Abstract
HIV-Selectest is a serodiagnostic enzyme immunoassay (EIA), containing p6 and gp41 peptides, designed to differentiate between vaccine-induced antibodies and true infections. A rapid test version of the HIV-Selectest was developed. Both assays detected HIV antibodies in men and women within 2 to 4 weeks of infection, with sensitivity similar to third-generation EIAs.
Most of the current HIV vaccine candidates contain multiple viral components. As a result, many vaccine recipients react positively in commercial diagnostic assays that detect HIV antibodies, including enzyme immunoassays (EIAs), Western blots (WB), and rapid tests. We have identified conserved sequences in Env gp41 and Gag p6 which are not included in most HIV vaccine candidates. A new HIV EIA termed HIV-Selectest was developed, which distinguishes between vaccine-induced antibodies and seroconversion due to true HIV infections (4, 5).
Before this assay can be implemented as part of the HIV detection algorithm during HIV vaccine trials, it was important to compare its sensitivity to those of currently available third-generation EIAs and rapid tests. In addition, attention must be given to detection of infections in women versus those in men, since women may experience HIV/AIDS differently from men (8, 10). We have recently developed a rapid test version of the HIV-Selectest in order to facilitate point-of-care testing during vaccine trials.
In the current studies, we evaluated serial samples obtained from the following multiple cohorts of men and women from the United States and Africa: plasma donors who acquired HIV-1 in the United States (Center for HIV/AIDS Vaccine Immunology [CHAVI]; clade B, predominantly males) (9); high-risk subjects identified with having acute HIV-1 infections at U.S. sites participating in the Acute Infection and Early Disease Research Program (AIEDRP; clade B, predominantly males); acutely infected subjects indentified in Africa by the Center for the AIDS Programme of Research in South Africa (CAPRISA; clade C infections, mainly women) (2); subjects participating in the Zambia-Emory HIV Research Project (ZEHRP; clade C discordant couple transmission pairs) (3, 6); early (3 to 6 months) postinfection time points in men enrolled in the U.S. Multicenter AIDS Cohort Study (MACS) (7); and women participating in the Women's Interagency HIV Study (WIHS) (11).
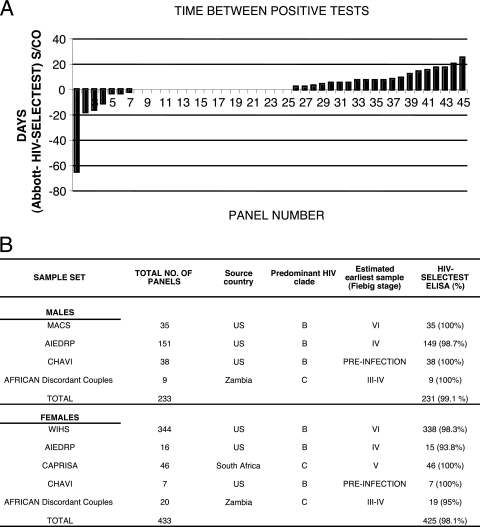
In collaboration with CHAVI, 87 plasma donor seroconversion panels were evaluated. In these panels, blood draws were very frequent (every 3 to 5 days), the date of initial PCR-confirmed infection was available, and viral loads were provided. The HIV-Selectest was performed as described in detail in references 4 and 5. Of the 87 CHAVI panels tested, only 45 reached seroconversion (38 males and 7 females) and could be used for comparison of antibody assay sensitivity. As seen in Fig. 1A, concordant results from the Abbott third-generation test and the HIV-Selectest EIA were seen in 18/45 panels (zero difference in detection day for panels 8 to 25). The Abbott EIA yielded positive results prior to the HIV-Selectest in 20/45 panels (positive columns for panels 26 to 45). Surprisingly, the HIV-Selectest EIA scored positive prior to the Abbott test in 7/45 panels (negative columns for panels 1 to 7). On average, the HIV-Selectest EIA detected anti-HIV antibodies 1.6 days (median = 0) after the commercial third-generation Abbott EIA.
FIG. 1.
Reactivity of the HIV-Selectest EIA, with samples obtained from acutely HIV-infected males and females. (A) Comparison of HIV-Selectest EIA performance with that of the Abbott kit during acute viremia (CHAVI panels). HIV-Selectest was performed as described in references 4 and 5. The difference between the first day of positive reactivity (signal-to-cutoff [S/CO] ratio > 1) in the Abbott test (third generation) and that in the HIV-Selectest EIA was plotted for individual panels. The bars under the horizontal line (zero difference) represent panels in which the HIV-Selectest EIA scored positive before the Abbott test, while bars above this line represent panels in which the Abbott test scored positive ahead of the HIV-Selectest EIA. (B) Summary of HIV-Selectest (EIA) reactivity with acutely HIV-infected males and females. HIV-Selectest shows high sensitivity for detection of HIV-1-infected male and female panels. HIV-Selectest reactivity is considered positive if an S/CO value of >1 is obtained in either the p6 or the gp41 peptide enzyme-linked immunosorbent assay (ELISA). Fiebig staging has been previously described (1). No statistical significant difference between males and females was observed (P value = 0.68).
It was important to expand our survey to include community clinic settings. Additional panels of confirmed early HIV infections in men and women with predominantly clade B infections were evaluated through collaboration with the AIEDRP, MACS, and WIHS. The HIV-Selectest EIA sensitivity ranged from 98.7 to 100% and from 93.8 to 98.3% in men and women, respectively, based on detection of the first seropositive specimen defined by reference assays in these studies (Fig. 1B).
Detection of clade C infections was evaluated in the CAPRISA acute infection and ZEHRP discordant couple panels from South Africa and Zambia, respectively. The CAPRISA project was established in South Africa in order to capture early heterosexual clade C infections and to establish the rate of viremia and disease progression. Most of the CAPRISA participants are women. HIV-1 diagnosis was based on several rapid tests and EIAs and supplemented by PCR-based viral load measurements, and acutely infected subjects were enrolled into a longitudinal follow-up protocol with weekly specimen collection. Samples tested with the HIV-Selectest EIA were the first specimens available after confirmed seroconversion by the serodiagnostic tests used in the CAPRISA study (i.e., within 1 to 4 weeks of estimated infection date). Overall, the HIV-Selectest scored positive no later than 1 sample after seroconversion in 46 panels.
Performance of the HIV-Selectest in capturing clade C infections was also evaluated in the ZEHRP discordant couple study. Early seroconversions from 9 female-to-male and 20 male-to-female transmissions were tested. The HIV-Selectest EIA scored positive with the first HIV infection-confirmed bleed from 9/9 males and 19/20 females in this panel (Fig. 1B).
Together, the data obtained in the current studies demonstrated that in spite of the fact that HIV-Selectest EIA is composed of a limited number of peptides from only two HIV-1 proteins (gp41 and p6), its sensitivity to detect early anti-HIV-1 IgG antibodies does not differ significantly from commercially available rapid tests and third-generation EIAs used in the HIV screening algorithms in many countries. Overall assay sensitivity was found to be 99.1% and 98.1% in early infection samples obtained from males and females, respectively (Fig. 1B). A two-tailed, unequal sample size, equal variance t test was performed on the combined HIV-Selectest reactivity for the HIV-1-positive males and females. Detection rates were not significantly different between the HIV-infected men and women, irrespective of the infecting HIV-1 viral strains (P value = 0.68).
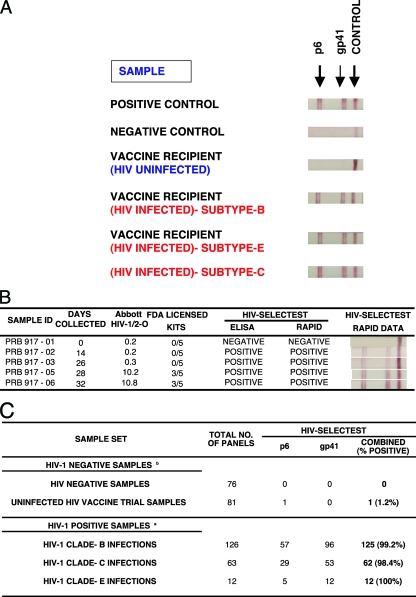
Point-of-care rapid testing is potentially of great value to resource-poor countries. During vaccine trials, a rapid test could help to capture breakthrough infections, which often occur between the 3-month scheduled clinic visits. To that end, we have adapted the HIV-Selectest to a lateral flow test based on immunochromatographic strip (ICS) technology. As seen in Fig. 2A, both the p6 and gp41 peptides were reactive with positive control plasma from SeraCare BioServices (Gaithersburg, MD) but not with negative controls obtained from uninfected individuals. Importantly, HIV vaccine recipients (Vaccine Research Center trials VRC-009 and VRC-010) that were not infected but gave false-positive results in several commercial HIV rapid tests scored negative in the HIV-Selectest rapid test, as previously demonstrated with the EIA version (4). On the other hand, breakthrough infections in HIV vaccine trial participants (VaxGen phase III trials in the United States [clade B] and Thailand [clade E]) gave positive reactivity in the rapid test. Clade C infections (from CAPRISA) also scored positive in the HIV-Selectest rapid test (Fig. 2A).
FIG. 2.
HIV-Selectest rapid test for detection of HIV-1 infections. (A) One positive control protein (goat anti-human IgG; Jackson Immunoresearch) and two sets of peptides from HIV-1 gp41 and p6 were striped onto 5-μm nitrocellulose membranes (Whatman) at 1 mg/ml per peptide. One-hundred-fold-diluted serum/plasma samples were allowed to migrate gradually through the membrane and react to HIV peptides, followed by detection with protein A-coated gold conjugate. Uninfected vaccine recipients were obtained from VRC-009 and VRC-010 (DNA/HIV prime, adenovirus type 5 [Ad5]/HIV boost). HIV-infected vaccine recipients were obtained from the VaxGen phase III trials in the United States (clade B) and Thailand (clade E). Clade C samples were obtained from the CAPRISA cohort. (B) Performance of the HIV-Selectest (EIA and rapid test) using the HIV-1 seroconversion panel (PRB 917; SeraCare BioServices, Inc.). Information on days of collection and results obtained with the Abbott HIV-1/2-O EIA and 5 FDA-licensed kits (including Abbott HIV-1, Abbott HIV-1/2, GenSys HIV-1, GenSys HIV-1/2, and OraTek HIV) were provided by SeraCare BioServices. (C) Summary of HIV-Selectest rapid test results with uninfected HIV samples (including HIV vaccine trial samples) and samples obtained from acutely HIV-infected individuals. HIV-1 clade B samples were obtained from SeraCare BioServices and NHLBI, clade C samples were obtained from the CAPRISA cohort, and clade E samples were obtained from the VaxGen vaccine trial VAX-003. HIV-negative samples were obtained from unexposed blood donors and SeraCare BioServices. The HIV vaccine trial samples were obtained from uninfected vaccine recipients from the HIV vaccine trials VRC-008, VRC-009, and VRC-010.
In order to compare the sensitivity of the HIV-Selectest rapid test and EIA with the performance of a third-generation EIA, we evaluated seroconversion panels from SeraCare BioServices, Inc. In the case of panel PRB 917 (Fig. 2B), the Abbott HIV-1/2-O test was reactive on day 28, while the HIV-Selectest EIA and rapid assay (both p6 and gp41) scored positive as early as day 14, with the bands becoming stronger on subsequent days (Fig. 2B).
In several proof-of-concept studies conducted with the HIV-Selectest rapid test, assay specificity is around 99.4% based on 76 seronegative healthy individuals and 81 samples obtained from HIV-uninfected vaccine trial participants (Fig. 2C, top). Assay sensitivity is currently in the range of 99.2 to 100% for confirmed infections with HIV-1 clade B, C, and E viruses (Fig. 2C, bottom), but individual samples may give different patterns of intensity with the peptide bands, in agreement with what was found in the EIA version of the HIV-Selectest (4).
The addition of the HIV-Selectest (EIA and rapid test) to the detection algorithm during ongoing vaccine trials could facilitate early diagnosis of breakthrough infections and reduce the number of questionable HIV-seropositive samples that need nucleic acid testing.
Acknowledgments
We do not have any conflicts of interest. This project was supported in part by NHLBI IAG (2005 to 2006), Center for HIV/AIDS Vaccine Immunology (CHAVI) grant AI-067854, by NIAID grants R37-AI51231 and R01-AI64060, by NIH grant AI51794, by the South African National Research Foundation (grant 67385), by IAVI, and by a grant from the Office of Women's Health, FDA (2006 to 2007). This study is partially funded by the FDA Office of Women's Health.
Vaccine samples were provided by Barney Graham, Vaccine Research Center, National Institutes of Allergy and Infectious Diseases, NIH, Bethesda, MD.
The Multicenter AIDS Cohort Study (MACS) includes the following: in Baltimore, MD, at The Johns Hopkins University Bloomberg School of Public Health, Joseph B. Margolick (Principal Investigator), Barbara Crain, Adrian Dobs, Homayoon Farzadegan, Joel Gallant, Lisette Johnson-Hill, Shenghan Lai, Ned Sacktor, Ola Selnes, James Shepard, and Chloe Thio; in Chicago, IL, at Howard Brown Health Center, Feinberg School of Medicine, Northwestern University, and Cook County Bureau of Health Services, John P. Phair (Principal Investigator), Steven M. Wolinsky (Co-Principal Investigator), Sheila Badri, Bruce Cohen, Craig Conover, Maurice O'Gorman, David Ostrow, Frank Palella, and Daina Variakojis; in Los Angeles, CA, at University of California, UCLA Schools of Public Health and Medicine, Roger Detels (Principal Investigator), Otoniel Martínez-Maza (Co-Principal Investigator), Aaron Aronow, Robert Bolan, Elizabeth Breen, Anthony Butch, Rita Effros, John Fahey, Beth Jamieson, Eric N. Miller, John Oishi, Harry Vinters, Barbara R. Visscher, Dorothy Wiley, Mallory Witt, Otto Yang, Stephen Young, and Zuo Feng Zhang; in Pittsburgh, PA, at University of Pittsburgh, Graduate School of Public Health, Charles R. Rinaldo (Principal Investigator), Lawrence A. Kingsley (Co-Principal Investigator), James T. Becker, Ross D. Cranston, Jeremy J. Martinson, John W. Mellors, Anthony J. Silvestre, and Ronald D. Stall; at the Data Coordinating Center, The Johns Hopkins University Bloomberg School of Public Health, Lisa P. Jacobson (Principal Investigator), Alvaro Muñoz (Co-Principal Investigator), Alison Abraham, Keri Althoff, Christopher Cox, Gypsyanber D'Souza, Stephen J. Gange, Elizabeth Golub, Janet Schollenberger, Eric C. Seaberg, and Sol Su; at the NIH, National Institute of Allergy and Infectious Diseases, Robin E. Huebner; and at the National Cancer Institute, Geraldina Dominguez. Funding includes grants UO1-AI-35042, 5-MO1-RR-00052 (GCRC), UO1-AI-35043, UO1-AI-35039, UO1-AI-35040, UO1-AI-35041, and U01 AI42532 (JM). The MACS website is located at http://www.statepi.jhsph.edu/macs/macs.html.
Data in this article were collected by the Women's Interagency HIV Study (WIHS) Collaborative Study Group, with centers (Principal Investigators) at the New York City/Bronx Consortium (Kathryn Anastos); Brooklyn, NY (Howard Minkoff); Washington, DC, Metropolitan Consortium (Mary Young); The Connie Wofsy Study Consortium of Northern California (Ruth Greenblatt); Los Angeles County/Southern California Consortium (Alexandra Levine); Chicago Consortium (Mardge Cohen); and Data Coordinating Center (Stephen Gange). The WIHS is funded by the National Institute of Allergy and Infectious Diseases (grants UO1-AI-35004, UO1-AI-31834, UO1-AI-34994, UO1-AI-34989, UO1-AI-34993, and UO1-AI-42590) and by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (grant UO1-HD-32632). The study is cofunded by the National Cancer Institute, the National Institute on Drug Abuse, and the National Institute on Deafness and Other Communication Disorders. Funding is also provided by the National Center for Research Resources (UCSF-CTSI grant UL1 RR024131).
The contents of this publication are solely our responsibility and do not necessarily represent the official views of the National Institutes of Health.
Footnotes
Published ahead of print on 11 November 2009.
REFERENCES
- 1.Fiebig, E. W., D. J. Wright, B. D. Rawal, P. E. Garrett, R. T. Schumacher, L. Peddada, C. Heldebrant, R. Smith, A. Conrad, S. H. Kleinman, and M. P. Busch. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 17:1871-1879. [DOI] [PubMed] [Google Scholar]
- 2.Gray, E. S., P. L. Moore, I. A. Choge, J. M. Decker, F. Bibollet-Ruche, H. Li, N. Leseka, F. Treurnicht, K. Mlisana, G. M. Shaw, S. S. Karim, C. Williamson, and L. Morris. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 81:6187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haaland, R. E., P. A. Hawkins, J. Salazar-Gonzalez, A. Johnson, A. Tichacek, E. Karita, O. Manigart, J. Mulenga, B. F. Keele, G. M. Shaw, B. H. Hahn, S. A. Allen, C. A. Derdeyn, and E. Hunter. 2009. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 5:e1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khurana, S., J. Needham, B. Mathieson, I. R. Rodriguez-Chavez, A. T. Catanzaro, R. T. Bailer, J. Kim, V. Polonis, D. A. Cooper, J. Guerin, M. L. Peterson, M. Gurwith, N. Nguyen, B. S. Graham, and H. Golding. 2006. Human immunodeficiency virus (HIV) vaccine trials: a novel assay for differential diagnosis of HIV infections in the face of vaccine-generated antibodies. J. Virol. 80:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khurana, S., J. Needham, S. Park, B. Mathieson, M. P. Busch, G. Nemo, P. Nyambi, S. Zolla-Pazner, S. Laal, J. Mulenga, E. Chomba, E. Hunter, S. Allen, J. McIntyre, I. Hewlett, S. Lee, S. Tang, E. Cowan, C. Beyrer, M. Altfeld, X. G. Yu, A. Tounkara, O. Koita, A. Kamali, N. Nguyen, B. S. Graham, D. Todd, P. Mugenyi, O. Anzala, E. Sanders, N. Ketter, P. Fast, and H. Golding. 2006. Novel approach for differential diagnosis of HIV infections in the face of vaccine-generated antibodies: utility for detection of diverse HIV-1 subtypes. J. Acquir. Immune Defic. Syndr. 43:304-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKenna, S. L., G. K. Muyinda, D. Roth, M. Mwali, N. Ng'andu, A. Myrick, C. Luo, F. H. Priddy, V. M. Hall, A. A. von Lieven, J. R. Sabatino, K. Mark, and S. A. Allen. 1997. Rapid HIV testing and counseling for voluntary testing centers in Africa. AIDS 11(Suppl. 1):S103-S110. [PubMed] [Google Scholar]
- 7.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 8.Napravnik, S., C. Poole, J. C. Thomas, and J. J. Eron, Jr. 2002. Gender difference in HIV RNA levels: a meta-analysis of published studies. J. Acquir. Immune Defic. Syndr. 31:11-19. [DOI] [PubMed] [Google Scholar]
- 9.Tomaras, G. D., N. L. Yates, P. Liu, L. Qin, G. G. Fouda, L. L. Chavez, A. C. Decamp, R. J. Parks, V. C. Ashley, J. T. Lucas, M. Cohen, J. Eron, C. B. Hicks, H. X. Liao, S. G. Self, G. Landucci, D. N. Forthal, K. J. Weinhold, B. F. Keele, B. H. Hahn, M. L. Greenberg, L. Morris, S. S. Karim, W. A. Blattner, D. C. Montefiori, G. M. Shaw, A. S. Perelson, and B. F. Haynes. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82:12449-12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Touloumi, G., N. Pantazis, A. G. Babiker, S. A. Walker, O. Katsarou, A. Karafoulidou, A. Hatzakis, and K. Porter. 2004. Differences in HIV RNA levels before the initiation of antiretroviral therapy among 1864 individuals with known HIV-1 seroconversion dates. AIDS 18:1697-1705. [DOI] [PubMed] [Google Scholar]
- 11.Wilson, T. E., Y. Barron, M. Cohen, J. Richardson, R. Greenblatt, H. S. Sacks, and M. Young. 2002. Adherence to antiretroviral therapy and its association with sexual behavior in a national sample of women with human immunodeficiency virus. Clin. Infect. Dis. 34:529-534. [DOI] [PubMed] [Google Scholar]