Abstract
A recent report on several cases of invasive aspergillosis caused by Neosartorya udagawae suggested distinctive patterns of disease progression between N. udagawae and Aspergillus fumigatus. This prompted us to characterize N. udagawae in comparison to A. fumigatus. Our findings showed that both species exist in two mating types at similar ratios and produce gliotoxin. However, the thermotolerance of the two species differs: while A. fumigatus is able to grow at 55°C but not at 10°C, N. udagawae is able to grow at 10°C but fails to grow at >42°C. Furthermore, compared to A. fumigatus, the conidia of N. udagawae require longer incubation periods to germinate at 37°C and are more susceptible to neutrophil attack as well as hydrogen peroxide; N. udagawae is also less virulent in gp91phox−/− mice. These findings suggest that growth and susceptibility to the host response might account for the reduced virulence of N. udagawae and the subtle distinction in the progression of the disease caused by the two species.
Neosartorya udagawae, a heterothallic ascomycete with an Aspergillus fumigatus-like anamorph, was described in 1995 (10). The species description was based on two strains with opposite mating types, strains CBS114217 and CBS114218, isolated from the soil of a Brazilian sugar cane plantation (10). N. udagawae has been emerging as a cause of invasive aspergillosis (IA) in humans and animals. It was first recognized as a human pathogen in a lung transplant recipient with a pulmonary infection. The fungus was initially identified simply as an Aspergillus sp. on the basis of the morphology of conidial structures (15). Its subsequent identification as N. udagawae was based on the sequences of genes encoding β-tubulin and rodlet A. A similar approach allowed the correct identification of 12 N. udagawae strains among 86 clinical isolates that had been identified as A. fumigatus (3, 31). Recently, we described four cases of IA caused by N. udagawae. Three were in patients with chronic granulomatous disease (CGD), and one was in a patient with myelodysplastic syndrome (MDS) (31). The median duration of illness was approximately seven times longer than that typically observed for illness caused by A. fumigatus. The MICs of various antifungals for the N. udagawae isolates were higher than those for A. fumigatus, and the disease caused by N. udagawae was refractory to standard therapy (3, 31).
The clinical differences between the infections caused by N. udagawae and A. fumigatus prompted us to compare the biological characteristics of the two species.
Although it is known that N. udagawae produces A. fumigatus-like conidial structures with colonies that are covered by more pronounced short, silky, aerial hyphae and the secondary metabolites fumigatin, fumagilin, tryptoquivaline, and tryptoquivalone (22), our understanding of N. udagawae is still limited. The study described here shows that N. udagawae resembles A. fumigatus in its mating type distribution and gliotoxin production but exhibits a distinctive growth behavior, sensitivity to host defenses in vitro, and virulence in mice with CGD. It is plausible that such differences may account for the distinct pattern of disease development observed in the clinical cases (31).
MATERIALS AND METHODS
Fungal strains.
A. fumigatus strains B-5233 (29) and AF293 (17) were maintained as described previously (29). The N. udagawae strains used are listed in Table 1. Germination, growth, and assays for the interaction with neutrophils and resistance to hydrogen peroxide were performed with N. udagawae strains CBS114217, CBS114218, M31, M34, and F41 and A. fumigatus strain B-5233. Virulence in mice was compared by using M31, CBS114218, and B-5233. The N. udagawae strains listed in Table 1 as well as A. fumigatus strains B-5233 and AF293 were used for all other assays. To obtain a conidial suspension, the strains were cultured on oatmeal agar (12) or Aspergillus minimal agar (29) for 7 days at 37°C. The conidia were harvested in 0.01% Tween 20-phosphate-buffered saline (PBS) (T/P) and washed with distilled water. Unless otherwise specified, the concentration of the conidial suspension was adjusted with RPMI 1640 medium buffered with 25 mM HEPES, pH 7.4 (R25). Typically, a dilution of 100-fold with R25 was required to reach the desired concentration of conidial suspension, and thus, the medium was considered to be 100% R25. The incubations were performed at 37°C in 5% CO2.
TABLE 1.
N. udagawae strains used in this study
| Strain | Other designations | Source | GenBank accession no. |
Reference or source | |
|---|---|---|---|---|---|
| benA | rodA | ||||
| CBS114217 | CBM FA-0702 (type strain) | Soil, Brazil | AB248302 | AB250101 | 10 |
| CBS114218 | CBM FA-0703 (type strain) | Soil, Brazil | AB248303 | AB250102 | 10 |
| M29 | Isolate, patient 4 | Lung, CGD patient | FJ830682 | FJ830678 | 31 |
| M31 | Isolate, patient 1 | Lung, CGD patient | FJ830683 | FJ830679 | 31 |
| M34 | Isolate, patient 3 | Lung, MDS patient | FJ830684 | FJ830680 | 31 |
| F41 | Isolate, patient 2 | Lung, CGD patient | FJ830685 | FJ830681 | 31 |
| FH47 | Lung, patient with hematological malignancy | DQ058393 | DQ058377 | This study | |
| FH82 | CDC57 | Patient with unknown disease | DQ058392 | DQ058376 | 3 |
| FH83 | CDC58 | Patient with unknown disease | DQ438529 | DQ439761 | 3 |
| FH102 | Lung, patient with hematological malignancy | DQ438535 | DQ439767 | This study | |
| FH103 | Lung, patient with hematological malignancy | DQ058395 | DQ058379 | 3 | |
| FH237 | UT1516 | Brain from patient with unknown disease | DQ058394 | DQ058378 | 3 |
Phylogeny.
Phylogenetic analysis was based on partial sequences of the genes encoding β-tubulin (benA) and rodlet A (rodA) (2). The GenBank accession numbers for benA and rodA of A. fumigatus are AY685150 and DQ439777, respectively. Table 1 lists the GenBank accession numbers of the benA and rodA sequences from all N. udagawae strains used in the study. The sequences from N. udagawae FH104, FH105, and FH106 (3) were excluded since they were isolated from the same patient from whom FH103 was isolated. Lasergene software (version 7.1.0; DNAStar, Inc., Madison, WI) was used for sequence assembly and alignment. Multiple-sequence alignment was performed by the Clustal W method, and percent similarity was calculated (9). The phylogenetic trees were constructed by the neighbor-joining method, and bootstrap analysis was performed with the same software and with 1,000 replications.
Growth conditions and media.
For growth on solid medium, the strains were inoculated onto Czapek-Dox or malt extract agar plates. The plates were incubated at 10, 30, 35, 37, 42, and 45°C for 5 to 7 days. For growth in liquid medium, 200 μl of 1 × 106 conidia/ml was added to the wells of an eight-well chambered slide (Nalge Nunc International, Rochester, NY). The chambered slides were incubated for 10 or 44 h, and 20 μl of 0.03% neutral red (Matheson Coleman & Bell, Cincinnati, OH) was added to detect metabolically active cells (27). Approximately 200 cells per strain were analyzed for germination. Microscopic observation was carried out with a Zeiss Axiovert microscope and Axiovision (version 4.0) software.
PCR primers.
The genotype of the mating locus, MAT, and analysis for the presence of the gliP sequence were determined by PCR. Primers located in the conserved regions of the MAT loci of Aspergillus and Neosartorya species were used to generate amplicons containing the MAT1-2 high-mobility-group (HMG) domain and the MAT1-1 alpha box. The primers were HMG1 (CTCTTGTGGCAGGATGCTCT), HMG2 (TTGCTGGTAGAGGGCAGTCT), alpha1 (CTGGAGGAGCTTCTGCAGTAC), and alpha2 (GGAGTACGCCTTCGCGAG). To detect the gliP gene in the N. udagawae genome, primers gliP1 (GGTATCCTGAAAGCC) and gliP2 (CAACGCAGAGAAAATCTC), located in conserved regions of gliP from A. fumigatus, A. niger, A. clavatus, A. terreus, A. oryzae, and N. fischerii, were constructed. The amplicons were sequenced in the Laboratory of Molecular Technology, SAIC—Frederick Inc., Frederick, MD.
Mating.
N. udagawae clinical isolates were paired with type strains CBS114217 and CBS114218 and among themselves in all possible combinations on oatmeal (without tomato paste) agar plates (11). The plates were sealed with Parafilm and were incubated at 30°C for >60 days in the dark.
Virulence study.
Six- to 13-week-old gp91phox−/− mice (Jackson Laboratories) (20) maintained at the animal facility of the National Institute of Allergy and Infectious Diseases, Bethesda, MD, were used. The conidia (5 × 103) of strains B-5233, M31, and CBS114218 suspended in 50 μl of T/P buffer were inoculated into mice (nine mice per fungal strain) by the pharyngeal aspiration technique (21). The inoculum size was determined from hemacytometer counts and conidial viability. Survival was monitored for 40 days, and the results were analyzed by log-rank test. Histopathological sections of the lung were stained with hematoxylin and eosin (H&E) and Gomori's methenamine-silver (GMS).
Neutrophil assay.
Blood samples from five healthy donors were collected according to protocol 99-CC-0168, which was approved by National Institutes of Health Internal Review Board, and neutrophils were isolated as described previously (33). For interaction with neutrophils, 100 μl of 1 × 106 conidia/ml was added to the wells of a 96-well plate. The plate was incubated for 4 h, before 100 μl of R25 containing 10 μl of autologous plasma or 100 μl of R25 containing 1 × 105 neutrophils plus 10 μl of autologous plasma was added. The experiments were performed in triplicate with blood from each donor. The plates were further incubated for 20 h, and the neutrophils were lysed by adding 0.05% Triton X-100 (50 μl/well). A modified 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) assay (13) was performed as follows: 100 μl of the supernatant was replaced by the XTT reagent (Dulbecco's PBS containing 0.5 mg/ml XTT and 40 μg/ml of coenzyme Q [Sigma, St. Louis, MO]), followed by incubation for 30 min at 37°C and an additional 2 h at room temperature. A total of 100 μl of the supernatant was transferred to a new plate, and the optical density at 450 nm (OD450) was determined. The OD450s of the control wells containing plasma or plasma plus neutrophils without conidia were subtracted from the OD450s of the respective samples containing conidia. Relative metabolic activity was determined by the equation [(OD450 conidia + neutrophils)/OD450 conidia] × 100. For microscopy, 200 μl of 5 × 105 conidia/ml was added to the wells of an eight-well chambered slide. The chambered slide was incubated for 4 h, followed by addition of 100 μl of R25 containing 15 μl of autologous plasma or 100 μl of R25 containing 1 × 105 neutrophils plus 15 μl of autologous plasma. The samples were incubated for an additional 20 h and observed by bright-field microscopy.
Hydrogen peroxide.
A total of 100 μl of 1 × 106 conidia/ml plus 100 μl of 0.62, 1.25, or 2.5 mM hydrogen peroxide solution was added to a 96-well plate or to an eight-well chambered slide and incubated for 16 to 20 h. For the plates, 100 μl of XTT reagent was added to determine the OD450, as described above. For the chambered slides, 20 μl of 0.03% neutral red was added to the wells before the samples were examined by bright-field microscopy.
Gliotoxin.
A total of 1 × 107 conidia were inoculated in 50 ml of Czapek-Dox broth or medium 199 and incubated for 3 days at 37°C. The gliotoxin in the supernatants was analyzed by high-performanc liquid chromatography (HPLC) (28).
RESULTS
Genetic and morphological characterization.
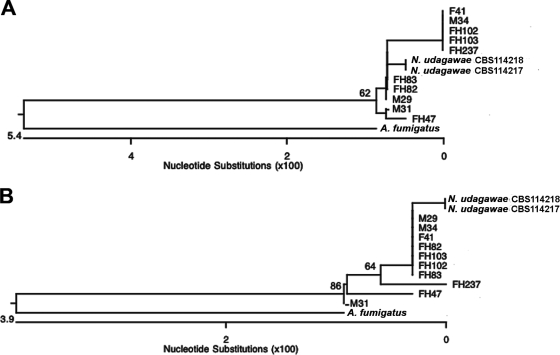
The genetic relatedness of 12 N. udagawae strains (Table 1) was characterized by constructing phylogenetic trees based on their benA and rodA sequences. N. udagawae strains formed a homogeneous monophyletic cluster with a significant distance from A. fumigatus (Fig. 1). For benA, the clinical isolates aligned into three subclusters (1st subcluster, isolates F41, M34, FH102, FH103, and FH237; 2nd subcluster, isolates FH82, FH83, and M29; 3rd subcluster, isolates M31 and FH47) (Fig. 1A). The similarities within each subcluster were 99.8% to 100%, 99.8%, and 99.8%, respectively. All clinical isolates demonstrated 99.1% to 99.8% similarity with both strain CBS114217 and strain CBS114218. For rodA, seven clinical isolates clustered with 100% similarity to each other (isolates M29, M34, F41, FH82, FH83, FH102, and FH103) (Fig. 1B). Isolates FH47, M31, and FH237 appeared as separate branches. Consistent with previous observations, the rodA sequence appeared to be less phylogenetically informative than benA (25). The sequences of all isolates except isolates FH47, FH237, and M31 showed 99.7% to 100% similarity compared with those of the type strains; the sequences of isolates FH47, FH237, and M31 showed similarities of 98.5%, 98.8%, and 99.1%, respectively. The low level of genetic diversity among the strains of N. udagawae suggests a relatively recent origin of the species.
FIG. 1.
Phylogenetic analysis. Neighbor-joining trees based on partial benA (A) and rodA (B) sequences of clinical and type strains of N. udagawae as well as A. fumigatus are shown. The numbers at the nodes represent bootstrap values (of 1,000 bootstrap replications)of >50%.
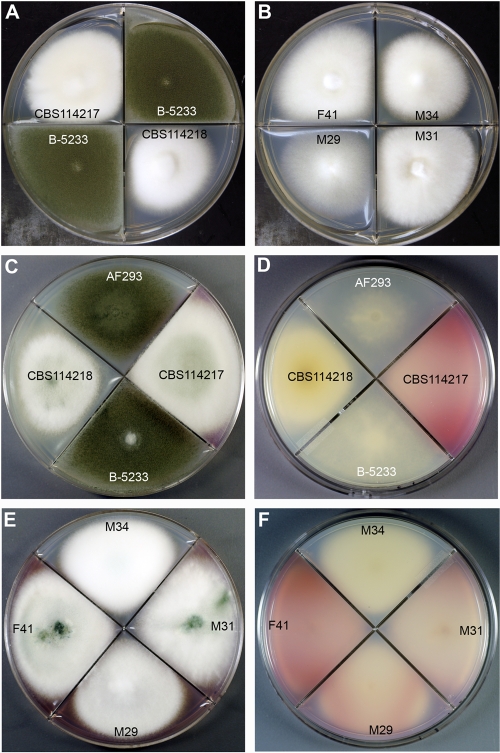
The biological phenotypes of N. udagawae and A. fumigatus, such as colony morphology and growth characteristics, were investigated to augment the phylogenetic data and further characterize the two species. Strains M29, M31, M34, and F41 were chosen for use in growth assays at temperatures ranging from 10 to 45°C for 7 days. N. udagawae grew minimally at 10°C and optimally at 30 to 35°C, but it did not grow at a temperature higher than 42°C. On the contrary, A. fumigatus failed to grow at 10°C but grew well at ≥45°C. Since the optimum growth temperature of A. fumigatus is 37°C, we compared the colony morphologies of the two species at this temperature (Fig. 2A,B). The colonies of A. fumigatus B-5233 grew rapidly, and they were covered with a lawn of bluish green conidia. The N. udagawae strains grew slowly for the first 2 days, but after 5 days the colonies reached a size similar to those of A. fumigatus. The colonies were mainly composed of white aerial hyphae, with conidia being sparsely produced. Unlike A. fumigatus, most of the N. udagawae strains secreted a pink-purple extrolite(s) on the agar plates, with the coloration being more evident on Aspergillus minimal medium (Fig. 2C to F). The colored extrolite(s) and growth at 10°C could serve as a marker to distinguish strains of N. udagawae from A. fumigatus.
FIG. 2.
(A and B) Growth characteristics of N. udagawae and A. fumigatus on Czapek-Dox agar after 5 days at 37°C. (A) A. fumigatus strain B-5233 and N. udagawae type strains CBS114217 and CBS114218; (B) N. udagawae clinical strains M29, M31, M34, and F41. (C to F) Growth characteristics of A. fumigatus and N. udagawae on Aspergillus minimal medium after 7 days at 37°C. (C and E) Colony surface; (D and F) reverse surface. A purplish pink compound is evident in the reverse of the colonies of all N. udagawae strains except CBS114218 and M34.
Mating types.
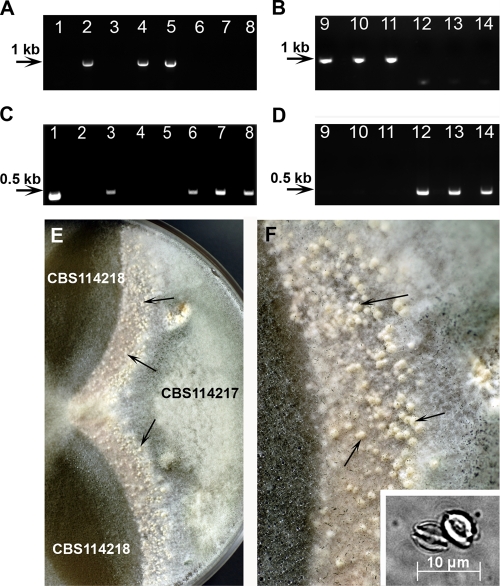
Although Horie et al. (10) described heterothallism in N. udagawae and designated CBS114217 and CBS114218 as the type strains of the two opposite mating types, their MAT genotype was not identified. Subsequently, Varga et al. arbitrarily assigned CBS114217 and CBS114218 to be mating type A and mating type a strains, respectively (30). In general, heterothallic filamentous ascomycetes have a single MAT locus represented by two idiomorphs, MAT1-1, which contains an alpha box domain, and MAT1-2, which contains an HGM domain (7). In A. fumigatus, strain AF293, which contains the HMG domain, is considered the MAT1-2 strain, whereas AF217, which bears the alpha box, is the MAT1-1 strain (19). In this study, the HMG domain was identified in N. udagawae type strain CBS114218 and four clinical isolates (isolates M29, FH47, FH82, and FH83), indicating that they are MAT1-2, like A. fumigatus AF293 (Fig. 3A and B). On the other hand, type strain CBS114217 and six clinical isolates (isolates M31, M34, F41, FH102, FH103, and FH237) produced an alpha box amplicon like A. fumigatus MAT1-1 strain B-5233 (Fig. 3C and D). Similar to A. fumigatus, the distribution of the MAT1-1 and MAT1-2 genotypes was nearly one to one among the N. udagawae clinical isolates.
FIG. 3.
Genotyping of MAT idiomorphs and mating assay. The mating type was designated on the basis of the presence of the alpha box (MAT1-1) or the HMG domain (MAT1-2) (A to D). (A and B) Amplicons from the HMG domain; (C and D) amplicons from the alpha box. Lanes 1 and 2, A. fumigatus strains B-5233 (MAT1-1) and AF293 (MAT1-2), respectively; lanes 3 and 4, N. udagawae type strains CBS14217 and CBS114218, respectively; lanes 5 to 14, N. udagawae clinical isolates M29 (lane 5), M31 (lane 6), M34 (lane 7), F41 (lane 8), FH47 (lane 9), FH82 (lane 10), FH83 (lane 11), FH102 (lane 12), FH103 (lane 13), and FH237 (lane 14). In the mating assay, strain CBS114217 was crossed with strain CBS114218 on an oatmeal agar plate, and the plate was incubated at 30°C for 18 days (E and F). Arrows point to the cleistothecia produced along the junction between the two strains. Panel F shows a higher magnification of panel E. The inset in panel E shows ascospores released from cleistothecia.
In the mating assays with N. udagawae, the only crosses producing cleistothecia containing ascospores were between CBS114217 MAT1-1 and CBS114218, M29, FH47, FH82, and FH83 (Fig. 3E and F and data not shown). The abundance of ascopsore-containing cleistothecia varied between the pairs. The most abundant number of ascospores was produced in the crosses between CBS114217 and clinical strains M29 and FH83. Ascospores were only sparsely produced in the crosses between CBS114217 and FH47 and between CBS114217 and FH82 (data not shown). Twenty-eight single ascospores from cleistothecia produced in the cross between CBS114217 and M29 were isolated on yeast glucose agar by micromanipulation and incubated at 25°C and 40°C. None of the spores germinated at these temperatures, suggesting that they were not viable. The inset of Fig. 3F shows typical ascospores released from the cleistothecia obtained from the cross between CBS114217 and CBS114218. Interestingly, while all four MAT1-2 clinical strains mated with CBS114217, only one of the MAT1-1 clinical strains, strain F41, formed cleistothecia when it was paired with CBS114218. The cleistothecia, however, remained empty even after prolonged incubation (data not shown).
Virulence in gp91phox−/− mice.
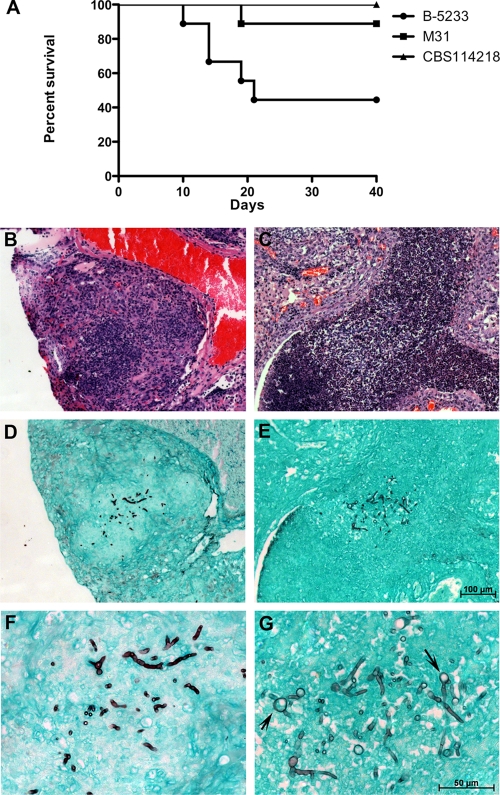
The low frequency of N. udagawae among the 86 clinical isolates (3, 31) suggests that the species may be less common in the environment, less amenable to culture from diagnostic specimens, or less virulent than A. fumigatus. To address the virulence hypothesis, A. fumigatus B-5233 and N. udagawae CBS114218 and M31 were inoculated into CGD gp91phox−/− mice. This mouse strain was chosen because CGD patients are known to be susceptible to IA caused by either species (23, 31). Fifty percent of the mice infected with A. fumigatus died by day 20, whereas only one mouse in the group infected with N. udagawae M31 died by day 40. No deaths occurred in the group inoculated with CBS114218 (Fig. 4A). The histological sections of the lungs from the mouse that succumbed to the infection caused by strain M31 showed granulomatous lesions containing fungal hyphae, which were also found in the lungs of mice infected with A. fumigatus (Fig. 4B to G). Interestingly, swollen round structures interspersed with hyphal filaments, similar to those seen in the lungs of patients with CGD infected with N. udagawae (31), were observed within several granulomatous foci in the lungs of the M31-infected mouse (Fig. 4G). The death of only 1 of 18 mice inoculated with N. udagawae supports the hypothesis that N. udagawae is less virulent than A. fumigatus.
FIG. 4.
Virulence assay in a murine model. gp91phox−/− mice with CGD were inoculated with A. fumigatus strain B-5233 or the N. udagawae strains CBS114218 and M31. (A) Survival curve; P ≤ 0.01 for CBS114218 compared to B-5233, and P ≤ 0.04 for M31 compared to B-5233 (log-rank test). (B to G). Histopathology of lung tissue from mice that succumbed to infection caused by strains B-5233 (B, D, and F) and M31 (C, E, and G). (B and C) Sections stained with H&E; (D to G) sections stained with GMS. Panels F and G are higher magnifications of panels D and E, respectively. The magnification in panels B to D is the same as that in panel E. The magnification in panel F is indicated in panel G. Arrows indicate swollen round structures formed in the hyphae of N. udagawae strain M31.
Germination and growth rate in vitro.
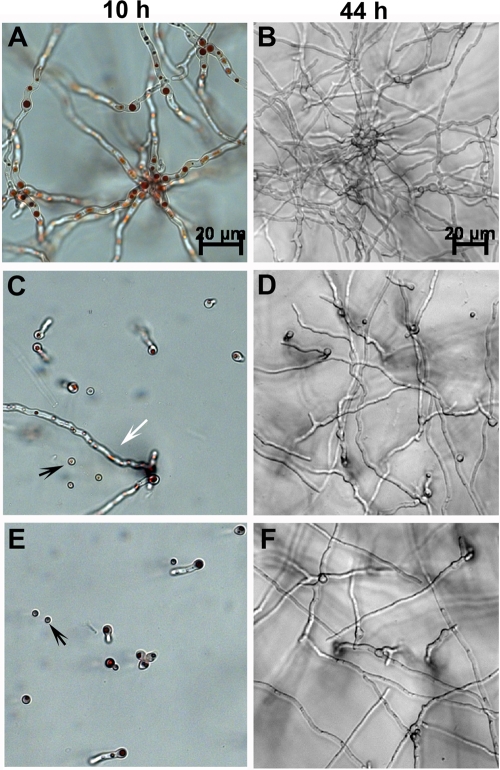
Since conidial germination is a prerequisite for invasive growth and, thus, for virulence, we first compared the rates of germination of A. fumigatus and N. udagawae. Conidia were inoculated in chambered glass slides, and the slides were assessed for germination at 10 and 44 h. At 10 h, 95% of the A. fumigatus conidia had germinated and developed elongated hyphae. At 44 h, extensive mycelial growth was observed (Fig. 5A and B). In N. udagawae, however, only 50 and 40% of the conidia from CBS114218 and M31, respectively, germinated at 10 h, and elongated hyphae were rarely observed (Fig. 5C and E). The neutral red dye staining patterns of both ungerminated and germinated conidia suggested that the majority of the cells were metabolically active. By 44 h, extensive hyphal growth was observed in both CBS114218 and M13 (Fig. 5D and F) and in other N. udagawae strains (data not shown). These results suggest not only that the germination of N. udagawae is slower than that of A. fumigatus but also that it has a more unsynchronized pattern of germination than A. fumigatus.
FIG. 5.
Growth of N. udagawae and A. fumigatus in RPMI 1640 medium. Resting conidia were inoculated in chambered slides containing RPMI 1640 and 25 mM HEPES, and the chambered slides were incubated at 37°C for 10 and 44 h. (A and B) A. fumigatus strain B-5233; (C and D) N. udagawae type strain CBS114218; (E and F) N. udagawae clinical strain M31. The magnification in panels C and E is the same as that in panel A. The magnification in panels D and F is the same as that in panel B. Black arrows, ungerminated conidia; white arrow, occasional conidia that germinated and formed long hyphal filament.
Response to neutrophils.
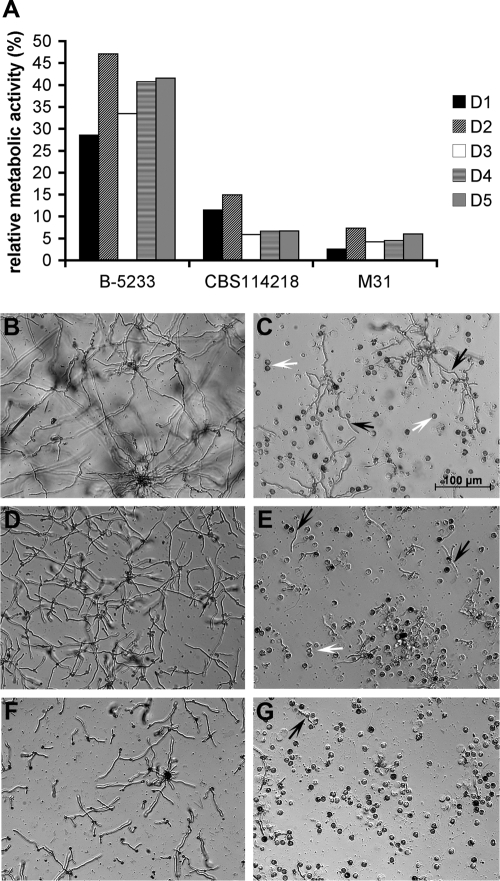
We next compared the pattern of the neutrophil-conidium interaction between the two species to determine if susceptibility to neutrophils contributed to the difference in virulence. The growth of N. udagawae exposed to neutrophils was markedly decreased compared to that of A. fumigatus, as evidenced by the differences in metabolic activity (Fig. 6A). This pattern was observed with neutrophils from six different donors, suggesting that the higher sensitivity of N. udagawae was a true phenotype and not due to the neutrophils from a particular donor. Microscopy confirmed that neutrophils reduced the growth of both species (Fig. 6B and G) but exerted a higher degree of growth arrest against the N. udagawae strains. These results suggest that N. udagawae might be more efficiently cleared by neutrophils than A. fumigatus, resulting in the higher survival rate of infected hosts.
FIG. 6.
Effect of human neutrophils on conidia of N. udagawae and A. fumigatus. (A) Metabolic activity of conidia after challenge with neutrophils relative to that of control conidia without neutrophils. Each bar represents a biological replicate carried out with neutrophils from a single donor. Neutrophils from five different donors (donors D1 to D5) were tested. The metabolic activity of the control conidia without neutrophils was considered to be 100%. (B to G) Bright-field microscopy images showing the growth of A. fumigatus B-5233 (B and C) and N. udagawae CBS114218 (D and E) and M31 (F and G). (B, D, and F) Growth in the absence of neutrophils; (C, E, and G) growth in the presence of neutrophils. White arrows, neutrophils; black arrows, hyphal filaments. The magnification indicated in panel C applies to all other panels.
Oxidative stress.
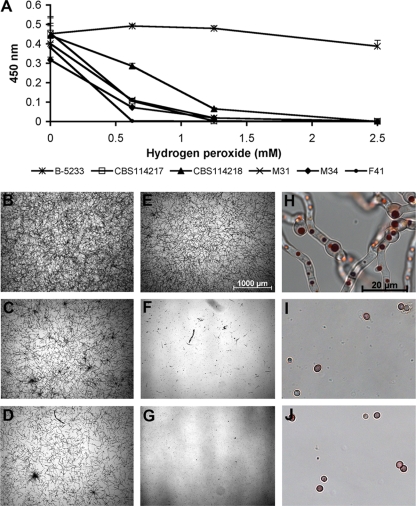
Although we mainly focused on N. udagawae strains isolated from patients with CGD, whose phagocytes are deficient in the generation of reactive oxygen species (ROS), not all the strains have been isolated from such patients. Since ROS are of primary importance in the host defense against microbial pathogens, we evaluated the effect of hydrogen peroxide, an ROS produced by phagocytes, on N. udagawae. Figure 7A shows that there were significant reductions in the metabolic activity of the five N. udagawae strains compared to that of A. fumigatus B-5233. Supporting these results, bright-field microscopy showed a slight reduction in the growth of B-5233 treated with 2.5 mM hydrogen peroxide (Fig. 7B and E), whereas the growth of N. udagawae conidial was completely abolished with the same concentration of hydrogen peroxide (Fig. 7C and F and Fig. 7D and G). The higher magnification of the samples in panels F and G of Fig. 7 revealed that hydrogen peroxide inhibited germination (Fig. 7I and J). In fact, the pattern of neutral red staining and the absence of growth even after an additional incubation of 24 h suggested that the conidia were not viable (data not shown). Similar results were obtained with strains CBS114217, M34, and F41.
FIG. 7.
Effect of hydrogen peroxide on N. udagawae and A. fumigatus. (A) Metabolic activity assayed by the XTT assay and measured at OD450. The conidia of A. fumigatus strain B-5233 and N. udagawae strains CBS114217, CBS114218, M31, M34, and F41 were incubated in the presence of hydrogen peroxide at concentrations of 0.625, 1.25, and 2.5 mM. (B to J) Bright-field microscopy images of B-5233 (B, E, and H), CBS114218 (C, F, and I), and M31 (D, G, and J). (B to D) Conidia incubated without hydrogen peroxide; (E to J) conidia incubated with 2.5 mM hydrogen peroxide. Samples were observed after 20 h incubation. The magnifications in panels H to J are 50 times those in panels E to G. The magnification of panels B to G is indicated in panel E. The magnification bar shown in panel H also applies to panels I and J.
Gliotoxin.
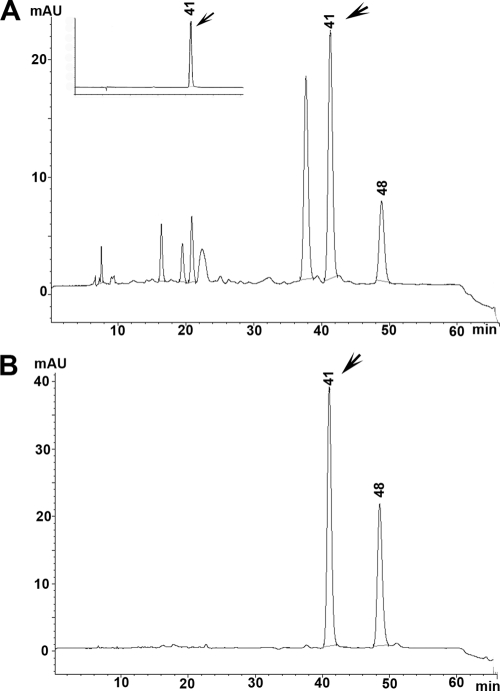
Although it has been reported that N. udagawae produces fumigatin, fumagilin, tryptoquivaline, and tryptoquivalone (22), there was no information regarding gliotoxin, one of the most potent toxins with immunosuppressive properties (8, 16, 26) produced by A. fumigatus. Because gliotoxin could be an important virulence factor for N. udagawae, as it is for A. fumigatus (24, 28), we assessed whether N. udagawae was also a gliotoxin producer. To verify whether N. udagawae has the capability of producing gliotoxin, we tested the N. udagawae isolates for the presence of the gliP gene, which encodes a nonribosomal peptide synthase that catalyzes the first step of gliotoxin biosynthesis. All N. udagawae strains generated an amplicon with a sequence that matched that of the gliP gene (data not shown), suggesting that N. udagawae is a potential gliotoxin producer. Gliotoxin production was confirmed by HPLC analysis of N. udagawae culture supernatants. Czapek-Dox broth and liquid medium 199, which have been shown to be suitable for gliotoxin production in A. fumigatus, were used as the culture media (1, 4). Gliotoxin was detected in the Czapek-Dox cultures of strains M31, F41, FH102, and FH103 but not those of strains CBS114218, M29, M34, FH82, FH83, and FH237, which required liquid medium 199. This toxin, however, was not detected in the cultures of strains CBS14217 and FH47, regardless of the type of culture medium used. Figure 8 shows representative chromatograms from the culture supernatants of A. fumigatus B-5233 and N. udagawae M31. The peak at 41 min was identified as gliotoxin on the basis of the retention time and the UV spectra of pure gliotoxin (Fig. 8A, inset, and data not shown). Attempts to perform quantitative analysis were hampered by highly variable levels of gliotoxin among the biological replicates (data not shown). Our findings suggest that N. udagawae is capable of producing gliotoxin and that gliotoxin biosynthesis may be dependent upon environmental conditions, such as the nutritional elements available.
FIG. 8.
Analysis of gliotoxin. HPLC analysis of culture supernatants from A. fumigatus strains B-5233 (A) and representative N. udagawae strain M31 (B). The inset in panel A shows the gliotoxin standard. Arrows, the peak that eluted at 41 min, which corresponds to gliotoxin. The identity of the peak eluting at 48 min is unknown. The results of a representative HPLC analysis are shown. mAU, milliabsorbance units.
DISCUSSION
This study focused on the biological characterization of N. udagawae to assess its differences from as well as its similarities to A. fumigatus, the species as which it has most often been misidentified. To date, 15 strains of N. udagawae from patients with different underlying conditions have been reported: 2 in this study (strains FH47 and FH102) and 13 in previous reports (3, 15, 31). Three of them were isolated from patients with CGD. Invasive aspergillosis in patients with CGD is most often caused by A. fumigatus, followed by A. nidulans and rarely by other Aspergillus species (6, 23). N. udagawae, therefore, may be the third most common Aspergillus species that causes invasive disease in CGD patients. It is likely that a close examination of clinical isolates previously identified as A. fumigatus or even N. fischeri in many medical centers worldwide will uncover more strains of N. udagawae.
Other pathogenic species of Aspergillus, such as A. lentulus, A. viridinutans, and A. fumisynnematus, have been misidentified as A. fumigatus solely on the basis of phenotypic characteristics. Among these species, A. viridinutans is more closely related to N. udagawae (3, 32), and both of these species can be distinguished from the other two A. fumigatus-like species by their inability to grow at 45°C. Unlike A. fumigatus, however, A. lentulus and A. fumisynnematus fail to grow at temperatures of ≥48°C (3, 32) but grow at 10°C. It is clear, therefore, that the ability to grow at temperatures of ≥50°C but not at 10°C can be used as a presumptive diagnostic criterion to distinguish A. fumigatus from the other pathogenic species in the section Fumigati. It is interesting to note that all pathogenic non-A. fumigatus species in the section Fumigati except A. viridinutans can grow at 10°C; no information is available on the minimum growth temperature of A. viridinutans (22).
Although N. udagawae is a heterothallic species, previous mating studies with clinical strains have been unsuccessful (3, 31). In our study, mating occurred on oatmeal agar but not on the conventional mycological media on which the heterothallism of N. udagawae was originally discovered (10) and on which other Neosartorya species are able to mate (12). Furthermore, N. udagawae required longer periods for ascospore formation than other Neosartorya, and the N. udagawae ascospores were nonviable. In general, the fertility of N. udagawae appears to be declining, and this appears to be more so for MAT1-1 strains than MAT1-2 strains. A decline in the level of sexual reproduction was also reported for A. fumigatus (Neosartorya fumigata) isolates collected in Dublin, Ireland (18). Ascopsore formation in the Dublin strains required exceptionally rigid environmental conditions, such as a constant temperature of 30°C for 6 months, which is not readily found in nature.
The reduced rate of mortality of mice with CGD infected with N. udagawae and the low frequency of occurrence of this species among clinical strains (3, 31) suggests that N. udagawae is less virulent than A. fumigatus. Since both species are capable of producing gliotoxin, it is unlikely that the difference in their virulence is attributable to this toxin. On the other hand, susceptibility to host defenses may be a key factor for the difference in virulence. It is generally believed that macrophages kill A. fumigatus conidia, whereas neutrophils clear hyphae. Recently, however, increasing attention has been paid to the importance of neutrophils against the conidia of A. fumigatus (5, 14, 33). In fact, a recent study suggested that neutrophils and not alveolar macrophages are essential for anticonidial defense (14). Together, this observation and our findings of the susceptibility of N. udagawae conidia to neutrophils suggest that the lower virulence of this fungus might be due to the more efficient clearance of the N. udagawae conidia by the host. In addition, the higher level of susceptibility of N. udagawae conidia to hydrogen peroxide, one of the ROS involved in host defense, and the low rate of germination are likely to contribute to more efficient killing of the pathogen. Collectively, these factors may render N. udagawae less fit than A. fumigatus as a cause of IA. The biological differences between the two species, such as the maximum and minimum growth temperatures, the production of a pink/purple extrolite(s), and susceptibility to neutrophils/hydrogen peroxide, should be useful for the discrimination of N. udagawae from A. fumigatus in clinical laboratories when use of the molecular approach is not feasible.
Acknowledgments
This study was supported by funds from the intramural program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. D.C.V. is supported by a Canadian Institutes of Health Research fellowship. K.A.M. is supported by NIAID grants R21 AI055928 and AI067971.
We thank Ashok Varma for critically reviewing the manuscript.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Amitani, R., G. Taylor, E. N. Elezis, C. Llewellyn-Jones, J. Mitchell, F. Kuze, P. J. Cole, and R. Wilson. 1995. Purification and characterization of factors produced by Aspergillus fumigatus which affect human ciliated respiratory epithelium. Infect. Immun. 63:3266-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balajee, S. A., J. L. Gribskov, E. Hanley, D. Nickle, and K. A. Marr. 2005. Aspergillus lentulus sp. nov., a new sibling species of A. fumigatus. Eukaryot. Cell 4:625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balajee, S. A., D. Nickle, J. Varga, and K. A. Marr. 2006. Molecular studies reveal frequent misidentification of Aspergillus fumigatus by morphotyping. Eukaryot. Cell 5:1705-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belkacemi, L., R. C. Barton, V. Hopwood, and E. G. Evans. 1999. Determination of optimum growth conditions for gliotoxin production by Aspergillus fumigatus and development of a novel method for gliotoxin detection. Med. Mycol. 37:227-233. [PubMed] [Google Scholar]
- 5.Bonnett, C. R., E. J. Cornish, A. G. Harmsen, and J. B. Burritt. 2006. Early neutrophil recruitment and aggregation in the murine lung inhibit germination of Aspergillus fumigatus conidia. Infect. Immun. 74:6528-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, M. S., R. E. Isturiz, H. L. Malech, R. K. Root, C. M. Wilfert, L. Gutman, and R. H. Buckley. 1981. Fungal infection in chronic granulomatous disease. The importance of the phagocyte in defense against fungi. Am. J. Med. 71:59-66. [DOI] [PubMed] [Google Scholar]
- 7.Debuchy, R., and B. G. Turgeon. 2006. Mating type structure, evolution and function in Euascomycetes, p. 293-323. In U. Kües and R. Fischer (ed.), The Mycota. 1. Growth, differentiation and sexuality, vol. 1. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 8.Eichner, R. D., M. Al Salami, P. R. Wood, and A. Mullbacher. 1986. The effect of gliotoxin upon macrophage function. Int. J. Immunopharmacol. 8:789-797. [DOI] [PubMed] [Google Scholar]
- 9.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 10.Horie, Y., M. Miyaji, K. Nishimura, M. F. Franco, and K. I. Coelho. 1995. Two new species of Neosartorya from Brazilian soil. Mycoscience 36:159-165. [Google Scholar]
- 11.Kwon-Chung, K. J., and J. E. Bennett. 1992. Medical mycology. Lea & Febiger, Philadelphia, PA.
- 12.Kwon-Chung, K. J., and S. J. Kim. 1974. A second heterothallic Aspergillus. Mycologia 56:628-638. [PubMed] [Google Scholar]
- 13.Meshulam, T., S. M. Levitz, L. Christin, and R. D. Diamond. 1995. A simplified new assay for assessment of fungal cell damage with the tetrazolium dye, (2,3)-bis-(2-methoxy-4-nitro-5-sulphenyl)-(2H)-tetrazolium-5-carboxanilide (XTT). J. Infect. Dis. 172:1153-1156. [DOI] [PubMed] [Google Scholar]
- 14.Mircescu, M. M., L. Lipuma, N. van Rooijen, E. G. Pamer, and T. M. Hohl. 2009. Essential role for neutrophils but not alveolar macrophages at early time points following Aspergillus fumigatus infection. J. Infect. Dis. 200:647-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moragues, M. D., S. Brena, I. Miranda, R. Laporta, J. P. Munoz, and A. Palacio. 2006. Abstr. VIII Congr. Nacional Micol., abstr. D15.
- 16.Mullbacher, A., and R. D. Eichner. 1984. Immunosuppression in vitro by a metabolite of a human pathogenic fungus. Proc. Natl. Acad. Sci. U. S. A. 81:3835-3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nierman, W. C., A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyo, M. Berriman, K. Abe, D. B. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. Garcia, M. J. Garcia, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jimenez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafon, J. P. Latge, W. Li, A. Lord, C. Lu, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Penalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. Rodriguez de Cordoba, J. M. Rodriguez-Pena, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. Vazquez de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell, and D. W. Denning. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151-1156. [DOI] [PubMed] [Google Scholar]
- 18.O'Gorman, C. M., H. T. Fuller, and P. S. Dyer. 2009. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 457:471-474. [DOI] [PubMed] [Google Scholar]
- 19.Paoletti, M., C. Rydholm, E. U. Schwier, M. J. Anderson, G. Szakacs, F. Lutzoni, J. P. Debeaupuis, J. P. Latge, D. W. Denning, and P. S. Dyer. 2005. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Biol. 15:1242-1248. [DOI] [PubMed] [Google Scholar]
- 20.Pollock, J. D., D. A. Williams, M. A. Gifford, L. L. Li, X. Du, J. Fisherman, S. H. Orkin, C. M. Doerschuk, and M. C. Dinauer. 1995. Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat. Genet. 9:202-209. [DOI] [PubMed] [Google Scholar]
- 21.Rao, G. V., S. Tinkle, D. N. Weissman, J. M. Antonini, M. L. Kashon, R. Salmen, L. A. Battelli, P. A. Willard, M. D. Hoover, and A. F. Hubbs. 2003. Efficacy of a technique for exposing the mouse lung to particles aspirated from the pharynx. J. Toxicol. Environ. Health A 66:1441-1452. [DOI] [PubMed] [Google Scholar]
- 22.Samson, R. A., S. Hong, S. W. Peterson, J. C. Frisvad, and J. Varga. 2007. Polyphasic taxonomy of Aspergillus section Fumigati and its teleomorph Neosartorya. Stud. Mycol. 59:147-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segal, B. H., E. S. DeCarlo, K. J. Kwon-Chung, H. L. Malech, J. I. Gallin, and S. M. Holland. 1998. Aspergillus nidulans infection in chronic granulomatous disease. Medicine 77:345-354. [DOI] [PubMed] [Google Scholar]
- 24.Spikes, S., R. Xu, C. K. Nguyen, G. Chamilos, D. P. Kontoyiannis, R. H. Jacobson, D. E. Ejzykowicz, L. Y. Chiang, S. G. Filler, and G. S. May. 2008. Gliotoxin production in Aspergillus fumigatus contributes to host-specific differences in virulence. J. Infect. Dis. 197:479-486. [DOI] [PubMed] [Google Scholar]
- 25.Staab, J. F., S. A. Balajee, and K. A. Marr. 2009. Aspergillus section Fumigati typing by PCR-restriction fragment polymorphism. J. Clin. Microbiol. 47:2079-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanzani, M., E. Orciuolo, R. Lewis, D. P. Kontoyiannis, S. L. Martins, L. S. St John, and K. V. Komanduri. 2005. Aspergillus fumigatus suppresses the human cellular immune response via gliotoxin-mediated apoptosis of monocytes. Blood 105:2258-2265. [DOI] [PubMed] [Google Scholar]
- 27.Sugui, J. A., H. S. Kim, K. A. Zarember, Y. C. Chang, J. I. Gallin, W. C. Nierman, and K. J. Kwon-Chung. 2008. Genes differentially expressed in conidia and hyphae of Aspergillus fumigatus upon exposure to human neutrophils. PLoS One 3:e2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugui, J. A., J. Pardo, Y. C. Chang, K. A. Zarember, G. Nardone, E. M. Galvez, A. Mullbacher, J. I. Gallin, M. M. Simon, and K. J. Kwon-Chung. 2007. Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot. Cell 6:1562-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsai, H. F., R. G. Washburn, Y. C. Chang, and K. J. Kwon-Chung. 1997. Aspergillus fumigatus arp1 modulates conidial pigmentation and complement deposition. Mol. Microbiol. 26:175-183. [DOI] [PubMed] [Google Scholar]
- 30.Varga, J., Z. Vida, B. Toth, F. Debets, and Y. Horie. 2000. Phylogenetic analysis of newly described Neosartorya species. Antonie Van Leeuwenhoek 77:235-239. [DOI] [PubMed] [Google Scholar]
- 31.Vinh, D. C., Y. R. Shea, J. A. Sugui, E. R. Parrilla-Castellar, A. F. Freeman, J. W. Campbell, S. Pittaluga, P. A. Jones, A. Zelazny, D. Kleiner, K. J. Kwon-Chung, and S. M. Holland. 2009. Invasive aspergillosis due to Neosartorya udagawae. Clin. Infect. Dis. 49:102-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yaguchi, T., Y. Horie, R. Tanaka, T. Matsuzawa, J. Ito, and K. Nishimura. 2007. Molecular phylogenetics of multiple genes on Aspergillus section Fumigati isolated from clinical specimens in Japan. Nippon Ishinkin Gakkai Zasshi 48:37-46. [DOI] [PubMed] [Google Scholar]
- 33.Zarember, K. A., J. A. Sugui, Y. C. Chang, K. J. Kwon-Chung, and J. I. Gallin. 2007. Human polymorphonuclear leukocytes inhibit Aspergillus fumigatus conidial growth by lactoferrin-mediated iron depletion. J. Immunol. 178:6367-6373. [DOI] [PubMed] [Google Scholar]