Abstract
Plasmodium falciparum is the main cause of human malaria and is one of the important pathogens causing high rates of morbidity and mortality. The total number of malaria patients in Vietnam has gradually decreased over the last decade. However, the spread of pathogens with drug resistance remains a significant problem. Defining the trend in genotypes related to drug resistance is essential for the control of malaria in Vietnam. We undertook a longitudinal survey of Plasmodium falciparum malaria in 2001, 2002, and 2005 to 2007. The pfcrt, pfmdr1, pfdhfr, and pfdhps genes were analyzed by sequencing; and correlations by study year, age, gender, and genotype were identified statistically. The ratio of the chloroquine resistance genotype pfcrt 76T was found to have decreased rapidly after 2002. High numbers of mutations in the pfdhfr and pfdhps genes were observed only in 2001 and 2002, while the emergence of parasites with a new K540Y mutation in the P. falciparum dihydropteroate synthetase (PfDHPS) was observed in 2002. For males and those in younger age brackets, a correlation between vulnerability to P. falciparum infection and strains with pfcrt 76K or strains with decreased numbers of mutations in pfdhfr and pfdhps was demonstrated. The parasites with pfcrt 76T exhibited a greater number of mutations in pfdhfr and pfdhps.
Plasmodium falciparum has long been one of the most important pathogens, causing severe illness and large numbers of deaths worldwide. In the 1990s, more than 1 million people living in Vietnam suffered from P. falciparum infections, resulting in thousands of deaths per year. Since then, the National Institute of Malariology, Parasitology, and Entomology (NIMPE) and the government of Vietnam have focused a great deal of time and effort on a malaria control program. As a result, the incidence of malaria reported in 2003 was only 12% of that reported in 1992 (2). However, the spread of drug-resistant isolates, including multidrug-resistant strains, has become a critical problem in Vietnam and has led to the significant failure of treatment. Thus, a further understanding of the incidence of malaria cases with detailed parasite genotype information and the identification of factors relating to the acquisition of drug-resistant isolates may prove important for the determination of effective and economical treatment choices in clinical settings.
The pfcrt gene is located on chromosome 7 and encodes the vacuolar membrane transporter protein P. falciparum chloroquine-resistant transport (PfCRT) (21). While several point mutations associated with chloroquine resistance have been determined previously, substitution of K for T in codon 76 has been shown to be specifically related to resistance in vitro (21, 23). The allelic variation of pfcrt-related drug resistance differs among various geographical areas. Variants with the sequences CVIET, CVIDT, and SVMNT at pfcrt positions 72 to 76 are prevalent in the Indochinese Peninsula (13, 21, 24). The pfmdr1 gene is located on chromosome 5 and encodes P-glycoprotein homologue 1 (Pgh1). This protein is localized to the digestive vacuole membrane, where it is thought to function in the import of solutes, including some antimalarial drugs, into the digestive vacuole (21). pfmdr1 mutations in codons 86, 184, 1034, 1042, and 1246 have been reported previously and have been shown to correlate with susceptibility to chloroquine, quinine, and mefloquine (23). Sulfadoxine-pyrimethamine (SP) resistance is thought to be due to specific point mutations in the pfdhfr and the pfdhps genes. The pfdhfr gene encodes dihydrofolate reductase (DHFR), the target enzyme of pyrimethamine and trimethoprim. Conformational changes in this enzyme due to point mutations result in the prevention of adequate drug access. The codon positions in the pfdhfr gene that are related to resistance include 16, 50, 51, 59, 108, 140, and 164 (23). The deduced pathway for the resistant mutants suggested that all multiple mutants emerged through stepwise selection from the single mutant with the S108N mutation (18). The pfdhps gene encodes the enzyme dihydropteroate synthetase (DHPS). Point mutations in this gene also lead to conformational changes in DHPS and result in resistance to sulfadoxine and sulfamethoxazole. The loci responsible for resistance have been identified at positions 436, 437, 540, 581, and 613 (23).
In Vietnam, chloroquine-resistant P. falciparum was reported for the first time in the 1960s (12, 14). Ngo et al. reported in 2003 that all of the isolates acquired from 18 adult rubber plantation workers residing in southern Vietnam demonstrated the pfcrt 76T mutation (14). In contrast, Phuc et al. reported that the prevalence of the mutant was only 38.5% when the strains from 39 malaria patients in the Quang Tri Province of central Vietnam were investigated (15). P. falciparum strains resistant to antifolates have also continued to increase in prevalence since the 1980s. Masimirembwa et al. analyzed 40 P. falciparum isolates obtained from malaria patients and reported that 97.5% of the isolates demonstrated a pfdhfr mutation that was related to pyrimethamine resistance, while 95.0% demonstrated a pfdhps mutation that was associated with sulfadoxine resistance (12).
In the 1990s, the treatment for malaria in Vietnam mainly involved monotherapy with artemisinin or single-dose combinations of mefloquine with artemisinin or artesunate. However, the rates of recrudescence after the use of these treatment regimens were as high as 25%. As a result, the Vietnamese Ministry of Health introduced CV8 treatment, which consisted of dihydroartemisinin, piperaquine, trimethoprim, and primaquine, as part of the National Malaria Control Program (NMCP) (6, 20).
Here we present the results of a longitudinal survey conducted in 2001, 2002, and 2005 to 2007 in the Binh Phuoc Province of Vietnam. The study was undertaken to investigate the incidence of malaria caused by P. falciparum and to document the changes in genotype that were related to drug resistance. From this study, we were able to identify the allelic and haplotype changes that occurred over the study years and deduce the factors associated with drug resistance.
MATERIALS AND METHODS
Study site and participants.
This study was preapproved by the Ethics and Scientific Committee of the NIMPE (Hanoi, Vietnam) and the Institute of Tropical Medicine, Nagasaki University (Nagasaki, Japan) and was performed in the Binh Phuoc Province of southern Vietnam. The study periods included June 2001, August and September 2002, May and October 2005, May and October 2006, and May and September 2007. We recruited 527 volunteers from among the residents of villages in the province in 2001, 687 in 2002, 1,070 in 2005, 899 in 2006, and 634 in 2007. Information regarding age and gender were recorded for nearly all participants in 2001 (507 of 527) and 2002 (682 of 687). This information was recorded only for P. falciparum-positive participants in 2005, 2006, and 2007. Informed consent was obtained from all participants or their parents or guardians prior to their entry into the study. Blood samples were obtained from each participant, and blood smears were prepared for the microscopic identification of P. falciparum infection. Blood samples collected on filter paper were used for the genetic investigation of P. falciparum. Exclusion criteria for the analysis were pregnancy, splenectomy, severe malnutrition, and reported prior treatment with an antimalarial drug. Patients who demonstrated P. falciparum infection microscopically were administered antimalarial drugs, according to the Vietnamese Ministry of Health policy.
PCR amplification and sequencing.
DNA was extracted from the blood samples blotted onto filter paper by a previously reported method (17). The primers used are presented in Table 1. Amplification was performed in 20 μl of reaction buffer containing 1 μl DNA, 0.5 μM of each primer, 200 μM of each deoxynucleoside triphosphate, 2 mM MgCl2, and 1 U Taq polymerase (Takara Bio, Inc., Ohtsu, Japan). For pfcrt and pfmdr1, the outer PCR was performed under the following conditions: after an initial denaturation step at 94°C for 2 min, the samples were subjected to 30 cycles of denaturation at 94°C for 20 s, hybridization at 52°C for 80 s, and DNA synthesis at 60°C for 4 min. The products of the outer PCR were then diluted eightfold and used as a template for the nested PCR, which was performed under the following conditions: after an initial step of denaturation at 94°C for 2 min, the samples were subjected to 35 cycles of denaturation at 94°C for 20 s, hybridization at 56°C for 80 s, and DNA synthesis at 60°C for 2 min. The PCR conditions for pfdhfr and pfdhps were those reported previously (11). The amplified products were then directly sequenced by use of a BigDye Terminator (version 1.1) cycle sequencing kit and a model 3730 genetic analyzer (Applied Biosystems) (16). The primers used for sequence analysis are presented in Table 2. In the case of the detection of new or rare mutations, two independent PCR products were subjected to sequence analysis. Additionally, the sequencing reactions were carried out from both the 5′ and the 3′ sides.
TABLE 1.
Primers used to genotype pfcrt, pfmdr1, pfdhfr, and pfdhps
| Primer | Sequence (5′ → 3′) | Purpose |
|---|---|---|
| Crt1F | CATTGTCTTCCACATATATGACATAAA | Outer PCR for pfcrt |
| Crt4R | GATCTCTATACCTTCAACATTATTCCT | Outer PCR for pfcrt |
| Crt2F | TTTCCCTTGTCGACCTTAACAGATGGC | Nested PCR for the former part of pfcrt |
| Crt12R | ATCCTATTTTACCTCTACGACTGT | Nested PCR for the former part of pfcrt |
| Crt11F | TTTCTTATAGGCTATGGTATCCTTT | Nested PCR for the latter part of pfcrt |
| Crt15R | TTTTAATTTCACACTTACCAAAGT | Nested PCR for the latter part of pfcrt |
| MDR2F | AAAGATGGTAACCTCAGTATCAAAGAAGA | Outer and nested PCRs for the former part of pfmdr1 |
| MDR8R | ATGATTCGATAAATTCATCTATAGCAGCAA | Outer and nested PCRs for the former part of pfmdr1 |
| MDR10R | TTTTTTGGACACATCAACAACATCAGAATC | Nested PCR for the former part of pfmdr1 |
| MDR5F | AGAAGATTATTTCTGTAATTTGATAGAAAAAGC | Nested PCR for the former part of pfmdr1 |
| DHFR1F | ATGATGGAACAAGTCTGCGACGTTTTCGAT | PCR for pfdhfr |
| DHFR2R | TTCATTTAACATTTTATTATTCGTTTTCTT | PCR for pfdhfr |
| DHPS1F | CCATTCCTCATGTGTATACAACAC | PCR for pfdhps |
| DHPS2R | GTTTTAATCACATGTTTGCACTTTC | PCR for pfdhps |
TABLE 2.
Primers used for sequencing
| Primer | Sequence (5′→3′) | Target gene | Objective loci |
|---|---|---|---|
| CRT2.2F | TTCGACCTTAACAGATGGCTCACGTTTA | pfcrt | 72-76, 97 |
| CRT9.2F | TATTTCTTATGACCTTTTTAGGAACGA | pfcrt | 144, 148, 160, 163 |
| CRT10.2F | ATTTATTTACTCCTTTTTAGATATCAC | pfcrt | 194, 220 |
| CRT11.2F | ATTCCTATAACGCATTATAATTATTTC | pfcrt | 271 |
| CRT16R | AAACATTCCCATATTTATTTCCTC | pfcrt | 326, 333 |
| CRT14F | TTATAGATTATCGACAAATTTTCT | pfcrt | 356 |
| CRT17F | GTACAACGTATCATATTTTATAAT | pfcrt | 371 |
| MDRp3F | GAGTACCGCTGAATTATTTAGA | pfmdr1 | 86, 147,153, 184 |
| MDR14F | ATGTTTATGTTAACAATTATCTTACCA | pfmdr1 | 363, 487 |
| MDR7R | GCACATTAATTTTCCAGCATAACTACCAGT | pfmdr1 | 1033, 1034 |
| MDR6F | AGAATTATTGTAAATGCAGCTTTATGGGGATTC | pfmdr1 | 1042, 1109 |
| MDR15F | ATGATCACATTATATTAAAAAATGATAT | pfmdr1 | 1246 |
| DHFR2R | TTCATTTAACATTTTATTATTCGTTTTCTT | pfdhfr | 16, 50, 51, 59, 108, 140, 164 |
| DHPS3F | TTTGTTGAACCTAAACGTGCTGTTCAAA | pfdhps | 436, 437 |
| DHPS4R | TTTATTTTCATTTTGTTGTTCATCATG | pfdhps | 540, 581, 613 |
Statistical analysis.
Statistical analysis was performed by using the Stat View program (version 5). The categorical variables were compared by the χ2 test or the Fisher exact test. Continuous variables were analyzed by the t test, analysis of variance, or Spearman's rank correlation test. Odds ratios (ORs) and 95% confidence intervals (CIs) were computed as an estimate of the relative risk. Multiple logistic regression analysis was undertaken for variables associated with the year of the study, age, gender, and genotypes related to drug resistance. A P value of <0.05 was defined as being statistically significant. Data for infections caused by isolates with mixed haplotypes were excluded from the haplotype statistical analysis. Correlations between genotype, age, and gender were performed only for the samples obtained between 2001 and 2002.
RESULTS
Characteristics of study participants.
The numbers of participants entered into this study were 527 in 2001, 687 in 2002, 1,070 in 2005, 899 in 2006, and 634 in 2007. The rates of positive cases diagnosed microscopically were 24.5% (n = 129), 24.0% (n = 165), 3.4% (n = 36), 5.0% (n = 45), and 3.5% (n = 22) for those years, respectively (P = 0.16). Among the participants, 52.5% (266 of 507) of the subjects investigated in 2001 were male, while 47.9% (327 of 682) of the subjects investigated in in 2002 were male. The average ages were 24.9 ± 17.9 years (age range, 1 to 78 years) in 2001 and 23.6 ± 17.1 years (age range, 0 to 89 years) in 2002. Among the participants observed in 2001 and 2002, the rates of positivity for infection were 26.8% (159 of 593) for males and 22.5% (134 of 596) for females (P = 0.08). The odds ratio for males was calculated to be 1.26, and the 95% CI was 0.97 to 1.64. The average age of the participants negative for P. falciparum infection in 2001 and 2002 was 25.4 ± 17.9 years, and that of the participants positive for infections was 20.5 ± 14.6 years (P < 0.001). The gender proportion and the average age of the positive participants for each year are presented in Table 3.
TABLE 3.
Characteristics of participants and parasite genotypes
| Characteristic | Position | Amino acidsa | 2001 | 2002 | 2005 | 2006 | 2007 | P valuec |
|---|---|---|---|---|---|---|---|---|
| Ageb | 22.1 ± 15.2 (2-75) | 19.3 ± 14.0 (0-72) | 23.1 ± 16.3 (1-65) | 14.3 ± 11.8 (0-48) | 9.63 ± 10.8 (2-46) | 0.05 | ||
| % Males | 52.7 | 55.5 | 66.7 | 38.1 | 25.0 | 0.07 | ||
| pfcrt | 74 | M/I/mix | 31/42/9 (37.8/51.2/11.0)d | 67/22/11 (67.0/22.0/11.0) | 33/0/0 (100/0/0) | 31/6/1 (81.6/15.8/2.6) | 11/3/2 (68.8/18.8/12.5) | <0.001 |
| 75 | N/E/D/mix | 31/28/13/10 (37.8/34.1/15.9/12.2) | 67/7/15/11 (67.0/7.0/15.0/11.0) | 33/0/0/0 (100/0/0/0) | 31/1/5/1 (83.8/2.6/13.2/2.6) | 11/1/2/2 (68.8/6.3/12.5/12.5) | <0.001 | |
| 76 | K/T/mix | 31/42/9 (37.8/51.2/11.0) | 67/22/11 (67.0/22.0/11.0) | 33/0/0 (100/0/0) | 31/6/1 (81.6/15.8/2.6) | 11/3/2 (68.8/18.8/12.5) | <0.001 | |
| 144 | A/F/mix | 63/12/7 (76.8/14.6/8.5) | 75/17/8 (75.0/17.0/8.0) | 36/0/0/ (100/0/0) | 35/3/0 (92.1/7.9/0) | 14/2/0 (87.5/12.5/0) | 0.07 | |
| 148 | L/I/mix | 63/12/7 (76.8/14.6/8.5) | 75/17/8 (75.0/17.0/8.0) | 36/0/0 (100/0/0) | 35/3/0 (92.1/7.9/0) | 14/2/0 (87.5/12.5/0) | 0.07 | |
| 194 | I/T/mix | 63/12/7 (76.8/14.6/8.5) | 75/17/8 (75.0/17.0/8.0) | 36/0/0 (100/0/0) | 35/3/0 (92.1/7.9/0) | 14/2/0 (87.5/12.5/0) | 0.07 | |
| 220 | A/S/mix | 31/42/9 (37.8/51.2/11.0) | 65/24/1 (65.0/24.0/11.0) | 36/0/0 (100/0/0) | 31/6/1 (81.6/15.8/2.6) | 11/4/1 (68.8/25.0/6.3) | <0.001 | |
| 271 | Q/E/mix | 31/42/9 (37.8/51.2/11.0) | 68/22/10 (68.0/22.0/10.0) | 36/0/0 (100/0/0) | 31/5/2 (81.6/13.2/5.3) | 11/4/1 (68.8/25.0/6.3) | <0.001 | |
| 326 | N/S/mix | 54/23/6 (65.9/28.0/7.3) | 93/3/4 (93.0/3.0/4.0) | 36/0/0 (100/0/0) | 36/2/0 (94.7/5.3/0) | 15/0/1 (92.1/0/6.3) | <0.001 | |
| 333 | T/S/mix | 59/17/7 (72.0/20.7/8.5) | 76/16/8 (76.0/16.0/8.0) | 36/0/0 (100/0/0) | 35/3/0 (92.1/7.9/0) | 13/2/1 (81.3/12.5/6.3) | 0.02 | |
| 356 | I/T/mix | 55/24/4 (67.1/29.3/4.9) | 90/6/4 (90.0/6.0/4.0) | 36/0/0 (100/0/0) | 36/2/0 (92.1/5.3/0) | 15/0/1 (93.8/0/6.3) | <0.001 | |
| 371 | R/I/mix | 49/28/6 (59.8/34.1/7.3) | 89/7/4 (89.0/7.0/4.0) | 36/0/0 (100/0/0) | 35/1/2 (92.1/2.6/5.3) | 13/1/2 (81.3/6.3/12.5) | <0.001 | |
| pfmdr1 | 130 | E/K | 66/1 (98.5/1.5) | 90/1 (98.9/1.1) | 33/0 (100/0) | 35/0 (100/0) | 13/1 (92.9/7.1) | |
| 184 | Y/F/mix | 53/8/5 (80.3/12.7/7.9) | 72/7/12 (79.1/7.7/13.2) | 33/0/0 (100/0/0) | 31/3/1 (88.6/8.6/2.9) | 13/1/0 (92.9/7.1/0) | 0.32 | |
| 1042 | N/D/mix | 55/8/4 (83.3/12.1/6.0) | 80/4/7 (87.9/4.4/7.7) | 33/0/0 (100/0/0) | 33/3/0 (91.7/8.3/0) | 12/1/1 (85.7/7.1/7.1) | 0.17 | |
| 1109 | V/I/mix | 62/0/4 (93.9/0/6.1) | 91/1/0 (98.9/1.1/0) | 33/0/0 (100/0/0) | 33/3/0 (91.7/8.3/0) | 13/0/0 (100/0/0) | ||
| pfdhfr | 51 | N/I/mix | 4/71/0 (5.3/94.7/0) | 6/39/5 (12.0/78.0/10.0) | 1/23/0 (4.2/95.8/0) | 10/24/0 (29.4/70.6/0) | 3/8/0 (27.3/72.7/0) | <0.001 |
| 59 | C/R/mix | 4/70/1 (5.3/93.3/1.3) | 3/43/4 (6.0/86.0/8.0) | 0/22/2 (0/91.7/8.3) | 0/34/0 (0/100/0) | 0/10/0 (0/100/0) | 0.39 | |
| 108 | S/N/mix | 4/71/0 (5.3/94.7/0) | 1/48/1 (2.0/96.0/2.0) | 0/24/0 (0/100/0) | 0/35/0 (0/100/0) | 0/11/0 (0/100/0) | 0.38 | |
| 164 | I/L/mix | 36/28/10 (48.6/37.8/13.5) | 40/5/5 (80.0/10.0/10.0) | 21/3/0 (87.5/12.5/0) | 32/3/0 (91.4/8.6/0) | 9/2/1 (75.0/16.7/8.3) | <0.001 | |
| pfdhps | 436 | S/A/F/mix | 21/25/10/7 (33.3/39.7/15.9/11.1) | 44/33/8/10 (46.3/34.7/8.4/10.7) | 10/19/1/0 (33.3/63.3/3.3/0) | 25/3/0/8 (69.4/8.3/0/22.3) | 13/0/1/2 (81.3/0/6.3/12.5) | <0.001 |
| 437 | A/G/mix | 7/56/0 (11.1/88.9/0) | 0/92/3 (0/96.8/3.2) | 1/29/0 (3.3/96.7/0) | 1/34/1 (2.8/94.4/2.8) | 0/17/0 (0/100/0) | <0.001 | |
| 540 | K/E/N/Y/mix | 19/27/6/0/11 (30.2/63.5/9.5/0/17.4) | 32/39/7/1/17 (33.3/40.6/7.3/1.0/17.7) | 7/19/1/0/3 (23.3/63.3/3.3/0/10.0) | 6/19/0/0/12 (16.2/51.4/0/0/32.4) | 1/9/0/0/7 (5.9/52.9/0/0/41.2) | 0.23 | |
| 581 | A/G/mix | 47/8/8 (74.6/12.7/12.7) | 61/20/16 (62.9/19.6/17.4) | 23/4/3 (76.7/13.3/10.0) | 8/16/13 (21.6/43.2/35.1) | 4/9/4 (23.5/52.9/23.5) | <0.001 | |
| 613 | A/T/S/mix | 49/1/8/5 (77.8/1.6/12.7/8.0) | 87/0/6/3 (90.6/0/6.3/3.1) | 30/0/0/0 (100/0/0/0) | 36/0/0/0 (100/0/0/0) | 15/0/0/0 (100/0/0/0) | <0.001 |
Boldface type indicates amino acid mutations.
Values are means ± standard deviations (minimum-maximum).
P values are for the correlation between each year. Data for mixed infections for each locus were excluded from the statistical analysis.
The data represent the number (percentage) of patients with the indicated genotype.
Genotype and timescale. (i) pfcrt.
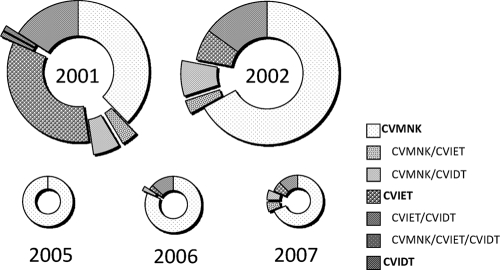
The substitution of K for T at codon 76, which encodes the critical change required for chloroquine resistance, was dominant in 2001 (42/82 [51.2%] isolates had T residues, and an additional 9/82 [11.0%] were mixed cases), a proportion that rapidly decreased by 2005 (0%). In 2006, however, this genotype appeared to have reemerged, demonstrating a prevalence of 15.8% (6/38 isolates had T residues) and then one of 18.8% (3/16 isolates had T residues) in 2007. With regard to the haplotype present in codons 72 to 76, the mutated sequences consisted of two types, CVIET and CVIDT. The CVIET sequence was observed at a high rate in 2001 (28/82 [34.1% isolates); however, the rate of observation of this sequence was lower subsequent to 2002. In 2001, 15.9% (13/82) of the isolates presented the CVIDT sequence, and this tendency appeared to be stable in all years except 2005 (Fig. 1).
FIG. 1.
Sequence polymorphism for pfcrt at positions 72 to 76. Each pie graph represents a sample year. The pieces in the pies reflect the positive ratio each year. The data for mixed infections are presented at the pop-up positions.
The mutated alleles identified at positions 220 and 271 demonstrated similar rates of occurrence, while the additional polymorphic loci showed different proportions (Table 3). The isolates with the CVIET sequence exhibited five haplotype forms on the downstream side of position 76, in which the vast majority of isolates (79.4% [27/34]) had the same sequence as the Dd2 strain (CVIET-A-L-I-S-E-S-T-T-I at positions 72 to 76, 144, 148, 194, 220, 271, 326, 333, 356, and 371, respectively [boldface indicates the mutated alleles]). The remaining four haplotype forms with the CVIET sequence demonstrated the same substitution at positions 220, 271, and 371, while the substitutions at positions 326, 333, and 356 were found to vary (N-T-I, N-S-I, N-S-T, and N-S-T at positions 326, 333, and 356, respectively). In contrast, the isolates with the CVIDT sequence demonstrated four haplotype forms on the downstream part of position 76. The majority of the isolates (90.9% [30/33]) had the same sequence as the Cambodian isolate reported previously (5) (CVIDT-F-I-T-S-E-N-S-I-R at positions 72 to 76, 144, 148, 194, 220, 271, 326, 333, 356, and 371, respectively). All other CVIDT variants demonstrated the same substitutions at positions 220 and 271, while the substitutions at positions 144, 148, 194, and 333 demonstrated various combinations (A-L-T-T, A-L-T-S, and F-I-S-S at positions 144, 148, 194, and 333, respectively). Cases of mixed infections were excluded from this statistical analysis of haplotypes.
(ii) pfmdr1.
The amino acids at positions 86N and 1246D, sites that have previously been shown to exhibit variations in other studies (23), demonstrated no polymorphisms in the current study. The amino acids at positions 130, 184, 1042, and 1109 did exhibit polymorphisms; however, the proportions identified over the study period were not statistically significantly different, irrespective of the uniform allelic pattern in 2005 (Table 3). The mutation at position 130 (E to K) reported recently was in a sample from a study performed in Cambodia (7).
(iii) pfdhfr and pfdhps.
In relation to the sequence of the P. falciparum DHFR, all isolates showed normal patterns at positions 16 and 50. Mutations at those positions are closely related to cycloguanil, pyrimethamine, and trimethoprim resistance (18). Isolates demonstrating a lack of mutation were obtained only in 2001 and 2002. All isolates obtained after 2005 possessed more than two mutations. A mutation at position 164, which is related to high-level resistance, was found at the highest percentage in 2001 (28/74 [37.8%] isolates had L residues), a result that then tended to decrease over time (Table 3). The mutation in P. falciparum DHPS (PfDHPS) at position 437, which is regarded as both the first and the most important change required for sulfadoxine and sulfamethoxazole resistance, showed a high proportion in every year. At position 436, two types of amino acid substitutions, S for A or F, were observed in this study.
Position 540 appeared to be more polymorphic in this study. We observed three types of substitutions: K for either E, N, or Y. To the best of our knowledge, the substitution of K for Y has not previously been reported. The substitution of A for G at position 581 was observed every year in this study and tended to increase in proportion, from 12.7% (8/63) in 2001 to 52.9% (9/17) in 2007. The codon responsible for generating G is generally found to be GGG; however, we identified the GGT codon in some of the 2002 isolate samples. One of these was identified as a single infection, while two were found to result from a mixed infection and expressed GCG (A) or GGG (Table 3).
Factors related to the special genotypes.
The positive samples from 2001 and 2002 were used for the analysis. Among males and females, 36.3% and 42.9%, respectively, exhibited the pfcrt 76T mutation (P = 0.40). The average age of the participants whose isolates exhibited the pfcrt 76T mutation was significantly greater than that of the participants whose isolates exhibited the pfcrt 76K mutation (25.7 ± 15.0 and 17.3 ± 14.5 years, respectively; P < 0.01) (Table 4). By using multivariate analysis to determine the risk that isolates would express the pfcrt 76T mutation, the occurrence of an infection in the year 2002 appeared to be accompanied by a lower risk (P < 0.01; 95% CI = 0.13 to 0.52), while an age of ≥20 years demonstrated a greater risk (P = 0.02; 95% CI = 1.19 to 4.80). The number of mutations in the pfdhfr and pfdhps genes individually and combined were compared with the ages of the infected participants by using Spearman's rank correlation test. The number of mutations was found to positively correlate with age for each gene and both genes combined (for pfdhfr, P = 0.02; for pfdhps, P = 0.03; for the two genes combined, P = 0.02) (Table 5). By multivariate analysis of the data for isolates with seven or more mutations, males demonstrated a reduced risk of disease infection (P = 0.04; 95% CI = 0.05 to 0.89), while an age of 20 years or older (P = 0.05; 95% CI = 1.01 to 16.59) and the presence of the pfcrt 76T mutation (P < 0.01; 95% CI = 2.92 to 59.32) appeared to represent independent risk factors. There were no significant associations between the study years (P = 0.07; 95% CI = 0.07 to 1.12).
TABLE 4.
Correlation between genotype and patient characteristicsa
| Characteristic | Amino acid(s)b or no. of mutations | Genderc |
Aged | No. of people | P value | ||
|---|---|---|---|---|---|---|---|
| No. (%) male | No. (%) female | P valueg | |||||
| pfcrt | 76K | 58 (63.7) | 40 (57.1) | 17.3 ± 14.5 | 98 | ||
| 76T | 33 (36.3) | 30 (42.9) | 0.4 | 25.7 ± 15.0 | 63 | <0.01h | |
| pfmdr1 | NYSNDe | 63 (86.3) | 55 (88.7) | 19.0 ± 15.1 | 118 | ||
| NFSND | 1 (1.4) | 4 (6.5) | 10.6 ± 6.5 | 5 | |||
| NYSDD | 2 (2.7) | 0 (0) | 22.5 ± 7.8 | 2 | |||
| NFSDD | 7 (9.6) | 3 (4.8) | 0.16 | 22.5 ± 14.5 | 10 | 0.1i | |
| pfdhfr + pfdhps | ≥5f | 31 (79.5) | 27 (81.8) | 0.8 | |||
| ≥6 | 18 (46.2) | 20 (60.0) | 0.22 | ||||
| ≥7 | 11 (28.2) | 14 (42.4) | 0.21 | ||||
Data for mixed infections were excluded.
Boldface type indicates amino acid mutations.
n = 91 for males, n = 70 for females.
The data are means ± standard deviations.
pfmdr1 positions 86, 184, 1034, 1042, and 1246, respectively.
The data represent the number of mutations for pfdhfr and pfdhps.
P values were obtained from analysis of relationships between gender and each genotype by the χ2 test.
Obtained from the t test for correlation between pfcrt genotypes and patient age.
Obtained by analysis of variance for correlation between pfmdr1 genotypes and patient age.
TABLE 5.
Numbers of mutations for pfdhfr and pfdhps and age of patients
| Gene | No. of mutations | Agea | No. of people | P value |
|---|---|---|---|---|
| pfdhfr | 0 | 24.6 ± 12.4 | 8 | |
| 2 | 19.5 ± 24.7 | 2 | ||
| 3 | 17.5 ± 15.9 | 61 | ||
| 4 | 27.8 ± 14.2 | 31 | 0.02 | |
| pfdhps | 0 | 25.4 ± 9.1 | 5 | |
| 1 | 18.0 ± 18.1 | 10 | ||
| 2 | 17.9 ± 15.6 | 36 | ||
| 3 | 21.1 ± 13.4 | 50 | ||
| 4 | 30.3 ± 15.6 | 14 | 0.03 | |
| pfdhfr + pfdhps | 0 | 27.0 ± 9.6 | 4 | |
| 2 | 16.3 ± 13.2 | 3 | ||
| 3 | 10.5 ± 12.0 | 2 | ||
| 4 | 18.3 ± 21.7 | 4 | ||
| 5 | 19.1 ± 15.5 | 21 | ||
| 6 | 17.6 ± 12.4 | 13 | ||
| 7 | 26.7 ± 13.8 | 19 | ||
| 8 | 32.8 ± 12.3 | 6 | 0.02 |
The data are means ± standard deviations.
The correlation between the pfcrt 76T and pfmdr1 haplotypes was also calculated (Table 6). We found that 93.2% of the pfcrt 76K isolates were wild type for pfmdr1, while 73.3% of the pfcrt 76T isolates were wild type for pfmdr1. None of the pfcrt 76K isolates demonstrated the haplotype with double mutations (NFSDD) for pfmdr1, while 22.2% of the pfcrt 76T isolates demonstrated the haplotype with double mutations (NFSDD) for pfmdr1 (P < 0.001). The total numbers of mutations in pfdhfr, pfdhps, and both genes combined were higher among the pfcrt 76T isolates than the pfcrt 76K isolates (P <0.001, 0.001, and <0.001, respectively) (Table 6). Among the pfmdr1 isolates with the NFSDD double mutations, the total number of mutations in the pfdhfr gene tended to be slightly higher (P = 0.09) (Table 6).
TABLE 6.
Relationship between different genotypes
| Gene | Amino acid(s)a |
pfcrt 76 |
pfdhfr |
pfdhps |
pfdhfr + pfdhps |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. (%) of isolates with: |
P value | Avg no. of mutations ± SD | No. of samples | P value | Avg no. of mutations ± SD | No. of samples | P value | Avg no. of mutations ± SD | No. of samples | P value | |||
| K | T | ||||||||||||
| pfcrt 76 | K | 2.7 ± 1.1 | 51 | 2.2 ± 1.1 | 60 | 4.6 ± 2.0 | 35 | ||||||
| T | 3.4 ± 1.0 | 43 | <0.001 | 2.8 ± 0.7 | 46 | <0.001 | 6.3 ± 1.5 | 34 | <0.001 | ||||
| pfmdr1 | NYSNDb | 69 (93.2) | 33 (73.3) | 3.1 ± 0.9 | 66 | 2.6 ± 0.9 | 80 | 4.0 ± 2.0 | 49 | ||||
| NFSND | 4 (5.4) | 1 (2.2) | 2.0 ± 1.7 | 3 | 2.3 ± 1.0 | 4 | 5.6 ± 1.6 | 3 | |||||
| NYSDD | 1 (1.4) | 1 (2.2) | 3 | 1 | 0 | 0 | |||||||
| NFSDD | 0 (0) | 10 (22.2) | <.001 | 3.6 ± 0.5 | 8 | 0.09 | 2.5 ± 1.0 | 6 | 6.0 ± 1.5 | 6 | |||
Boldface type indicates amino acid mutations.
pfmdr1 positions 86, 184, 1034, 1042, and 1246, respectively.
DISCUSSION
Through the analysis of P. falciparum gene sequences in samples from malaria patients residing in southern Vietnam in 2001 and 2002, we observed a tendency for males to be more vulnerable than females to P. falciparum infection and that aging may be a protective factor. Although gender does not appear to be directly related to vulnerability to malaria, except during pregnancy, socioeconomic conditions may alter this tendency. In areas where malaria is highly endemic, the risk of infection with P. falciparum is greatest at 1 to 5 years of age. This risk gradually decreases as effective immunity develops. In areas with moderate levels of malaria transmission, a peak age of infection is observed later in childhood. In areas with low levels of transmission, vulnerability to infection does not appear to vary among the different ages, as immunity is not long lasting (3). In this study, the level of transmission in 2001 and 2002 was moderate, so the immunity accompanying aging may partially work in preventing P. falciparum infection.
The number of isolates with the pfcrt 76T mutation was found to rapidly decrease between 2001 and 2005. We hypothesized that this was the result of a reduction in pressure from the use of chloroquine due to the preferred use of artemisinin derivatives in clinical settings. In Malawi, the efficacy of chloroquine for the treatment of P. falciparum malaria was calculated to be less than 50% in 1993; however, after the replacement of chloroquine treatment in 2001, the efficacy was increased to 99% (9). An additional analysis also demonstrated that the chloroquine resistance mutation pfcrt 76T was detected in 85% of isolates in 1992, which was reduced to 13% in 2000 (8). We did not detect parasites expressing the pfcrt 76T mutation in 2005; however, the mutant isolates were found to have reemerged in 2006 and 2007. One possible explanation for this finding may be the existence of lasting chloroquine pressure, as P. vivax remains endemic in this area and, thus, chloroquine is still prescribed.
The variation in the haplotypes observed in pfcrt may be explained in part by the combination of the CVIET Dd2 parental type and the reported CVIDT Cambodia type. These findings suggest that recombination occurs frequently in pfcrt, including regions within exons, and that these recombinations produce animate descendants. It remains unclear whether these variants demonstrate an infectious ability similar to that of their parental variants. Further investigations in vitro and in vivo may be useful in clarifying this situation. In addition, analysis with several microsatellite markers located both inside and around the pfcrt locus may aid with the definition of genetic diversity (4, 13).
In the Cambodian study conducted between 2001 and 2002, Khim et al. reported variations in pfmdr1 haplotypes that were similar to those reported in this study. They observed for the first time mutations located at positions 130 and 1109 (7). We also identified these mutations in our samples. Since the site of the Cambodian study was within close proximity to our study site, these isolates may have been generated by low-level clonal spread across these areas. In Vietnam, Ngo et al. investigated the pfmdr1 genotype in Binh Phuoc Province and detected three patterns of pfmdr1 haplotypes (N-S-N-D, Y-S-N-D, and N-S-D-D at positions 86, 1034, 1042, and 1242, respectively) (14).
We also identified polymorphisms in the pfdhfr gene at positions 51, 59, 108, and 164 but not at position 16 or 50. The number of substitutions observed is known to correlate with the level of pyrimethamine resistance. In our study, strains with four mutations were prevalent in 2001 (44.4%); however, the number was found to rapidly decrease. In addition, the proportions of isolates without mutations also tended to decrease.
Substitutions at the loci of the pfdhps gene (S436A/F, A437G, K540E/N, A581G, and A613T/S) have been implicated in the development of resistance by decreasing the binding affinity of the enzyme (23). In this study, we observed all of these mutations and identified one novel type of Y mutation at position 540. The K540N substitution has recently been reported on Car Nicobar Island in India. The parasite harboring this mutation was isolated in 2005, and the authors indicated that the mutation was caused by the additional selective pressure from co-trimoxazole (sulfamethoxazole-trimethoprim) with SP following the 2004 tsunami. Computer modeling and molecular docking analysis also demonstrated that the sulfadoxine binding affinity of the new PfDHPS K540N mutant was similar to that of K540E (10).
Although the substitution of 581A for G was originally reported in South America (23), it is now also widespread among Asian countries. The report from Cambodia documented that 65.5% of the total isolates also contained this mutation (7). In Vietnam, a few isolates expressing this amino acid substitution have been reported. The studies by Masimirembwa et al. (12) and Phuc et al. (15) independently demonstrated that 1 in 40 and 3 in 40 isolates, respectively, expressed these mutations. The substitutions reported are encoded by the GGG codon, and a codon that we newly found was synonymously changed with GGT.
Using a rodent Plasmodium chabaudi model, Wargo et al. demonstrated that antifolate-resistant parasites were competitively suppressed by sensitive parasites in the absence of drug pressure (22). This finding suggests the possibility that the lasting antifolate drug pressure in this area is due to the prescription of either SP or the trimethoprim contained in CV8 or from the prescription of co-trimoxazole as an antibiotic for the treatment of bacterial infectious diseases and antipneumocystis infections in HIV-infected patients. The rapid spread of co-trimoxazole-resistant Streptococcus pneumoniae in Vietnam has been described in the Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. That report documented that in Vietnam the rate of non-co-trimoxazole-susceptible Streptococcus pneumoniae isolates was as high as 91.3%. This high rate indicated the strong pressure from co-trimoxazole in Vietnam (19).
Interestingly, we also demonstrated a weak correlation between female gender and two genetic makers of drug resistance, pfcrt 76T and a high number of mutations in both pfdhfr and pfdhps combined. By multivariate analysis, pfcrt 76T did not appear to demonstrate a statistically significant difference in correlation with female gender, while the antifolate resistance genotype, which had a higher correlation, did. In addition, we observed a strong correlation between these genetic makers and aging. Regarding the results that the patients in the older age groups demonstrated a reduced vulnerability to P. falciparum infection in this study, we can make hypothesis that females and older people may be more compliant with taking antimalarial or antibiotic medications, which might cause low rates of infection among these people.
In this study, the isolates with pfcrt 76T demonstrated a greater number of mutations in pfdhfr and pfdhps. A similar result was reported in a study undertaken in Sudan, where a correlation between the CVIET sequence in pfcrt at positions 72 to 76 and four mutations in pfdhfr and pfdhps was observed (1). The authors suggested that the administration of sulfadoxine-pyrimethamine for the treatment of infections harboring chloroquine-resistant isolates induces pfdhfr and pfdhps mutations. The situation in Vietnam, however, may prove to be different, and thus, further investigation by the use of microsatellite marker analysis may be required to define genetic movement. Such findings may aid with the development of strategies for the eradication of malaria from sites where it is endemic.
Acknowledgments
We are grateful to all the participants and the staff members of Local Binh Phuoc Clinics for their kind assistance. We thank K. Tanabe and N. Sakihama of Osaka University for their advice on blood collection and parasite DNA isolation.
This work was supported by a Grant-in-Aid for Scientific Research (KAKENHI, 19406011 to H.U.) from the Japan Society for Promotion of Science and a Core University Program of Japan Society for Promotion of Science, the Collaborative Study on Emerging and Re-emerging Infectious Diseases in Vietnam.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.A-Elbasit, I. E., I. F. Khalil, M. I. Elbashir, E. M. Masuadi, I. C. Bygbjerg, M. Alifrangis, and H. A. Giha. 2008. High frequency of Plasmodium falciparum CICNI/SGEAA and CVIET haplotypes without association with resistance to sulfadoxine/pyrimethamine and chloroquine combination in the Daraweesh area, in Sudan. Eur. J. Clin. Microbiol. Infect. Dis. 27:725-732. [DOI] [PubMed] [Google Scholar]
- 2.Barat, L. M. 2006. Four malaria success stories: how malaria burden was successfully reduced in Brazil, Eritrea, India, and Vietnam. Am. J. Trop. Med. Hyg. 74:12-16. [PubMed] [Google Scholar]
- 3.Bates, I., C. Fenton, J. Gruber, D. Lalloo, A. Medina Lara, S. B. Squire, S. Theobald, R. Thomson, and R. Tolhurst. 2004. Vulnerability to malaria, tuberculosis, and HIV/AIDS infection and disease. Part 1. Determinants operating at individual and household level. Lancet Infect. Dis. 4:267-277. [DOI] [PubMed] [Google Scholar]
- 4.DaRe, J. T., R. K. Mehlotra, P. Michon, I. Mueller, J. Reeder, Y. D. Sharma, M. Stoneking, and P. A. Zimmerman. 2007. Microsatellite polymorphism within pfcrt provides evidence of continuing evolution of chloroquine-resistant alleles in Papua New Guinea. Malar. J. 6:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durrand, V., A. Berry, R. Sem, P. Glaziou, J. Beaudou, and T. Fandeur. 2004. Variations in the sequence and expression of the Plasmodium falciparum chloroquine resistance transporter (Pfcrt) and their relationship to chloroquine resistance in vitro. Mol. Biochem. Parasitol. 136:273-285. [DOI] [PubMed] [Google Scholar]
- 6.Giao, P. T., P. J. de Vries, Q. Hung le, T. Q. Binh, N. V. Nam, and P. A. Kager. 2004. CV8, a new combination of dihydroartemisinin, piperaquine, trimethoprim and primaquine, compared with atovaquone-proguanil against falciparum malaria in Vietnam. Trop. Med. Int. Health 9:209-216. [DOI] [PubMed] [Google Scholar]
- 7.Khim, N., C. Bouchier, M. T. Ekala, S. Incardona, P. Lim, E. Legrand, R. Jambou, S. Doung, O. M. Puijalon, and T. Fandeur. 2005. Countrywide survey shows very high prevalence of Plasmodium falciparum multilocus resistance genotypes in Cambodia. Antimicrob. Agents Chemother. 49:3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kublin, J. G., J. F. Cortese, E. M. Njunju, R. A. Mukadam, J. J. Wirima, P. N. Kazembe, A. A. Djimde, B. Kouriba, T. E. Taylor, and C. V. Plowe. 2003. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J. Infect. Dis. 187:1870-1875. [DOI] [PubMed] [Google Scholar]
- 9.Laufer, M. K., P. C. Thesing, N. D. Eddington, R. Masonga, F. K. Dzinjalamala, S. L. Takala, T. E. Taylor, and C. V. Plowe. 2006. Return of chloroquine antimalarial efficacy in Malawi. N. Engl. J. Med. 355:1959-1966. [DOI] [PubMed] [Google Scholar]
- 10.Lumb, V., M. K. Das, P. Mittra, A. Ahmed, M. Kumar, P. Kaur, A. P. Dash, S. S. Singh, and Y. D. Sharma. 2009. Emergence of an unusual sulfadoxine-pyrimethamine resistance pattern and a novel K540N mutation in dihydropteroate synthetase in Plasmodium falciparum isolates obtained from Car Nicobar Island, India, after the 2004 tsunami. J. Infect. Dis. 199:1064-1073. [DOI] [PubMed] [Google Scholar]
- 11.Maiga, O., A. A. Djimde, V. Hubert, E. Renard, A. Aubouy, F. Kironde, B. Nsimba, K. Koram, O. K. Doumbo, J. Le Bras, and J. Clain. 2007. A shared Asian origin of the triple-mutant dhfr allele in Plasmodium falciparum from sites across Africa. J. Infect. Dis. 196:165-172. [DOI] [PubMed] [Google Scholar]
- 12.Masimirembwa, C. M., N. Phuong-dung, B. Q. Phuc, L. Duc-Dao, N. D. Sy, O. Skold, and G. Swedberg. 1999. Molecular epidemiology of Plasmodium falciparum antifolate resistance in Vietnam: genotyping for resistance variants of dihydropteroate synthase and dihydrofolate reductase. Int. J. Antimicrob. Agents 12:203-211. [DOI] [PubMed] [Google Scholar]
- 13.Mehlotra, R. K., G. Mattera, M. J. Bockarie, J. D. Maguire, J. K. Baird, Y. D. Sharma, M. Alifrangis, G. Dorsey, P. J. Rosenthal, D. J. Fryauff, J. W. Kazura, M. Stoneking, and P. A. Zimmerman. 2008. Discordant patterns of genetic variation at two chloroquine resistance loci in worldwide populations of the malaria parasite Plasmodium falciparum. Antimicrob. Agents Chemother. 52:2212-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ngo, T., M. Duraisingh, M. Reed, D. Hipgrave, B. Biggs, and A. F. Cowman. 2003. Analysis of pfcrt, pfmdr1, dhfr, and dhps mutations and drug sensitivities in Plasmodium falciparum isolates from patients in Vietnam before and after treatment with artemisinin. Am. J. Trop. Med. Hyg. 68:350-356. [PubMed] [Google Scholar]
- 15.Phuc, B. Q., S. R. Caruana, A. F. Cowman, B. A. Biggs, N. V. Thanh, N. T. Tien, and K. Thuan le. 2008. Prevalence of polymorphisms in dhfr, dhps, pfmdr1 and pfcrt genes of Plasmodium falciparum isolates in Quang Tri Province, Vietnam. Southeast Asian J. Trop. Med. Public Health 39:959-962. [PubMed] [Google Scholar]
- 16.Reeder, J. C., K. H. Rieckmann, B. Genton, K. Lorry, B. Wines, and A. F. Cowman. 1996. Point mutations in the dihydrofolate reductase and dihydropteroate synthetase genes and in vitro susceptibility to pyrimethamine and cycloguanil of Plasmodium falciparum isolates from Papua New Guinea. Am. J. Trop. Med. Hyg. 55:209-213. [DOI] [PubMed] [Google Scholar]
- 17.Sakihama, N., T. Mitamura, A. Kaneko, T. Horii, and K. Tanabe. 2001. Long PCR amplification of Plasmodium falciparum DNA extracted from filter paper blots. Exp. Parasitol. 97:50-54. [DOI] [PubMed] [Google Scholar]
- 18.Sirawaraporn, W., T. Sathitkul, R. Sirawaraporn, Y. Yuthavong, and D. V. Santi. 1997. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc. Natl. Acad. Sci. U. S. A. 94:1124-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song, J. H., N. Y. Lee, S. Ichiyama, R. Yoshida, Y. Hirakata, W. Fu, A. Chongthaleong, N. Aswapokee, C. H. Chiu, M. K. Lalitha, K. Thomas, J. Perera, T. T. Yee, F. Jamal, U. C. Warsa, B. X. Vinh, M. R. Jacobs, P. C. Appelbaum, and C. H. Pai. 1999. Spread of drug-resistant Streptococcus pneumoniae in Asian countries: Asian Network for Surveillance of Resistant Pathogens (ANSORP) Study. Clin. Infect. Dis. 28:1206-1211. [DOI] [PubMed] [Google Scholar]
- 20.Tran, T. H., C. Dolecek, P. M. Pham, T. D. Nguyen, T. T. Nguyen, H. T. Le, T. H. Dong, T. T. Tran, K. Stepniewska, N. J. White, and J. Farrar. 2004. Dihydroartemisinin-piperaquine against multidrug-resistant Plasmodium falciparum malaria in Vietnam: randomised clinical trial. Lancet 363:18-22. [DOI] [PubMed] [Google Scholar]
- 21.Valderramos, S. G., and D. A. Fidock. 2006. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol. Sci. 27:594-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wargo, A. R., S. Huijben, J. C. de Roode, J. Shepherd, and A. F. Read. 2007. Competitive release and facilitation of drug-resistant parasites after therapeutic chemotherapy in a rodent malaria model. Proc. Natl. Acad. Sci. U. S. A. 104:19914-19919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wongsrichanalai, C., A. L. Pickard, W. H. Wernsdorfer, and S. R. Meshnick. 2002. Epidemiology of drug-resistant malaria. Lancet Infect. Dis. 2:209-218. [DOI] [PubMed] [Google Scholar]
- 24.Wootton, J. C., X. Feng, M. T. Ferdig, R. A. Cooper, J. Mu, D. I. Baruch, A. J. Magill, and X. Z. Su. 2002. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418:320-323. [DOI] [PubMed] [Google Scholar]