Abstract
We reviewed pneumococcal serotype 3 cases reported from 2000 through 2005 to a laboratory-based surveillance system for invasive pneumococcal disease in South Africa. The prevalence of serotype 3 invasive isolates was compared to their prevalence in carriage isolates to determine the odds of invasiveness due to serotype 3 among South African children. Three groups of serotype 3 strains were characterized by pulsed-field gel electrophoresis (PFGE) or Box element PCR (BOX-PCR), randomly selected invasive isolates from one province, isolates from a carriage study involving children in the same province, and antimicrobial-resistant invasive isolates collected nationally. Examples of the PFGE types identified were further characterized by multilocus sequence typing. In total, 15,980 viable isolates causing invasive disease were submitted, of which 661 (4%) were serotype 3, mostly from adults (85% [489/575]). Fewer serotype 3 isolates were nonsusceptible to antimicrobial agents tested (40/661 [6%]) than non-serotype 3 isolates (8,480/15,319 [55%]) (P < 0.001). Compared to non-serotype 3 cases, there was no association with HIV coinfection (2,212/2,569 [86%] versus 72/78 [92%]; P = 0.1) or increased case fatality ratio (1,190/4,211 [28%] versus 54/154 [35%]; P = 0.7). Serotype 3 in children had a low but statistically insignificant invasive disease potential (odds ratio [OR] of 0.15; 95% confidence interval [CI] of 0.01 to 1.06). Strains were grouped into 3 PFGE clusters, with the largest, cluster A, representing 54% (84/155), including 14 isolates confirmed as sequence type 458 (ST458). It was confirmed that 3 isolates from cluster B, which represented only 12% (18/155) of the isolates, were the serotype 3 global strain, ST180. We have therefore identified ST458 as predominating in South Africa, but with an invasive potential similar to that of the predominant global clone ST180.
Pneumococci with serotype 3 capsules are associated with invasive pneumococcal disease (IPD) in older children and adults (13, 14). Serotype 3 pneumococci have been associated with higher case fatality ratios compared to other serotypes (14). In South Africa, the importance of serotype 3 was highlighted when it was shown to be the major cause of intensive care admissions of patients with pneumococcal pneumonia in a tertiary care hospital in Johannesburg from January 1984 to December 1985 (10). Among this group of patients, serotype 3 infections had the highest complication rate and mortality compared with infections caused by other serotypes.
In young children, serotype 3 has been shown however to be associated with low invasive potential and higher carriage rates (4, 31). Despite the association with carriage, serotype 3 strains generally exhibit low levels of antibiotic resistance (10, 17-19); however, a fatal multidrug-resistant (MDR) serotype 3 strain was isolated from the blood of a South African 17-year-old boy in 1987 (20).
The establishment of the pneumococcal multilocus sequence type (MLST) database in 2003 has made it possible to monitor the spread of pneumococcal clones within and between countries. Sequence type 180 (ST180) is known to be predominant among invasive and carriage serotype 3 strains from several countries (2, 4, 6). Little is known about pneumococcal serotype 3 causing invasive disease in developing countries.
The 7-valent pneumococcal conjugate vaccine (PCV-7) is effective in reducing disease in vaccinated individuals as well as in unvaccinated individuals through a herd effect (23). There are, however, reports of the emergence of serotypes not included in the vaccine, including serotype 3 (2, 24). A recent study from Utah reported that more children with serotype 3 pneumonia had received at least one dose of PCV-7 compared to other serotypes (3). To detect serotype changes as a result of universal use of a new vaccine, surveillance of isolates prior to and following the introduction of the vaccine is required. In South Africa, PCV-7 was registered in 2005 but was initially only available in the private health care sector. The vaccine was implemented nationally as part of the routine childhood immunization program in April 2009.
The aim of this study was to review invasive pneumococcal serotype 3 isolates in South Africa over a 6-year period (2000 to 2005) prior to the introduction of PCV-7 and to compare the prevalence of serotype 3 in children to the prevalence of serotype 3 carriage to assess invasive disease potential in children. Further genotypic characterization was performed to describe the molecular epidemiology of these isolates.
MATERIALS AND METHODS
Invasive pneumococcal disease.
National laboratory-based surveillance for IPD in South Africa was initiated in July 1999 (15) and is ongoing. In 2003, surveillance was enhanced to include additional data, such as outcome and HIV serological status from sentinel sites in all nine provinces. HIV testing was requested by admitting physicians when clinically indicated according to standard practice. In Gauteng Province, South Africa, this included HIV enzyme-linked immunosorbent assay (ELISA) testing with confirmation by ELISA on a second specimen for all patients ≥18 months of age (30). For children of <18 months of age, qualitative HIV PCR testing was performed for the diagnosis of HIV. IPD case data and viable pneumococcal isolates were submitted to the National Institute for Communicable Diseases (NICD) in Johannesburg, South Africa (15). For the purposes of this study, a case of IPD was defined as the isolation of a pneumococcus from a specimen from a normally sterile site (e.g., blood culture, cerebrospinal fluid [CSF], pleural fluid, or joint fluid) from individuals of all ages throughout South Africa, identified from 2000 through 2005. In addition, cases included patients with invasive specimens testing positive by bacterial latex agglutination and supported either by Gram stain microscopy and/or PCR (25). If pneumococci were isolated from multiple specimens from the same patient, a primary specimen was determined according to a hierarchy in the following order: CSF, blood, pleural fluid, or other specimens. Only one isolate from each patient was included. All isolates were serotyped and if the same serotype was isolated more than once within 21 days, only the first isolate was included. Children and adults were defined as <15 years and ≥15 years of age, respectively.
A random sample of invasive serotype 3 strains was selected from Gauteng Province in South Africa for molecular characterization. Gauteng is the most densely populated province of nine provinces in South Africa (530 people per km2 in 2005) (8) with an estimated midyear population of 9 million in 2005 (Statistics South Africa [http://www.statssa.gov.za/publications/P0302/P03022005.pdf]). Invasive serotype 3 strains demonstrating reduced antimicrobial susceptibility to at least one agent tested and submitted from all nine provinces were randomly selected for further characterization. A serotype 3 MDR strain isolated in 1987 in South Africa was also included to determine whether this clone was circulating among recent isolates (20).
Carriage study.
A carriage study was conducted at Chris Hani Baragwanath Hospital in Soweto, Gauteng Province, South Africa, from 2007 to 2008. The study enrolled 251 healthy children who tested negative for HIV, who were born to 126 HIV-seropositive and 125 HIV-seronegative mothers. Children were tested for HIV using Roche Amplicor HIV-1 DNA PCR (version 1.5) (Roche Diagnostics, Branchburg, NJ). The children had nasopharyngeal swab samples taken at nine visits: at 1.5 months of age, thereafter approximately every 6 weeks until 7 months of age, at 9 months, 12 months, 16 months, and 24 months of age. In the carriage study, only species identity and serotype were determined. As the unit of measurement for the formula was serotype, we only counted any serotype once for any given child, irrespective of the possibility that the same serotype may have been a newly acquired different strain of the same serotype. If a particular serotype was isolated on more than one visit from the same child, therefore, this serotype was only counted once. The study was approved by the Human Research Ethics Committee (protocol CIPRA 4-MTC PNEUMO; ethics no. 050705), University of the Witwatersrand, South Africa. All mothers gave written consent for their children to participate in the study.
Capsular serotyping and antimicrobial susceptibility testing.
Serotypes of isolates were confirmed by the quellung method using type-specific antisera (Statens Serum Institut, Copenhagen, Denmark) (1). All strains were initially screened for susceptibility to oxacillin (for penicillin), tetracycline, chloramphenicol, erythromycin, clindamycin, rifampin, and trimethoprim-sulfamethoxazole by disk diffusion (Mast Diagnostics Group Ltd., Merseyside, United Kingdom). All isolates testing nonsusceptible on disk screening had MICs determined using agar dilution or Etest (AB Biodisk, Solna, Sweden). Testing and interpretation of all results were done according to Clinical and Laboratory Standards Institute guidelines and breakpoints (7). Isolates were considered to be nonsusceptible to penicillin at MICs of ≥0.12 mg/liter using the oral penicillin breakpoint. For other antimicrobials, isolates were defined as nonsusceptible if they were intermediately resistant or resistant to an agent tested, and isolates were classified as multidrug resistant if they were nonsusceptible to three or more classes of antimicrobial agents tested.
Comparison of serotype 3 and non-serotype 3 disease.
The Mantel-Haenszel χ2 test or Fisher's exact test was used to assess differences between patients with serotype 3 disease and those with disease caused by other serotypes (non-serotype 3 disease). Analysis was performed with Epi Info software, version 6.04d. We calculated incidence rates using midyear population estimates for South Africa supplied by Statistics South Africa. Analyses were carried out for all data collected from 2000 through 2005, except when including outcome and HIV serological status, which was restricted to data collected from enhanced sentinel sites from 2003 through 2005.
Estimation of the invasiveness of serotype 3.
Carriage and invasive disease data were used to estimate the probability of invasiveness due to serotype 3 by calculating an empirical odds ratio (OR) (31). Invasive case data of children aged 0 to 2 years from all sites in Gauteng Province, South Africa, submitted from January 2000 through December 2008 as part of national surveillance for IPD, were compared to data from the carriage study conducted in 2007 through 2008 that included children of similar ages, and from the same province. The OR and two-tailed Fisher's exact test were calculated using Epi Info version 6.04d. The OR was interpreted as previously described (4). A P value of <0.05 was considered statistically significant.
Molecular characterization.
Pulsed-field gel electrophoresis (PFGE) was performed on randomly selected serotype 3 isolates from Gauteng Province (n = 140) and eleven carriage isolates from Chris Hani Baragwanath Hospital, Soweto, Gauteng Province, South Africa, as described previously using SmaI restriction enzyme (Roche Diagnostics GmbH, Mannheim, Germany) (21). In addition, all serotype 3 isolates that were determined to be nonsusceptible to any of the antimicrobial agents tested were characterized by PFGE (n = 40). Box element PCR (BOX-PCR) was used to analyze those isolates that repeatedly did not yield PFGE patterns (n = 38) (16). UPGMA (unweighted-pair group method with arithmetic means) dendrograms were constructed using the GelCompar software version 4.1 (Applied Maths, Kortrijk, Belgium). PFGE and BOX-PCR clusters were defined as five or more isolates with ≥80% and ≥90% genetic relatedness on the dendrogram, respectively (11). An optimization value of 2% and position tolerance of 1% were used for the analysis. At least one isolate was arbitrarily selected for MLST (n = 30) from each of the PFGE and BOX-PCR clusters and from unrelated isolates. The procedure for MLST was performed as described previously (9).
RESULTS
National surveillance for invasive pneumococcal disease.
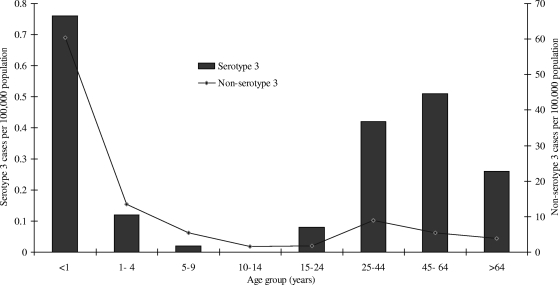
A total of 17,488 cases of IPD were reported from January 2000 through December 2005, and of these, 15,980 (91%) had viable isolates available for further testing. IPD cases were mainly reported from Gauteng province in South Africa (10,393/17,488 [59%]). Four percent (661/15,980) of the viable isolates were identified as serotype 3, the majority of which originated from Gauteng Province (467/661 [71%]). The proportion of serotype 3 disease differed by year (P < 0.001): 87/1,828 (4.8%), 116/2,120 (5.5%), 101/2,009 (5.5%), 108/2,894 (3.7%), 123/3,473 (3.5%), and 126/3,656 (3.4%) for the 6 years, respectively (Table 1). The majority of serotype 3 isolates were recovered from blood culture specimens (509/661 [77%]), but this was not significantly different from non-serotype 3 isolates (2,177/3,530 [61%]) (P = 0.2). Serotype 3 isolates were also isolated from CSF (113/661 [17%]), pleural fluid (32/661 [5%]), and other specimens from normally sterile sites (7/661 [1%]). The age of the individual was known in 575 of 661 (87%) serotype 3 cases, with the majority isolated from adults (489/575 [85%]). Age-specific incidence rates for 2005 were highest in children younger than 1 year of age, with a second peak in the 25- to 64-year-old age group (Fig. 1). In 2005, serotype 3 disease was more likely to be identified in adults (104/118 [88%]) than in children compared with non-serotype 3 disease (1,811/3,347 [54%]) (P < 0.001). A similar pattern was observed for the other years (data not shown).
TABLE 1.
Antibiograms of serotype 3 isolates causing invasive pneumococcal disease in South Africa from 2000 to 2005
| Antibiotic profilea or parameter | No. of serotype 3 isolates collected in the following yrb: |
|||||
|---|---|---|---|---|---|---|
| 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | |
| Antibiotic profile | ||||||
| Fully susceptible | 86 | 109 | 101 | 94 | 116 | 115 |
| M | 0 | 5 | 0 | 8 | 2 | 3 |
| R | 0 | 0 | 0 | 4 | 2 | 2 |
| T | 0 | 1 | 0 | 0 | 1 | 0 |
| EL | 1 | 0 | 0 | 0 | 0 | 0 |
| TM | 0 | 0 | 0 | 0 | 0 | 1 |
| PM | 0 | 0 | 0 | 1 | 1 | 1 |
| TEL | 0 | 0 | 0 | 0 | 0 | 1 |
| PRM | 0 | 1 | 0 | 0 | 0 | 0 |
| PTM | 0 | 0 | 0 | 1 | 0 | 2 |
| TELM | 0 | 0 | 0 | 0 | 1 | 0 |
| PTELM | 0 | 0 | 0 | 0 | 0 | 1 |
| Parameters | ||||||
| Total no. of serotype 3 cases | 87 | 116 | 101 | 108 | 123 | 126 |
| Total no. of non-serotype 3 cases | 1,741 | 2,004 | 1,908 | 2,786 | 3,350 | 3,530 |
| No. of cases with no isolate available | 175 | 110 | 127 | 333 | 311 | 451 |
| Total no. of cases reported | 2,003 | 2,230 | 2,137 | 3,227 | 3,784 | 4,107 |
Antibiotic abbreviations: P, penicillin G; R, rifampin; M, trimethoprim-sulfamethoxazole; T, tetracycline; E, erythromycin; L, clindamycin.
The values for the antibiotic profile are the number of isolates of serotype 3 isolates collected in the years 2000 to 2005. For the parameters, the values are the number of isolates or cases as specified.
FIG. 1.
Annual age-specific incidence rates of non-serotype 3 invasive pneumococcal disease (n = 3,347) and serotype 3 disease (n = 118) in South Africa in 2005.
For the period 2003 to 2005, HIV seroprevalence was known for 78 of the 154 (51%) serotype 3 cases presenting to enhanced surveillance sites, the majority of whom were HIV seropositive (72/78 [92%]). HIV coinfection in serotype 3 patients was not significantly different from that in non-serotype 3 cases (2,212/2,569 [86%]) (P = 0.1). The proportion of HIV-seropositive serotype 3 cases did not change over the 3-year period (∼92%) (P = 0.5). The case fatality ratio among serotype 3 cases remained stable for 2003 (18/47 [38%]), 2004 (18/48 [38%]), and 2005 (18/59 [31%]) (P = 0.4). Overall, the case fatality ratio did not differ significantly for serotype 3 cases (54/154 [35%]) compared with non-serotype 3 cases (1,190/4,211 [28%]) (P = 0.7).
Serotype 3 antimicrobial susceptibility.
Serotype 3 isolates were less likely to show nonsusceptibility to any of the antimicrobial agents tested (40/661 [6%]) than non-serotype 3 isolates (8,480/15,319 [55%]) (P < 0.001) (Table 1). Resistance to trimethoprim-sulfamethoxazole was most common (28/40 [70%]), and 6 of 40 (15%) isolates were MDR.
Invasive disease potential.
A total of 1,761 swabs were obtained from 251 children, and pneumococcal carriage was documented in 1,024 of 1,761 swabs (58%). Serotypes isolated from the same child more than once were excluded (n = 380). Fourteen serotype 3 strains were identified among the remaining 644 serotypes. Since only HIV-negative children were included in the carriage study, we compared these carriage data only to IPD data of children with HIV-seronegative results. For IPD cases reported for children aged 0 to 2 years during 2000 to 2008 from all sites in Gauteng province, South Africa (n = 3,735), 1,156 had both HIV and serotype results. The proportion of serotype 3 in HIV-uninfected children with serotype results was 1/309. Compared to other serotypes, serotype 3 had a low but statistically insignificant probability of causing invasive disease (OR of 0.15; 95% confidence interval [CI] of 0.01 to 1.06) (P = 0.05).
Molecular characterization.
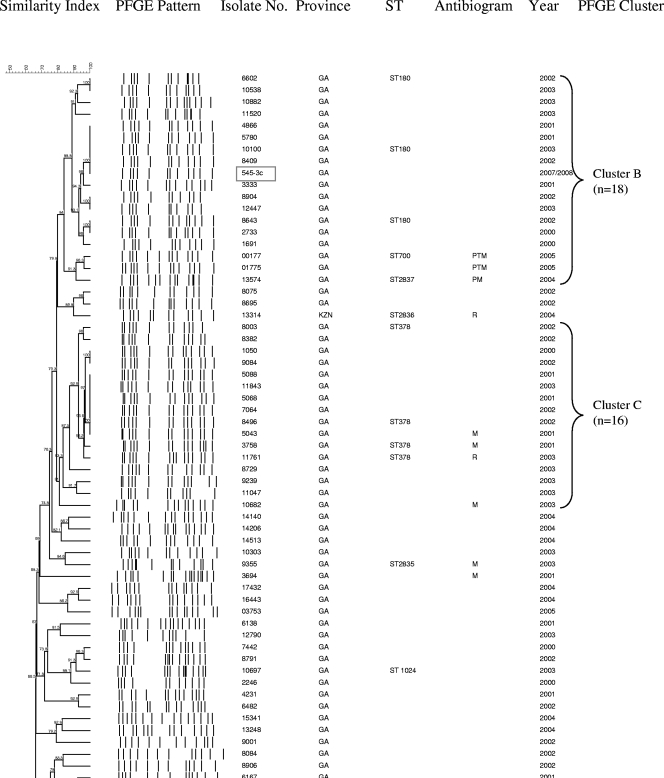
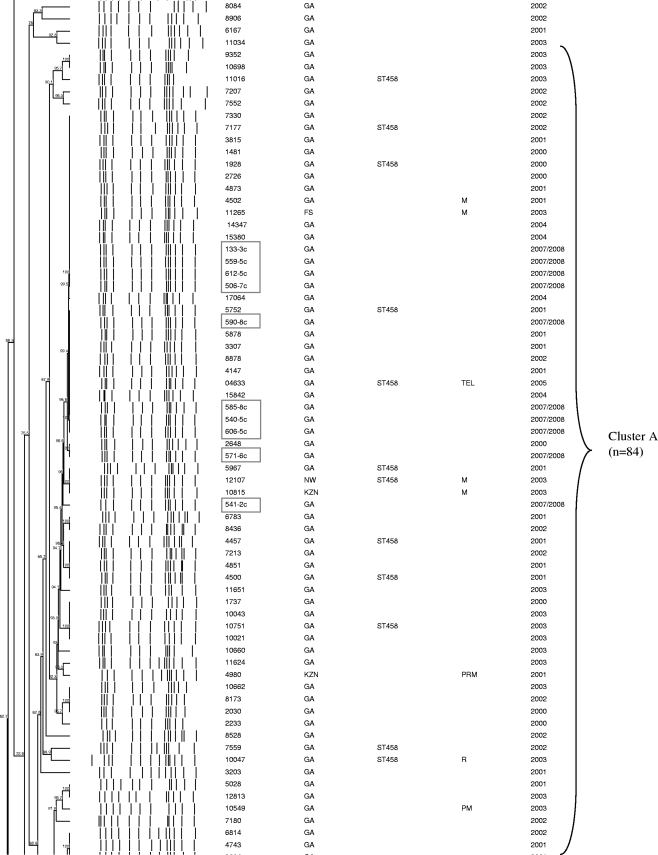
Of 467 serotype 3 isolates from Gauteng Province, South Africa, 140 (30%) were randomly selected for PFGE characterization. Two isolates were nonsusceptible to trimethoprim-sulfamethoxazole. PFGE results were available for 86% (120/140) of the serotype 3 invasive isolates from Gauteng Province (including 2 nonsusceptible isolates), 63% (25/40) of national antimicrobial nonsusceptible isolates, the MDR strain isolated in 1987, and 11 of the 14 carriage isolates. The isolates were grouped into 3 PFGE clusters and 36 unrelated isolates (Fig. 2). The largest PFGE cluster, cluster A, represented 54% (84/155) of all isolates, including 52% (62/120) of invasive isolates randomly selected from Gauteng Province and 91% (10/11) of carriage isolates from the same province. Fourteen isolates from cluster A, including five nonsusceptible isolates, were characterized as ST458. One isolate was novel ST1765, which shares four alleles with ST458. A large proportion of nonsusceptible isolates, including one MDR strain was also included in cluster A (14/25 [56%]). Comparison of data from nasopharyngeal carriage isolates of children from Gauteng province and data from IPD isolates of children from the same province and age group (0 to 2 years) revealed that carriage isolates were more likely to belong to cluster A than IPD isolates were (10/11 [91%] versus 8/18 [44%]; P = 0.019).
FIG. 2.
Schematic dendrogram showing pulsed-field gel electrophoresis (PFGE) clusters of 118 isolates randomly selected from serotype 3 isolates causing invasive pneumococcal disease in Gauteng Province, South Africa, in 2000 to 2005; 25 national antibiotic-resistant serotype 3 strains (2000 to 2005); a 1987 multidrug-resistant serotype 3 isolate (isolate C4023, which is located second from the bottom on the dendrogram); and 11 nasopharyngeal isolates from Gauteng Province in 2007 to 2008 (shown in boxes). Province abbreviations: GA, Gauteng; KZN, KwaZulu-Natal; NW, North-West; MP, Mpumalang. Antibiotic abbreviations used in the Antibiogram column: P, penicillin G; R, rifampin; M, trimethoprim-sulfamethoxazole; T, tetracycline; E, erythromycin: L, clindamycin.
Cluster B represented 12% (18/155), 17 of which were invasive isolates from Gauteng Province in South Africa and 1 was a carriage isolate. Three isolates of this cluster were identified as ST180. Cluster B also included three nonsusceptible isolates, two of which were MDR. Two of these nonsusceptible isolates were characterized as ST700 and ST2837, respectively, which differ from each other by one allele and share only two common alleles with ST180. Cluster C represented 10% (16/155) of the isolates, of which 4 were nonsusceptible. Four isolates were identified as ST378. The MDR strain from 1987 was unrelated to any other isolate and was identified as a new sequence type (ST2834).
Twenty (14%) of the 140 serotype 3 strains from Gauteng Province in South Africa and 15/40 (38%) nonsusceptible strains did not yield PFGE fingerprint patterns despite repeated attempts to perform characterization using this method. BOX-PCR showed that 19 isolates from Gauteng Province were unrelated, and only three were clustered with isolates identified as ST458 (data not shown). Compared to all of the Gauteng isolates with PFGE results, Gauteng isolates characterized by BOX-PCR were less likely to belong to cluster A (3/20 versus 62/120; P < 0.001]). Seven of 12 antimicrobial-resistant strains characterized by BOX-PCR were identical and were grouped together with three susceptible isolates identified as ST458.
DISCUSSION
The burden of pneumococcal disease is known to be highest in young children less than 5 years old and in the elderly (29). Similarly, our data showed the highest incidence to be in children younger than 1 year old. However, a second peak was observed in young adults, and serotype 3 strains were more likely than other serotypes to cause disease in this age group. This may be explained, in part, by the high incidence of HIV-infected individuals in this age group. Data collected over a 2-year period (1984 and 1985) from a South African hospital in Gauteng Province in South Africa showed that serotype 3 caused higher case fatality ratios than other serotypes (10). High mortality rates have also been observed in serotype 3 cases from other countries, including Spain and the United States (14). We found high case fatality ratios in this population dominated by HIV-infected individuals, with no difference between serotype 3 compared with non-serotype 3 cases.
Serotype 3 isolates from a number of countries, including Taiwan, Japan, and Switzerland, have been shown to have lower rates of antimicrobial resistance (17, 19, 26). The majority of South African isolates displayed a similar trend by showing susceptibility to most of the antimicrobial agents tested.
Three independent studies that used odds ratios to estimate the invasiveness of serotypes and clones found serotype 3 prevalence to be higher in carriage isolates in children than in invasive isolates, but not significantly so (4, 12, 31). Our data showed a similar trend. ST458, as inferred by PFGE clusters, was predominant among both invasive and carriage isolates from South African children.
Serotype 3 has been shown to be clonal, with ST180 predominating among serotype 3 isolates from many countries, including the United States, Scotland, and Poland (2, 6, 28). In contrast, three clones were found among serotype 3 isolates from South Africa, and ST180 was not the most common sequence type. ST458 was identified in selected isolates from the predominant PFGE cluster, which also included MDR strains and 91% of carriage isolates. Similarly, a study in Ghana showed that all seven serotype 3 strains isolated from meningitis patients from two different districts in 2001 were ST458 (22). This clone was also identified among four serotype 3 strains causing meningitis from 1998 to 2003 in Egypt (33) (Pneumococcal Molecular Epidemiology Network website [http://www.sph.emory.edu/PMEN/pmen_ww_spread_clones.html]). Recently, Porat et al. (27), reported 21% (11/52) of serotype 3 isolates from Israeli pediatric patients with noninvasive infections during 2003 to 2005 to be ST458. The same study showed that carriage and noninvasive serotype 3 isolates from Costa Rica and Lithuania were unrelated to this clone. These findings suggest that isolates belonging to ST458 may be more prevalent among serotype 3 isolates from African and Middle Eastern countries than from other parts of the world.
The documentation of strain identity within serotypes is important to monitor capsular switching, particularly in the era of conjugate vaccines. The present study provides genetic characterization of serotype 3 isolates circulating before the routine use of PCV-7. None of the MLST clones identified have previously been found to be associated with other serotypes (pneumococcal MLST database [http://spneumoniae.mlst.net/sql/burstspadvanced.asp]), indicating a lack of evidence for capsular switching among these strains.
Some isolates did not yield PFGE patterns after at least two attempts to perform characterization and were therefore characterized by BOX-PCR to try and establish the reason for the failed PFGE. This was also verified by performing BOX-PCR on some isolates for which both a sequence type and PFGE pattern was available. For isolates with no available PFGE pattern, BOX-PCR showed diversity among the isolates from Gauteng Province in South Africa; however, the nonsusceptible isolates were clonal, and cluster A was predominant. The reason for this observation and the failure to generate PFGE patterns for some isolates is unclear, but it may be attributed to variations in the amount of capsule expressed in these isolates. Serotype 3 capsules exhibit a mucoid phenotype on blood agar and are occasionally difficult to lyse in vitro.
Carrico and colleagues analyzed the relationship between PFGE clusters and MLST when using an 80% similarity cutoff by calculating the Wallace coefficient, which measures the probability that two strains sharing a genotype by one method also share the same genotype by another method (5). In their study, serotype 3 had the lowest Wallace coefficient value, indicating an absence of any correspondence. Overall, we observed good correlation between PFGE clusters and sequence types; however, this may account for the discrepancy observed between PFGE and MLST results for 3 isolates, where unrelated sequence types were grouped together by PFGE. PFGE profiles for these strains differed by three or less bands, indicating relatedness according to the Tenover criteria (32).
The higher proportion of serotype 3 isolates related to ST458, compared to the global clone ST180 predominating among serotype 3 isolates from many Northern Hemisphere countries, highlights the geographic differences in serotype distribution and emphasizes the need for surveillance of isolates from South Africa and other African countries.
Acknowledgments
We are grateful to the Respiratory and Meningeal Pathogens Research Unit (RMPRU) laboratory staff for their technical input, all clinical and laboratory staff throughout South Africa for submitting case reports and isolates for national surveillance, and all patients whose isolates were used in the study. We acknowledge the use of the pneumococcal MLST database, which is located at Imperial College London.
This work was funded by the Medical Research Council of South Africa, the National Institute for Communicable Disease (NICD) of the National Health Laboratory Service (NHLS), and the University of Witwatersrand and was supported in part by funds from the United States Agency for International Development's Antimicrobial Resistance Initiative, transferred via a cooperative agreement (number U60/CCU022088) from the Centers for Disease Control and Prevention (CDC), Atlanta, Georgia. From 2006 onwards, this work was supported in part by the HHS Centers for Disease Control and Prevention (CDC) and National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention (NCHHSTP) Global AIDS Program (GAP) cooperative agreement U62/PSO022901. The pneumococcal MLST database is funded by the Wellcome Trust.
The contents are solely the responsibility of the authors and do not necessarily represent the official views of the CDC.
The members of the Group for Enteric Respiratory, and Meningeal Disease Surveillance in South Africa (GERMS-SA) follow: Sandeep Vasaikar at the University of Transkei, Mthatha, Eastern Cape province; Nolan Janse van Rensberg, André Möller, Anne-Marie Pretorius, and Peter Smith at the University of the Free State, Bloemfontein, Free State province; Khatija Ahmed, Anwar Hoosen, Ruth Lekalakala, Donald Ngwira, and Pyu Pyu Sein at the University of Limpopo-Medunsa Campus, Garankuwa, Gauteng province; Heather Crewe-Brown, Charles Feldman, Alan Karstaedt, Olga Perovic, and Jeannette Wadula at the University of the Witwatersrand, Johannesburg, Gauteng; Mike Dove, Kathy Lindeque, Linda Meyer, and Gerhard Weldhagen at the University of Pretoria, Pretoria, Gauteng; Prathna Bhola, Prashini Moodley, Sharona Seetharam, Sindiswe Sithole, Wim Sturm, and Trusha Vanmali at the University of KwaZulu Natal, Durban, KwaZulu Natal; Ken Hamese at Polokwane/Mankweng Hospital Complex, Polokwane, Limpopo; Keith Bauer, Greta Hoyland, Jacob Lebudi, and Charles Mutanda at the National Health Laboratory Service, Mpumalanga; Rena Hoffmann, Lynne Liebowitz, and Elizabeth Wasserman at the University of Stellenbosch, Stellenbosch, Western Cape; Denise Roditi, John Simpson, and Andrew Whitelaw at the University of Cape Town, Cape Town, Western Cape; Adrian Brink at AMPATH Laboratories, Johannesburg, Gauteng; Claire Heney at Lancet Laboratories, Johannesburg, Gauteng; Marthinus Senekal at PathCare Laboratories, Cape Town, Western Cape; and Cheryl Cohen, Mireille Cheyip, Linda de Gouveia, John Frean, Nelesh Govender, Karen Keddy, Kerrigan McCarthy, Susan Meiring, Elizabeth Prentice, Vanessa Quan, Koshika Soma, and Anne von Gottberg at the National Institute for Communicable Diseases, Johannesburg, Gauteng.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Austrian, R. 1976. The quellung reaction, a neglected microbiologic technique. Mt. Sinai J. Med. 43:699-709. [PubMed] [Google Scholar]
- 2.Beall, B., M. C. McEllistrem, R. E. Gertz, Jr., S. Wedel, D. J. Boxrud, A. L. Gonzalez, M. J. Medina, R. Pai, T. A. Thompson, L. H. Harrison, L. McGee, and C. G. Whitney. 2006. Pre- and postvaccination clonal compositions of invasive pneumococcal serotypes for isolates collected in the United States in 1999, 2001, and 2002. J. Clin. Microbiol. 44:999-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, J. M., K. Ampofo, K. Korgenski, J. Daly, A. T. Pavia, E. O. Mason, and C. L. Byington. 2008. Pneumococcal necrotizing pneumonia in Utah: does serotype matter? Clin. Infect. Dis. 46:1346-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brueggemann, A. B., D. T. Griffiths, E. Meats, T. Peto, D. W. Crook, and B. G. Spratt. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424-1432. [DOI] [PubMed] [Google Scholar]
- 5.Carrico, J., F. Pinto, H. Zemlickova, V. Jakubu, S. Nunes, N. Frazao, R. Sa-Leao, H. de Lencastre, K. Kristinsson, T. Gunnarsdottir, A. Brueggemann, S. Dhillon, B. Spratt, S. Aguilar, I. Serrano, J. Melo-Cristino, and M. Ramirez. 2008. Quantitative analysis of congruence of Streptococcus pneumoniae typing methods: relations between serotype, multi-locus sequence typing and pulsed field gel electrophoresis type assignments, poster P2-027. 6th Int. Symp. Pneum. Pneum. Dis. (ISPPD-6). Reykjavik, Iceland, 8 to 12 June 2008.
- 6.Clarke, S. C., K. J. Scott, and S. M. McChlery. 2004. Serotypes and sequence types of pneumococci causing invasive disease in Scotland prior to the introduction of pneumococcal conjugate polysaccharide vaccines. J. Clin. Microbiol. 42:4449-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute/NCCLS. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement, vol. 28, p. 66, 67-126, 127. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 8.Day, C., and A. Gray. 2006. Health and related indicators, p. 369-506. In P. Ijumba and A. Padarath (ed.), South African health review 2006. Health Systems Trust, Durban, South Africa. http://www.hst.org.za/publications/item.php?item_id=697.
- 9.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 10.Feldman, C., H. Sacho, H. Levy, J. M. Kallenbach, and H. J. Koornhof. 1988. Pneumococcal bacteraemia in adults in a low socio-economic urban population. Q. J. Med. 69:961-971. [PubMed] [Google Scholar]
- 11.Gertz, R. E., Jr., M. C. McEllistrem, D. J. Boxrud, Z. Li, V. Sakota, T. A. Thompson, R. R. Facklam, J. M. Besser, L. H. Harrison, C. G. Whitney, and B. Beall. 2003. Clonal distribution of invasive pneumococcal isolates from children and selected adults in the United States prior to 7-valent conjugate vaccine introduction. J. Clin. Microbiol. 41:4194-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanage, W. P., T. H. Kaijalainen, R. K. Syrjanen, K. Auranen, M. Leinonen, P. H. Makela, and B. G. Spratt. 2005. Invasiveness of serotypes and clones of Streptococcus pneumoniae among children in Finland. Infect. Immun. 73:431-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hausdorff, W. P., J. Bryant, P. R. Paradiso, and G. R. Siber. 2000. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part I. Clin. Infect. Dis. 30:100-121. [DOI] [PubMed] [Google Scholar]
- 14.Henriques, B., M. Kalin, A. Ortqvist, L. B. Olsson, M. Almela, T. J. Marrie, M. A. Mufson, A. Torres, M. A. Woodhead, S. B. Svenson, and G. Kallenius. 2000. Molecular epidemiology of Streptococcus pneumoniae causing invasive disease in 5 countries. J. Infect. Dis. 182:833-839. [DOI] [PubMed] [Google Scholar]
- 15.Huebner, R. E., K. P. Klugman, U. Matai, R. Eggers, and G. Hussey. 1999. Laboratory surveillance for Haemophilus influenzae type b, meningococcal, and pneumococcal disease. Haemophilus Surveillance Working Group. S. Afr. Med. J. 89:924-925. [PubMed] [Google Scholar]
- 16.Johnson, C. N., J. W. Benjamin, Jr., S. A. Moser, S. K. Hollingshead, X. Zheng, M. J. Crain, M. H. Nahm, and K. B. Waites. 2003. Genetic relatedness of levofloxacin-nonsusceptible Streptococcus pneumoniae isolates from North America. J. Clin. Microbiol. 41:2458-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasahara, K., K. Maeda, K. Mikasa, K. Uno, K. Takahashi, M. Konishi, E. Yoshimoto, K. Murakawa, E. Kita, and H. Kimura. 2005. Clonal dissemination of macrolide-resistant and penicillin-susceptible serotype 3 and penicillin-resistant Taiwan 19F-14 and 23F-15 Streptococcus pneumoniae isolates in Japan: a pilot surveillance study. J. Clin. Microbiol. 43:1640-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kronenberg, A., P. Zucs, S. Droz, and K. Muhlemann. 2006. Distribution and invasiveness of Streptococcus pneumoniae serotypes in Switzerland, a country with low antibiotic selection pressure, from 2001 to 2004. J. Clin. Microbiol. 44:2032-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauderdale, T. L., M. M. Wagener, H. M. Lin, I. F. Huang, W. Y. Lee, K. S. Hseih, J. F. Lai, and C. C. Chiou. 2006. Serotype and antimicrobial resistance patterns of Streptococcus pneumoniae isolated from Taiwanese children: comparison of nasopharyngeal and clinical isolates. Diagn. Microbiol. Infect. Dis. 56:421-426. [DOI] [PubMed] [Google Scholar]
- 20.Lawrenson, J. B., K. P. Klugman, J. I. Eidelman, A. Wasas, S. D. Miller, and J. Lipman. 1988. Fatal infection caused by a multiply resistant type 3 pneumococcus. J. Clin. Microbiol. 26:1590-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lefevre, J. C., G. Faucon, A. M. Sicard, and A. M. Gasc. 1993. DNA fingerprinting of Streptococcus pneumoniae strains by pulsed-field gel electrophoresis. J. Clin. Microbiol. 31:2724-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leimkugel, J., F. A. Adams, S. Gagneux, V. Pfluger, C. Flierl, E. Awine, M. Naegeli, J. P. Dangy, T. Smith, A. Hodgson, and G. Pluschke. 2005. An outbreak of serotype 1 Streptococcus pneumoniae meningitis in northern Ghana with features that are characteristic of Neisseria meningitidis meningitis epidemics. J. Infect. Dis. 192:192-199. [DOI] [PubMed] [Google Scholar]
- 23.Lexau, C. A., R. Lynfield, R. Danila, T. Pilishvili, R. Facklam, M. M. Farley, L. H. Harrison, W. Schaffner, A. Reingold, N. M. Bennett, J. Hadler, P. R. Cieslak, and C. G. Whitney. 2005. Changing epidemiology of invasive pneumococcal disease among older adults in the era of pediatric pneumococcal conjugate vaccine. JAMA 294:2043-2051. [DOI] [PubMed] [Google Scholar]
- 24.McEllistrem, M. C., J. M. Adams, K. Patel, A. B. Mendelsohn, S. L. Kaplan, J. S. Bradley, G. E. Schutze, K. S. Kim, E. O. Mason, and E. R. Wald. 2005. Acute otitis media due to penicillin-nonsusceptible Streptococcus pneumoniae before and after the introduction of the pneumococcal conjugate vaccine. Clin. Infect. Dis. 40:1738-1744. [DOI] [PubMed] [Google Scholar]
- 25.Morrison, K. E., D. Lake, J. Crook, G. M. Carlone, E. Ades, R. Facklam, and J. S. Sampson. 2000. Confirmation of psaA in all 90 serotypes of Streptococcus pneumoniae by PCR and potential of this assay for identification and diagnosis. J. Clin. Microbiol. 38:434-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muhlemann, K., H. C. Matter, M. G. Tauber, and T. Bodmer. 2003. Nationwide surveillance of nasopharyngeal Streptococcus pneumoniae isolates from children with respiratory infection, Switzerland, 1998-1999. J. Infect. Dis. 187:589-596. [DOI] [PubMed] [Google Scholar]
- 27.Porat, N., C. Soley, M. M. Marengolciene, D. Greenberg, N. Givon-Lavi, R. Trefler, A. Arguedas, and R. Dagan. 2008. An international serotype 3 clone causing pediatric noninvasive infections in Israel, Costa Rica, and Lithuania. Pediatr. Infect. Dis. J. 27:709-712. [DOI] [PubMed] [Google Scholar]
- 28.Sadowy, E., A. Skoczynska, J. Fiett, M. Gniadkowski, and W. Hryniewicz. 2006. Multilocus sequence types, serotypes, and variants of the surface antigen PspA in Streptococcus pneumoniae isolates from meningitis patients in Poland. Clin. Vaccine Immunol. 13:139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott, J. A., A. J. Hall, R. Dagan, J. M. Dixon, S. J. Eykyn, A. Fenoll, M. Hortal, L. P. Jette, J. H. Jorgensen, F. Lamothe, C. Latorre, J. T. Macfarlane, D. M. Shlaes, L. E. Smart, and A. Taunay. 1996. Serogroup-specific epidemiology of Streptococcus pneumoniae: associations with age, sex, and geography in 7,000 episodes of invasive disease. Clin. Infect. Dis. 22:973-981. [DOI] [PubMed] [Google Scholar]
- 30.Singh, E., C. Cohen, N. Govender, and S. Meiring. 2008. A description of HIV testing strategies at 21 laboratories in South Africa. Commun. Dis. Surveill. Bull. 6:16-17. [Google Scholar]
- 31.Smith, T., D. Lehmann, J. Montgomery, M. Gratten, I. D. Riley, and M. P. Alpers. 1993. Acquisition and invasiveness of different serotypes of Streptococcus pneumoniae in young children. Epidemiol. Infect. 111:27-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wasfy, M. O., G. Pimentel, M. Abdel-Maksoud, K. L. Russell, C. P. Barrozo, J. D. Klena, K. Earhart, and R. Hajjeh. 2005. Antimicrobial susceptibility and serotype distribution of Streptococcus pneumoniae causing meningitis in Egypt, 1998-2003. J. Antimicrob. Chemother. 55:958-964. [DOI] [PubMed] [Google Scholar]