Abstract
Parvovirus B19 comprises three distinct genotypes (1, 2, and 3). The distribution of B19 genotypes has not before been examined in South Africa. Two hundred thirty-nine laboratory samples submitted to a diagnostic virology laboratory for parvovirus DNA detection were analyzed retrospectively. Of the 53 PCR-positive samples investigated, 40 (75.4%) were identified as genotype 1 by genotype-specific PCR or consensus NS1 PCR and sequencing and 3 (5.7%) as genotype 2 and 10 (18.9%) as genotype 3 by analysis of NS1 sequences. Furthermore, phylogenetic analysis identified two genotype 1 sequences which were distinct from the previously described genotypes 1A and 1B. Interestingly, a genotype 2 virus was detected in the serum of an 11-year-old child, providing evidence for its recent circulation. This is the first study to demonstrate the concurrent circulation of all three genotypes of B19 in South Africa and the provisional identification of a novel subtype of genotype 1. The implications of parvovirus B19 variation are discussed.
Parvovirus B19 is a member of the genus Erythrovirus of the family Parvoviridae (32). Until 2005, parvovirus B19 and adeno-associated viruses in the genus Dependovirus were the only known parvoviruses to infect humans (35). Recently two novel human parvoviruses have been discovered, namely, PARV4 and human bocavirus, with the latter associated with respiratory tract infections and PARV4, as yet, an orphan virus (1, 10, 18).
Parvovirus B19 targets erythroid progenitor cells, and infection in humans is associated with a spectrum of clinical manifestations ranging from the mild erythema infectiosum in children to pure red cell aplasia due to persistent infection in immunocompromised persons. Chronic anemia due to persistent parvovirus infection is not uncommonly seen in HIV-infected individuals, particularly where access to highly active antiretroviral therapy (HAART) is delayed (23).
While most individuals experience transient infection in childhood, there is mounting evidence that the virus persists following acute infection in certain tissue types, including liver, synovium, and skin (8, 15, 17, 19, 20, 26, 25, 31). The persistence of B19 DNA in tissue is thought to be lifelong and of uncertain clinical significance (25, 26). Norja et al. have termed this persistence of viral DNA in tissues the “bioportfolio” and are now utilizing this information to determine the evolution and molecular epidemiology of the virus (25, 26).
The genome of parvovirus B19 consists of a single strand of linear DNA of about 5,600 nucleotides which encodes three proteins of known function, the nonstructural protein NS1 and the two structural proteins viral protein 1 (VP1) and viral protein 2 (VP2) (6, 7). Genetic variation among B19 strains is very low, with <2% divergence across the genome; however, certain genes such as the VP1 unique region (VP1-u) gene have greater sequence variation (up to 4%) (11, 14). Recently, several strains with considerable sequence diversity were discovered, resulting in the identification of three distinct genetic clusters.
Three genotypes of erythrovirus are now recognized. Parvovirus B19 is the prototype of genotype 1 and is responsible for the majority of human infections worldwide (30). Genotypes 2 and 3 display more than 10% nucleotide divergence compared to reference B19 strains (30). Genotype 2 (prototypes LaLi and A6) has been identified at very low frequency in viremic individuals in Europe, Brazil, and Vietnam (16, 21, 24, 29, 33). In central and northern Europe, genotype 2 DNA has been found at much higher frequency in tissue samples of older individuals, and it is believed to be an ancestral virus that circulated in humans in this region up to the 1970s but was replaced by genotype 1 (26). Genotype 3 (prototypes V9 and D91.1) has been identified in French and Brazilian patients as well as in blood donors from Ghana, where it is though to be endemic (4, 29, 30).
In addition to the three main genotypes, two subgroups of genotype 1 strains in Vietnamese patients and two subgroups of genotype 3 strains from Ghana, Europe, and Brazil have been described (4, 27, 33). Recent studies show that parvovirus B19, in contrast to other DNA viruses, has an inherent rate of genetic drift similar to that of RNA viruses, which in part explains the observed diversity (25, 26).
To our knowledge, there have been no publications reporting on the diversity of B19 strains in South Africa. Of major relevance to the diagnostic laboratory is that both commercial and “in-house” PCR assays may fail to detect variants due to mismatches at primer binding sites (2, 5, 16). Indeed, this study was prompted by the identification of a patient with classic parvovirus-induced pure red cell aplasia in which the viral sequence was not amplified by a genotype 1-specific PCR assay. The aim of this retrospective study was to reanalyze samples that had been submitted to our diagnostic virology laboratory for parvovirus investigation using an assay able to detect all known parvovirus genotypes, with a view to determining the distribution of parvovirus variants in South Africa.
MATERIALS AND METHODS
Study samples.
Over a 2-year period (August 2006 to August 2008), 239 samples were submitted to the diagnostic virology laboratory for the detection of parvovirus infection by PCR. To avoid skewing of the data, multiple samples from the same patient were excluded. The sample types submitted for testing included blood (n = 233, 97.5%), bone marrow (n = 3, 1.3%), amniotic fluid (n = 2, 0.8%), and a single tissue sample. The majority of the samples were from immunocompromised patients in the Western Cape Province of South Africa who were suspected of having chronic parvovirus B19 infection.
Screening for parvovirus DNA in these samples had been performed using a genotype 1-specific VP2 nested PCR as described by Heegaard et al. (12). Sufficient sample was available from 141 PCR-negative and 11 PCR-positive serum and bone marrow specimens for further analysis. Samples had been stored at −20°C after initial testing.
Viral DNA isolation.
Depending on the sample volume, viral DNA was extracted from between 200 μl and 1 ml of serum or bone marrow using the Nuclisens EasyMAG platform (bioMérieux, Boxtel, the Netherlands) according to the manufacturer's instructions. DNA was eluted in 50 μl elution buffer and stored at −20°C.
Consensus NS1 PCR for the detection of all three parvovirus genotypes.
A consensus parvovirus B19 nested PCR was used to screen the samples in this study using primer sequences located within the NS1 gene as published by Candotti et al. (4). Primer positions refer to the parvovirus V9 sequence (GenBank accession number AX003421) (4). The first-round PCR was performed with a 50-μl reaction mixture containing 10 μl extracted DNA, 15 mM Tris-HCl (pH 8), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM deoxynucleotide triphosphates (ABgene, Epsom, United Kingdom), 20 pmol each of primers PVB-1 (5′-CACTATGAAAACTGGGCAATAAAC-3′; positions 1747 to 1770) and PVB-2 (5′-AATGATTCTCCTGAACTGGTCC-3′; positions 1988 to 1967), and 1.5 U Supertherm Taq polymerase (JMR Holdings, Kent, United Kingdom). Amplification was performed on a Thermo Hybaid PxE 0.2 thermal cycler (Thermo Scientific, Waltham, MA), with the following conditions: 1 cycle of 94°C for 2 min; 40 cycles of 94°C for 20 s, 55°C for 30 s, and 72°C for 45 s; and a final elongation step at 72°C for 7 min. The second-round PCR was performed using the same basic master mix ingredients containing 50 pmol of each inner primer, namely, PVB-3 (5′-ATAAACTACACTTTTGATTTCCCTG-3′; positions 1765 to 1789) and PVB-4 (5′-TCTCCTGAACTGGTCCCG-3′; positions 1982 to 1965) and 2 μl of first-round PCR product. Cycling conditions were as for the first-round PCR, although the annealing temperature was increased to 58°C. Amplified products were separated by electrophoresis in 2% agarose gel and visualized under UV irradiation after staining with ethidium bromide. The expected sizes of the outer and inner PCR products were 241 bp and 217 bp, respectively. All work was performed in an ISO-15189 accredited molecular laboratory which employs strict precautions to prevent contamination.
Parvovirus sequence and genotype analysis.
Amplified products from samples that were positive with the consensus NS1 PCR were sequenced. The PCR products were purified using a QIAquick PCR purification kit (Qiagen, Hilden, Germany) and sequenced on a ABI 3130XL genetic analyzer with a fluorescent dye terminator kit in accordance with the manufacturer's instructions (Applied Biosystems, Foster City, CA).
The nucleotide sequences were aligned using CLUSTALG software (version 1.4), and a phylogenetic tree was constructed with the Kimura 2-parameter neighbor-joining method with 1,000 bootstrap resamplings using the Treecon software program, version 1.3b (34). Comparative reference sequences of genotype 1A (M24682, M13178, NC_000883, DQ225150, DQ225151, DQ408301, Z68146, DQ225149, AF162273, and AY504945), genotype 1B (DQ357065 and DQ357064), genotype 2 (AY064476, AY064475, AY044266, DQ333428, DQ333426, and DQ333427), genotype 3A (DQ234771, AY582125, DQ234769, NC_004295, AX003421, and AJ249437), and genotype 3B (DQ408304, DQ234778, DQ234779, AY647977, AY582124, AY083234, and DQ408303) were obtained from GenBank. The phylogenetic tree was rooted with simian parvovirus (GenBank accession number U26342).
Confirmation of parvovirus genotypes determined by phylogenetic analysis of the NS1 PCR sequence.
To confirm the genotypes determined by analysis of the NS1 product, a larger PCR product spanning the NS1-VP1-u junction was amplified and sequenced. Primer sequences used in this seminested PCR were as published by Candotti et al. (4). First-round PCR was performed using the same reaction mixture and cycling conditions as the consensus NS1 first-round PCR, except for using 20 pmol of primers PVB-1 and B19SR (5′-CCAGGCTTGTGTAAGTCTTC-3′; positions 2691 to 2672) and extending at 72°C for 1 min 30 s. Second-round PCR was performed using 2 μl of the first-round product and the same reaction mixture containing 50 pmol of primers PVB-3 and B19SR and cycling conditions as for the first round. The 926-bp second-round PCR products were analyzed by electrophoresis in a 2% agarose gel stained with ethidium bromide. Amplified products were sequenced and aligned as for the NS1 PCR products using the following reference sequences for comparison: genotype 1A, M24682, M13178, NC_000883, DQ225151, DQ408301, Z68146, DQ225149, AF162273, and AY504945; genotype 1B, DQ357065 and DQ357064; genotype 2, AY064476, AY064475, AY044266, DQ333428, DQ333426, and DQ333427; and genotype 3, DQ408302, DQ408305, AY083234, AX003421, AJ249437, DQ408303, and DQ408304. The phylogenetic tree was rooted with simian parvovirus (GenBank accession number U26342).
Nucleotide sequence accession numbers.
The GenBank accession numbers of the nucleotide sequences analyzed in this study were FJ904084 to FJ904121 and GQ337718 to GQ337722.
RESULTS
Detection of parvovirus DNA in patient samples.
Of the 239 samples originally tested for B19 DNA over the 2-year period by means of the genotype 1 VP2 nested PCR, 26 samples (10.9%) were initially found to be PCR positive. The 11 PCR-positive serum samples that were available for subsequent amplification with the consensus NS1 PCR were confirmed positive with the consensus NS1 PCR. Twenty-seven (19.1%) of the 141 samples that were originally PCR negative were found to be PCR positive with the consensus NS1 PCR. Of these, 26 were serum samples, while the remaining parvovirus-positive sample was bone marrow.
Sequence analysis of the parvovirus variants.
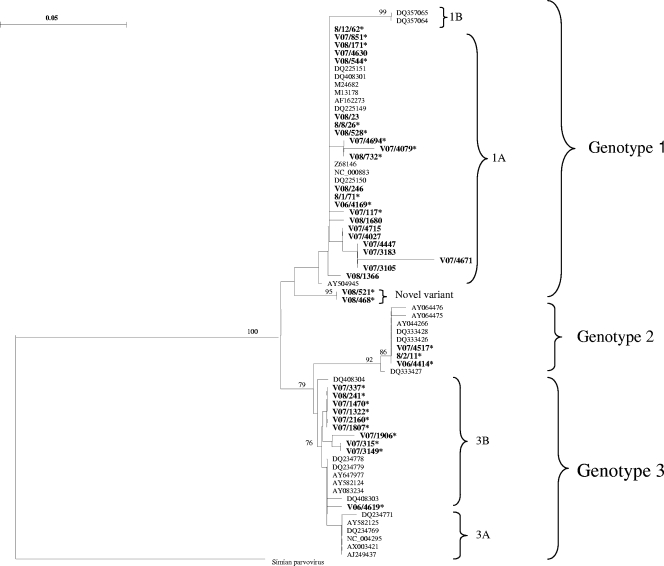
All 11 samples that amplified with both the VP2 and NS1 primers clustered with genotype 1A (Fig. 1) when the NS1 sequences were analyzed. Of the 27 samples that amplified only with the NS1 primers, 12 grouped with genotype 1A. Two samples, V08/521 and V08/468, formed a separate cluster within genotype 1 and distinct from genotype 1B, with high bootstrap values (Fig. 1). Three samples clustered with genotype 2 and 10 with genotype 3B.
FIG. 1.
Phylogenetic tree constructed from partial parvovirus B19 NS1 sequences rooted with simian parvovirus (GenBank accession number U26342). Thirty-eight B19-positive samples (boldface type) were aligned with comparative reference sequences identified by their GenBank accession numbers. B19 DNA-positive samples from this study clustered with genotypes 1A, 2, and 3B. Two samples clustered with genotype 1 but were separate from 1A and 1B and are indicted as a “novel variant.” Only significant bootstrap values (≥70%) are shown. Bar, 0.05 nucleotide substitutions per site. Asterisks refer to samples that were B19 DNA positive with the consensus NS1 PCR but negative with the genotype 1 VP2 PCR.
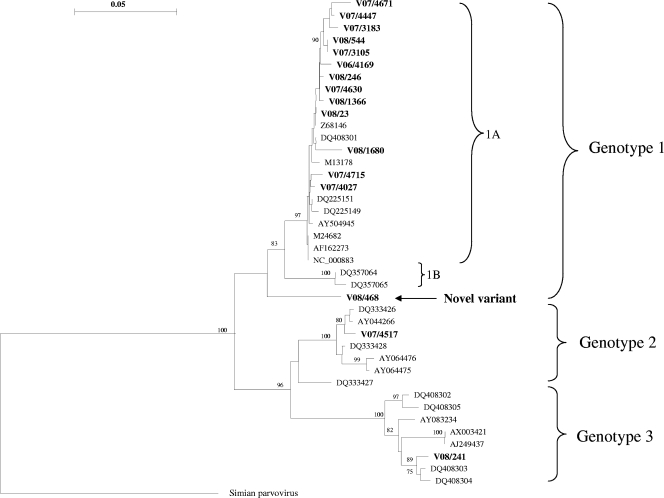
Sixteen samples were analyzed with the NS1-VP1-u PCR, as insufficient specimen volume precluded analysis of all samples positive with the NS1 PCR. Nevertheless, the presence of all three genotypes was confirmed. Phylogenetic analysis of the NS1-VP1-u product revealed that 13 samples clustered with genotype 1A (Fig. 2). The existence of a novel genotype 1 variant was confirmed when analyzing the NS1-VP1-u PCR product from sample V08/468. In addition, one sample containing genotype 2 and one containing genotype 3 were confirmed with the longer PCR product (Fig. 2).
FIG. 2.
Phylogenetic tree constructed from NS1-VP1-u junction sequences rooted with simian parvovirus (GenBank accession number U26342). Sixteen B19-positive samples (boldface type) were aligned with comparative reference sequences identified by their GenBank accession numbers. The presence of all three genotypes and a novel variant of genotype 1 was confirmed with this phylogenetic analysis. Only significant bootstrap values (≥70%) are shown. Bar, 0.05 nucleotide substitutions per site.
Clinical diagnoses of patients infected with parvovirus B19.
Comprehensive clinical details of the patients infected with parvovirus in this study were not available, as many of the samples had been referred from outlying health care facilities. From the limited details available, no particular genotype was found to exclusively or to predominantly infect a particular patient group.
Of the 38 B19 genotype 1-infected patients, 16 were HIV infected, with chronic anemia, 4 had a hematological malignancy, 4 had a cardiomyopathy, 2 had congenital parvovirus infections, 1 had a primary parvovirus infection, and 1 was a renal transplant patient with anemia. Clinical diagnoses in the remaining 10 genotype 1-infected patients were not available. One of the patients infected with the novel genotype 1 variant was HIV infected with pure red cell aplasia (V08/468), and the other had systemic lupus erythematosus with a pancytopenia (V08/521).
Of the genotype 2-infected patients, 1 was an 11-year-old HIV-infected child with anemia. No clinical diagnoses were available for the remaining two patients with genotype 2 infections. Of those infected with genotype 3, two were HIV infected with anemia, 2 had a hematological malignancy, 1 had congenital parvovirus infection, and 1 had a primary B19 infection. No diagnoses were available for the remaining 4 patients.
DISCUSSION
Published data on parvovirus genotypes in South Africa are limited to the study by Candotti et al., who reported the presence of B19 DNA in 2 out of a total of 360 blood donor plasma samples (4). Both of these B19-positive samples clustered with genotype 1A reference sequences following sequencing of an 863-bp product of the NS1-VP1-u junction (4). Of the 239 samples selected for this study, a total of 53 B19 PCR-positive samples were found, 26 by a genotype 1-specific PCR targeting the VP2 region and an additional 27 by a consensus PCR targeting the NS1 region. Of these, 40 (75.5%) were identified as genotype 1 by genotype 1-specific VP2 PCR or sequencing and phylogenetic analysis of the NS1 PCR product, 3 (5.6%) as genotype 2, and 10 (18.9%) as genotype 3. These results reflect the distribution of B19 genotypes in a largely immunocompromised group of patients and may not necessarily mirror the genotypes circulating in the general population. Unknown selective pressures acting on the study population might increase the proportion of unusual genotypes in this group.
In this study we identified 2 novel genotype 1 viruses (V08/521 and V08/468), which formed a separate cluster within genotype 1 distinct from 1A and 1B. The existence of this putative novel subtype of genotype 1 was confirmed by phylogenetic analysis of a larger NS1-VP1-u PCR product (926 bp), which also clustered separately from 1A and 1B. Further work to determine the full genome sequences of these two viruses is ongoing.
Fourteen genotype 1-containing samples were negative on initial screening with the genotype 1-specific VP2 PCR, but viral DNA was detected on retesting with the consensus NS1 PCR. The reason for this is not clear. Failure to detect the 2 viruses provisionally identified as a novel genotype 1 subtype is likely to be due to primer mismatches. However, the remaining 12 samples contained virus typical of genotype 1. A difference in analytical sensitivity between the VP2 and NS1 PCR is unlikely to explain this either, as both assays are known to have a limit of detection of approximately 100 IU/ml of sample.
Another interesting finding in this study is that it provides evidence of contemporary circulation of genotype 2 viruses in South Africa. One of the genotype 2 isolates in this study was detected in a serum sample from an 11-year-old patient who had no history of receiving blood or blood products. This is in contrast with studies in central and northern Europe that suggest that circulation of genotype 2 viruses ceased in the early 1970s and that these viruses were replaced by genotype 1 viruses (22, 25, 26).
The spectrum of clinical manifestations associated with genotypes 2 and 3 has so far been found to be similar to that of genotype 1 infections (11, 30). However, in both this and other studies, patients were selected for parvovirus investigation on the basis of clinical presentations typical of B19 infection (11, 30). Further studies are needed to determine whether infections with genotypes 2 and 3 are associated with novel clinical presentations. The existence of B19 variants has indeed complicated the laboratory diagnosis. PCR-based qualitative and quantitative assays may fail to amplify non-genotype 1 viruses, and PCR results should be interpreted with caution, particularly in geographical areas where variants are known to circulate. Commercial enzyme-linked immunosorbent assays (ELISAs) for the detection of IgG and IgM responses to B19 utilize recombinant genotype 1 VP2 proteins as the capture antigens. Despite significant diversity at the nucleotide level between the genotypes, there is a high degree of homology of the VP2 capsid protein at an amino acid level. Thus, there is likely to be significant immunological cross-reactivity between virus variants, and this has been demonstrated in previous studies (9, 13, 28). In addition, the existence of variants may have implications for treating chronically infected patients with immunoglobulin. Commercial preparations of immunoglobulin are likely to contain neutralizing antibodies predominantly to genotype 1 viruses. Neutralizing antibody responses are directed to epitopes located within the N terminus of VP1, and amino acid variability between the three genotypes is approximately 15% within this region (4). How effective parvovirus antibodies contained in standard immunoglobulin preparations are at neutralizing variant virus remains to be determined (3).
In summary, this is the first study to investigate the distribution of parvovirus B19 genotypes in South Africa. In this cohort, all three genotypes were detected, with genotype 1 predominating as anticipated. Furthermore, we provide evidence for the recent circulation of genotype 2 viruses and the presence of novel genotype 1 viruses in South Africa. The diversity observed within parvovirus B19 viruses has important implications for the laboratory diagnosis and possibly treatment of these infections. In addition it remains to be shown whether there are novel clinical manifestations associated with non-genotype 1 infections.
Acknowledgments
No financial support was received for this work.
There are no conflicts of interest for any of the authors.
Footnotes
Published ahead of print on 18 November 2009.
REFERENCES
- 1.Allander, T., M. T. Tammi, A. Eriksson, A. Bjerkner, A. Tiveljung-Lindell, and B. Andersson. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 102:12891-12896. (Author's correction, 102:15712.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baylis, S. A., N. Shah, and P. D. Minor. 2004. Evaluation of different assays for the detection of parvovirus B19 DNA in human plasma. J. Virol. Methods 121:7-16. [DOI] [PubMed] [Google Scholar]
- 3.Blümel, J., A. M. Eis-Hübinger, A. Stühler, C. Bönsch, M. Gessner, and J. Löwer. 2005. Characterization of parvovirus B19 genotype 2 in KU812Ep6 cells. J. Virol. 79:14197-14206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Candotti, D., N. Etiz, A. Parsyan, and J. P. Allain. 2004. Identification and characterization of persistent human erythrovirus infection in blood donor samples. J. Virol. 78:12169-12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen, B. J., J. Gandhi, and J. P. Clewley. 2006. Genetic variants of parvovirus B19 identified in the United Kingdom: implications for diagnostic testing. J. Clin. Virol. 36:152-155. [DOI] [PubMed] [Google Scholar]
- 6.Cotmore, S. F., V. C. McKie, L. J. Anderson, C. R. Astell, and P. Tattersall. 1986. Identification of the major structural and nonstructural proteins encoded by human parvovirus B19 and mapping of their genes by procaryotic expression of isolated genomic fragments. J. Virol. 60:548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotmore, S. F., and P. Tattersall. 1984. Characterization and molecular cloning of a human parvovirus genome. Science 226:1161-1165. [DOI] [PubMed] [Google Scholar]
- 8.Eis-Hübinger, A. M., U. Reber, T. Abdul-Nour, U. Glatzel, H. Lauschke, and U. Pütz. 2001. Evidence for persistence of parvovirus B19 DNA in livers of adults. J. Med. Virol. 65:395-401. [DOI] [PubMed] [Google Scholar]
- 9.Ekman, A., K. Hokynar, L. Kakkola, K. Kantola, L. Hedman, H. Bondén, M. Gessner, C. Aberham, P. Norja, S. Miettinen, K. Hedman, and M. Söderlund-Venermo. 2007. Biological and immunological relations among human parvovirus B19 genotypes 1 to 3. J. Virol. 81:6927-6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fryer, J. F., A. Kapoor, P. D. Minor, E. Delwart, and S. A. Baylis. 2006. Novel parvovirus and related variant in human plasma. Emerg. Infect. Dis. 12:151-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallinella, G., S. Venturoli, E. Manaresi, M. Musiani, and M. Zerbini. 2003. B19 virus genome diversity: epidemiological and clinical correlations. J. Clin. Virol. 28:1-13. [DOI] [PubMed] [Google Scholar]
- 12.Heegaard, E. D., I. P. Jensen, and J. Christensen. 2001. Novel PCR assay for differential detection and screening of erythrovirus B19 and erythrovirus V9. J. Med. Virol. 65:362-367. [DOI] [PubMed] [Google Scholar]
- 13.Heegaard, E. D., K. Qvortrup, and J. Christensen. 2002. Baculovirus expression of erythrovirus V9 capsids and screening by ELISA: serologic cross-reactivity with erythrovirus B19. J. Med. Virol. 66:246-252. [DOI] [PubMed] [Google Scholar]
- 14.Hemauer, A., A. von Poblotzki, A. Gigler, P. Cassinotti, G. Siegl, H. Wolf, and S. Modrow. 1996. Sequence variability among different parvovirus B19 isolates. J. Gen. Virol. 77:1781-1785. [DOI] [PubMed] [Google Scholar]
- 15.Hokynar, K., J. Brunstein, M. Söderlund-Venermo, O. Kiviluoto, E. K. Partio, Y. Konttinen, and K. Hedman. 2000. Integrity and full coding sequence of B19 virus DNA persisting in human synovial tissue. J. Gen. Virol. 81:1017-1025. [DOI] [PubMed] [Google Scholar]
- 16.Hokynar, K., P. Norja, H. Laitinen, P. Palomäki, A. Garbarg-Chenon, A. Ranki, K. Hedman, and M. Sönderlund-Venermo. 2004. Detection and differentiation of human parvovirus variants by commercial quantitative real-time PCR tests. J. Clin. Microbiol. 42:2013-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hokynar, K., M. Söderlund-Venermo, M. Pesonen, A. Ranki, O. Kiviluoto, E. K. Partio, and K. Hedman. 2002. A new parvovirus genotype persistent in human skin. Virology 302:224-228. [DOI] [PubMed] [Google Scholar]
- 18.Jones, M. S., A. Kapoor, V. V. Lukashov, P. Simmonds, F. Hecht, and E. Delwart. 2005. New DNA viruses identified in patients with acute viral infection syndrome. J. Virol. 79:8230-8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerr, J. R., J. P. Cartron, M. D. Curran, J. E. Moore, J. R. M. Elliott, and R. A. B. Mollan. 1995. A study of the role of parvovirus B19 in rheumatoid arthritis. Br. J. Rheumatol. 34:809-813. [DOI] [PubMed] [Google Scholar]
- 20.Kühl, U., M. Pauschinger, M. Noutsias, B. Seeberg, T. Bock, D. Lassner, W. Poller, R. Kandolf, and H.-P. Schultheiss. 2005. High prevalence of viral genomes and multiple viral infections in the myocardium of adults with “idiopathic” left ventricular dysfunction. Circulation 111:887-893. [DOI] [PubMed] [Google Scholar]
- 21.Liefeldt, L., A. Plentz, B. Klempa, O. Kershaw, A.-S. Endres, U. Raab, H.-H. Neumayer, H. Meisel, and S. Modrow. 2005. Recurrent high level parvovirus B19/genotype 2 viremia in a renal transplant recipient analyzed by real-time PCR for simultaneous detection of genotypes 1 to 3. J. Med. Virol. 75:161-169. [DOI] [PubMed] [Google Scholar]
- 22.Manning, A., S. J. Willey, J. E. Bell, and P. Simmonds. 2007. Comparison of tissue distribution, persistence, and molecular epidemiology of parvovirus B19 and novel human parvoviruses PARV4 and human bocavirus. J. Infect. Dis. 195:1345-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morelli, P., G. Bestetti, E. Longhi, C. Parravicini, M. Corbellino, and L. Meroni. 2007. Persistent parvovirus B19-induced anemia in an HIV-infected patient under HAART. Case report and review of literature. Eur. J. Clin. Microbiol. Infect. Dis. 26:833-837. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen, Q. T., S. Wong, E. D. Heegaard, and K. E. Brown. 2002. Identification and characterization of a second novel human erythrovirus variant, A6. Virology 301:374-380. [DOI] [PubMed] [Google Scholar]
- 25.Norja, P., A. M. Eis-Hübinger, M. Söderlund-Venermo, K. Hedman, and P. Simmonds. 2008. Rapid sequence change and geographical spread of human parvovirus B19: comparison of B19 virus evolution in acute and persistent infections. J. Virol. 82:6427-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norja, P., K. Hokynar, L.-M. Aaltonen, R. Chen, A. Ranki, E. K. Partio, O. Kiviluoto, I. Davidkin, T. Leivo, A. M. Eis-Hübinger, B. Schneider, H.-P. Fischer, R. Tolba, O. Vapalahti, A. Vaheri, M. Söderlund-Venermo, and K. Hedman. 2006. Bioportfolio: lifelong persistence of variant and prototypic erythrovirus DNA genomes in human tissue. Proc. Natl. Acad. Sci. USA 103:7450-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parsyan, A., C. Szmaragd, J.-P. Allain, and D. Candotti. 2007. Identification and genetic diversity of two human parvovirus B19 genotype 3 subtypes. J. Gen. Virol. 88:428-431. [DOI] [PubMed] [Google Scholar]
- 28.Parsyan, A., S. Kerr, S. Owusu-Ofori, G. Elliott, and J.-P. Allain. 2006. Reactivity of genotype-specific recombinant proteins of human erythrovirus B19 with plasmas from areas where genotype 1 or 3 is endemic. J. Clin. Microbiol. 44:1367-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanabani, S., W. K. Neto, J. Pereira, and E. C. Sabino. 2006. Sequence variability of human erythroviruses present in bone marrow of Brazilian patients with various parvovirus B19-related hematological symptoms. J. Clin. Microbiol. 44:604-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Servant, A., S. Laperche, F. Lallemand, V. Marinho, G. De Saint Maur, J. F. Meritet, and A. Garbarg-Chenon. 2002. Genetic diversity within human erythroviruses: identification of three genotypes. J. Virol. 76:9124-9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Söderlund, M., R. von Essen, J. Haapasaari, U. Kiistala, O. Kiviluoto, and K. Hedman. 1997. Persistence of parvovirus B19 DNA in synovial membranes of young patients with and without chronic arthropathy. Lancet 349:1063-1065. [DOI] [PubMed] [Google Scholar]
- 32.Tattersall, P., M. Bergoin, M. E. Bloom, K. E. Brown, R. M. Linden, N. Muzyczka, C. R. Parrish, and P. Tijssen. 2005. Family Parvoviridae, p. 353-369. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 33.Toan, N. L., A. Duechting, P. G. Kremsner, L. H. Song, M. Ebinger, S. Aberle, V. Q. Binh, D. N. Duy, J. Torresi, R. Kandolf, and C.-T. Bock. 2006. Phylogenetic analysis of human parvovirus B19, indicating two subgroups of genotype 1 in Vietnamese patients. J. Gen. Virol. 87:2941-2949. [DOI] [PubMed] [Google Scholar]
- 34.Van de Peer, Y., and R. De Wachter. 1997. Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput. Appl. Biosci. 13:227-230. [DOI] [PubMed] [Google Scholar]
- 35.Young, N. S., and K. E. Brown. 2004. Parvovirus B19. N. Engl. J. Med. 350:586-597. [DOI] [PubMed] [Google Scholar]