes occurred. A/Cambodia/S220/2008 (H1N1) was separated from the other Cambodian sequences obtained from viruses sampled during the same year by branches with high bootstrap values and long branches. A similar observation was made in 2005, as A/Cambodia/P105/2005 (H3N2) and A/Cambodia/P106/2005 (H3N2) HA1 sequences were significantly different from that of A/Cambodia/P038/2005 (H3N2) at the amino acid level. Indeed, A/Cambodia/P038/2005 (H3N2) did not cluster with other 2005 strains, suggesting the occurrence of another multiple introduction event, confirming that genetically different viruses can cocirculate, as previously suggested (5). Although A/Cambodia/S220/2008 (H1N1) HA1 differs from the other Cambodian isolates by two amino acids at the Sb antigenic site, these isolates could still have similar antigenic properties, as amino acid variation within a single antigenic cluster can be up to 19 substitutions while the intercluster variation can be as little as one substitution (8). Antigenic properties are therefore difficult to predict based solely on genetic analyses, and antigenic tests using a hemagglutination inhibition assay should ideally always be performed.
Nucleotide sequence accession numbers.
Sequences are available from GenBank under accession numbers FJ865225 through FJ865351.
Acknowledgments
We thank Aeron Hurt (WHOCC Melbourne) for his kind assistance, the staff of the NIC at the Virology Unit of the Institut Pasteur in Cambodia for their technical work, and Monica Naughtin for her editorial assistance. We are also grateful to the Ministry of Health (Communicable Disease Department) and to the WHO office in Cambodia and Manila for their collaboration in the Influenza-Like Illness surveillance system.
This study was supported by the French Ministry of Health, the French Agency for Development (SISEA project), and the Office of the Assistant Secretary for Preparedness and Response within the U.S. Department of Health and Human Services.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Barr, I. G., A. C. Hurt, P. Iannello, C. Tomasov, N. Deed, and N. Komadina. 2007. Increased adamantane resistance in influenza A(H3) viruses in Australia and neighbouring countries in 2005. Antivir. Res. 73:112-117. [DOI] [PubMed] [Google Scholar]
- 2.Boni, M. F. 2008. Vaccination and antigenic drift in influenza. Vaccine 26(Suppl. 3):C8-C14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bragstad, K., L. P. Nielsen, and A. Fomsgaard. 2008. The evolution of human influenza A viruses from 1999 to 2006: a complete genome study. Virol. J. 5:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 5.Holmes, E. C., E. Ghedin, N. Miller, J. Taylor, Y. Bao, K. St George, B. T. Grenfell, S. L. Salzberg, C. M. Fraser, D. J. Lipman, and J. K. Taubenberger. 2005. Whole-genome analysis of human influenza A virus reveals multiple persistent lineages and reassortment among recent H3N2 viruses. PLoS Biol. 3:e300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurt, A. C., J. Ernest, Y. M. Deng, P. Iannello, T. G. Besselaar, C. Birch, P. Buchy, M. Chittaganpitch, S. C. Chiu, D. Dwyer, A. Guigon, B. Harrower, I. P. Kei, T. Kok, C. Lin, K. McPhie, A. Mohd, R. Olveda, T. Panayotou, W. Rawlinson, L. Scott, D. Smith, H. D'Souza, N. Komadina, R. Shaw, A. Kelso, and I. G. Barr. 2009. Emergence and spread of oseltamivir-resistant A(H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antivir. Res. 83:90-93. [DOI] [PubMed] [Google Scholar]
- 7.Ikonen, N., R. Pyhälä, M. Toivonen, and H. Korpela. 2006. Influenza A/Fujian/411/02(H3N2)-lineage viruses in Finland: genetic diversity, epidemic activity and vaccination-induced antibody response. Arch. Virol. 151:241-254. [DOI] [PubMed] [Google Scholar]
- 8.Koelle, K., S. Cobey, B. Grenfell, and M. Pascual. 2006. Epochal evolution shapes the phylodynamics of interpandemic influenza A (H3N2) in humans. Science 314:1898-1903. [DOI] [PubMed] [Google Scholar]
- 9.Lavenu, A., M. Leruez-Ville, M. L. Chaix, P. Y. Boelle, S. Rogez, F. Freymuth, A. Hay, C. Rouzioux, and F. Carrat. 2006. Detailed analysis of the genetic evolution of influenza virus during the course of an epidemic. Epidemiol. Infect. 134:514-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson, M. I., L. Simonsen, C. Viboud, M. A. Miller, J. Taylor, K. S. George, S. B. Griesemer, E. Ghedin, E. Ghedi, N. A. Sengamalay, D. J. Spiro, I. Volkov, B. T. Grenfell, D. J. Lipman, J. K. Taubenberger, and E. C. Holmes. 2006. Stochastic processes are key determinants of short-term evolution in influenza A virus. PLoS Pathog. 2:e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson, M. I., C. Viboud, L. Simonsen, R. T. Bennett, S. B. Griesemer, K. St George, J. Taylor, D. J. Spiro, N. A. Sengamalay, E. Ghedin, J. K. Taubenberger, and E. C. Holmes. 2008. Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog. 4:e1000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rambaut, A., O. G. Pybus, M. I. Nelson, C. Viboud, J. K. Taubenberger, and E. C. Holmes. 2008. The genomic and epidemiological dynamics of human influenza A virus. Nature 453:615-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheu, T. G., V. M. Deyde, M. Okomo-Adhiambo, R. J. Garten, X. Xu, R. A. Bright, E. N. Butler, T. R. Wallis, A. I. Klimov, and L. V. Gubareva. 2008. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob. Agents Chemother. 52:3284-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith, G. J., D. Vijaykrishna, J. Bahl, S. J. Lycett, M. Worobey, O. G. Pybus, S. K. Ma, C. L. Cheung, J. Raghwani, S. Bhatt, J. S. Peiris, Y. Guan, and A. Rambaut. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459:1122-1125. [DOI] [PubMed] [Google Scholar]
- 15.Tang, J. W., K. L. K. Ngai, W. Y. Lam, and P. K. S. Chan. 2008. Seasonality of influenza A(H3N2) virus: a Hong Kong perspective (1997-2006). PLoS ONE 3:e2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.World Health Organization. 2003. Fact sheet 211. Influenza. World Health Organization, Geneva, Switzerland. http://www.who.int/mediacentre/factsheets/fs211/en/index.html.
- 17.World Health Organization. 2008. Influenza A(H1N1) virus resistance to oseltamivir. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/disease/influenza/H1N1webupdate20090318%20ed_ns.pdf.