Abstract
Pneumococci of serotype 19A are increasingly found to be the cause of infection in various geographic regions. We have characterized the serotype 19A isolates (n = 288) found among pneumococci responsible for infections (n = 1,925) and pneumococci recovered from asymptomatic carriers (n = 1,973) in Portugal between 2001 and 2006. We show that despite the existence of serotype 19A clones that have a greater potential to cause invasive disease or an enhanced colonization capacity, the lineage that is increasing as a cause of infection in Portugal is a multiresistant clone that is competent at both. The expanding Denmark14-230 clone found in Portugal is disseminated in other Mediterranean countries, where it is also increasingly responsible for invasive infections in both children and adults. The lineages driving the rise of serotype 19A infections in Asia and the United States (sequence type 320 [ST320] and ST199) are either absent or account for only a small proportion of isolates in Portugal. These data highlight the importance of locally circulating clones with the ability to compete in the nasopharyngeal niche in the emergence of the serotype 19A lineages which are an increasing cause of infection in various geographic regions.
In the United States, following the introduction of the seven-valent pneumococcal conjugate vaccine (PCV7), a significant decline in the number of invasive pneumococcal infections caused by vaccine types was observed. This phenomenon occurred not only among children targeted for vaccination but also in other age groups, including the elderly, who are known to have a high burden of pneumococcal disease. This indirect herd effect is, in fact, responsible for most of the observed reduction in the incidence of disease (9). Vaccination has also been shown to have a broader impact on pneumococcal populations, as demonstrated by a reduction in the incidence of otitis media caused by vaccine serotypes (14), a reduction in the incidence of antimicrobial-resistant pneumococcal invasive infections (20), and a declining proportion of vaccine serotypes asymptomatically carried in the nasopharynges of children (12, 16).
Recent studies from European countries such as Spain and Portugal, where the vaccine is available but is not part of the national vaccination plan, have shown that even relatively modest vaccination coverage rates can have a profound effect on the serotypes responsible for invasive infections in children and adults (2, 4, 27).
In addition, all the studies mentioned above documented an increase in the number of cases caused by nonvaccine serotypes and, in particular, the emergence of serotype 19A as an increasingly important cause of invasive infections in all age groups (2, 27, 28). Although vaccination with PCV7 has frequently been implicated as the cause for the increase in the incidence of serotype 19A infections (7), recent reports documented the same trend in geographic areas where PCV7 was not available (11, 19). Taken together, these observations suggest that vaccination may have simply reinforced or accelerated an ongoing temporal trend. In particular, in South Korea, the rise in the incidence of serotype 19A infections in the absence of PCV7 was tentatively explained by the emergence of a multidrug-resistant lineage of sequence type (ST) 320 (ST320). Antibiotic pressure was likely to be a major factor in selecting for this lineage (19).
A study of the recent increase in the incidence of serotype 19A in the United States identified several lineages currently in circulation, among which the preexisting clonal complexes, clonal complex (CC) 199 (CC199) and CC320, were found to be expanding (28). The capsular switch in STs usually associated with vaccine serotypes and the appearance of multiple-drug resistant clonal complexes were also implicated in the rise in the incidence of serotype 19A (26, 28). Detailed analysis of the capsular loci of some of these lineages showed that the concurrent acquisition of the entire capsular locus and the flanking pbp genes occurred (7), confirming the emergence of vaccine-escape recombinant strains. On the other hand, a recent study conducted with individuals from the Alaska native population found that the genetic diversity of circulating serotype 19A isolates was reduced. In that setting the increase in the incidence of serotype 19A was due to the expansion of a single CC, CC172 (35). In contrast, in Europe, CC230 has consistently been identified as a major serotype 19A lineage causing invasive infections before vaccine introduction. This clonal complex has increased in the era of PCV7 both in children (22) and in adults (4).
We have previously shown that in Portugal, where the vaccine has been available since 2001, the conditional relative risk of acquiring an invasive infection caused by serotype 19A increased significantly in the postvaccination period in both children and adults (2). In the same study, we reported that isolates expressing this serotype were frequently antimicrobial resistant (2). In Portugal, CC230 was one of the lineages expressing serotype 19A before vaccination (33). In the study described here, we compared the clonal composition of the population expressing serotype 19A responsible for invasive infections in both adults and children to that recovered from children who were asymptomatic carriers and who had noninvasive infections in Portugal.
MATERIALS AND METHODS
Bacterial isolates.
Three Streptococcus pneumoniae collections were examined for the presence of isolates expressing serotype 19A. Each collection contained isolates from different sources: isolates responsible for invasive infections, isolates causing noninvasive infections, and isolates recovered in asymptomatic carriage studies.
Isolates causing infections.
Since 1999 the Portuguese Surveillance Group for the Study of Respiratory Pathogens has monitored invasive and noninvasive pneumococcal infections in Portugal. This is a laboratory-based surveillance system in which 30 microbiology laboratories throughout Portugal are asked to identify all cases of pneumococcal infection and to send the isolates to a central laboratory for characterization. A case of invasive disease is defined by the recovery of an S. pneumoniae isolate from a normally sterile body site.
A collection of 1,480 S. pneumoniae isolates responsible for invasive pneumococcal infections from 2001 to 2006 was characterized. Among this collection, 122 strains were isolated from children <2 years of age, 102 strains were isolated from children ≥2 and <6 years of age, 52 strains were obtained from children and adolescents between ≥6 and <18 years of age, and 1,204 strains were obtained from adults (≥18 years of age). The collection of noninvasive pneumococci (mainly recovered from lower respiratory tract specimens and associated with a diagnosis of pneumonia) was composed of 445 isolates, all of which were obtained from children <6 years of age.
The results of serotyping and the antimicrobial resistance for isolates responsible for invasive disease up to 2005 were reported previously (2, 34), as were the results of the genetic analysis for isolates recovered up to 2002 (33).
Carriage isolates.
In 2001, 2002, 2003, and 2006, surveys of pneumococcal carriage were conducted with healthy children (age range, 6 months to 6 years) attending day care centers (DCCs) in the Lisbon, Portugal, area. Between 2001 and 2003, during the months from January to March, 2,314 nasopharyngeal swab specimens were obtained from children attending 13 DCCs. In 2006, during the same months, 571 nasopharyngeal swab specimens were recovered from children attending 11 DCCs. A total of 1,973 pneumococcal isolates were obtained. A description of the resistant strains recovered between 2001 and 2003 was published previously (23, 37), as were the results of serotyping and the antimicrobial resistance of the isolates recovered in 2006 (31).
Serotyping and antimicrobial susceptibility testing.
Serotyping was performed by the standard capsular reaction test with a chessboard system (36) and specific sera (Statens Serum Institut, Copenhagen, Denmark). Etest strips (AB Biodisk, Solna, Sweden) were used to determine the MICs for penicillin, as described previously (25), and the Clinical and Laboratory Standards Institute (CLSI) recommended breakpoints (10) were used to interpret the MICs. The isolates were further characterized by determining their susceptibilities to co-trimoxazole, levofloxacin, erythromycin, clindamycin, tetracycline, and chloramphenicol by the Kirby-Bauer disk diffusion technique, according to the CLSI recommendations and interpretative criteria (10).
PFGE and MLST.
Profiling by pulsed-field gel electrophoresis (PFGE) was performed for all serotype 19A isolates included in this study. The preparation of chromosomal DNA, digestion with SmaI, separation by PFGE, and analysis of the PFGE patterns were performed as described previously (33). Multilocus sequence typing (MLST) analysis (13) was undertaken with a third of the isolates of each PFGE cluster with four or more isolates. The isolates within each PFGE cluster were selected to represent isolates from the three collections analyzed and, as much as possible, isolates collected in all the years studied. Lineage assignment was done by using the goeBURST program (15) and the complete S. pneumoniae database available at spneumoniae.mlst.net.
Statistical analysis.
Simpson's index of diversity (SID) was used to measure the diversity of the population (8). Clustering comparison coefficients, i.e., the adjusted Rand and Wallace values, were used to compare two sets of partitions (8, 29).
The statistical associations between PFGE clones and population type or antimicrobial resistance were characterized by determination of the odds ratios (ORs) with 95% Wald confidence intervals (CIs) (3). For null ORs, the 95% CIs were computed by the Fisher method implemented in the epitools module for the R language. The significance of the ORs was tested by use of the chi-square statistic. The resulting P values were corrected for multiple testing by controlling the false discovery rate (FDR) under or equal to 0.05 through the linear procedure of Benjamini and Hochberg (5).
The ORs for the enhanced invasive potential of the main serotype 19A PFGE clusters identified were calculated by using the number of invasive and carriage isolates in reference to the numbers of all other serotype 19A isolates from these two sources. The ORs measuring the association of particular PFGE clusters with resistance were calculated by reference to the resistance found in all other PFGE clusters expressing serotype 19A identified in the study.
Temporal trends in proportions were evaluated by the Cochran-Armitage test (1). The chi-square statistic or the Fisher exact test was also used to evaluate whether there were associations between population type and antimicrobial resistance. The P values were considered significant if they were less than 0.05.
RESULTS
Proportion of isolates expressing serotype 19A.
From the collections of pneumococcal isolates responsible for infections, 178 isolates presented serotype 19A. This set was composed of 121 isolates (of which 45 were from children less than 6 years of age) recovered from normally sterile sites and 57 isolates obtained from noninvasive infections (all from children less than 6 years of age). Among the collection of isolates recovered from asymptomatic carriers, 110 isolates presented serotype 19A. Among all pneumococcal isolates responsible for infection, the proportion of isolates expressing serotype 19A increased over time, being 4.1% (n = 8) in 2001 and 10.2% (n = 48) in 2006 (P < 10−3), reflecting an increase in the incidence of serotype 19A isolates among both invasive and noninvasive isolates. Although an increase in the incidence of serotype 19A isolates was also noted among isolates from asymptomatic carriers, from 4.3% (n = 20) in 2001 to 6.5% (n = 27) in 2006, it did not reach significance (P = 0.124).
Antimicrobial susceptibility.
The proportions of resistant isolates are summarized in Table 1. Although there were differences in the proportions of resistant isolates in each of the three populations analyzed, none were statistically significantly supported.
TABLE 1.
Antibiotic resistance of serotype 19A S. pneumoniae isolates (n = 288)
| Antibiotic | No. (%) of resistant isolatesa |
||
|---|---|---|---|
| Invasive (n = 121) | Noninvasive (n = 57) | Carriage (n = 110) | |
| Penicillinb | 56 (46.3)c | 23 (40.4) | 43 (39.1) |
| Erythromycind | 70 (57.9) | 23 (40.4) | 59 (53.6) |
| Clindamycin | 69 (57.0) | 21 (36.8) | 58 (52.7) |
| Tetracycline | 71 (58.7) | 23 (40.4) | 67 (60.9) |
| Co-trimoxazole | 40 (33.1) | 20 (35.1) | 20 (18.2) |
| Chloramphenicol | 17 (14.0) | 3 (5.5) | 9 (8.2) |
| Levofloxacin | 0 | 0 | 0 |
A total of 288 isolates were tested.
The data for isolates nonsusceptible to penicillin (MIC ≥ 0.12 μg/ml) are indicated.
Five isolates were fully resistant (MIC ≥ 2 μg/ml).
The majority (148/152) of macrolide-resistant isolates presented the macrolide-lincosamide-streptogramin B resistance (MLSB) phenotype, characterized by simultaneous resistance to erythromycin and clindamycin. Only four isolates (2.6%) presented the macrolide resistance (M) phenotype, characterized by resistance to erythromycin only.
Nevertheless, when the proportion of multidrug-resistant isolates (resistance to three or more different classes of antimicrobials) is considered, it was higher among invasive isolates (49%) than among noninvasive isolates (36%) and carriage isolates (30%) (P = 0.004).
Genotypic analysis and evaluation of invasive disease potential.
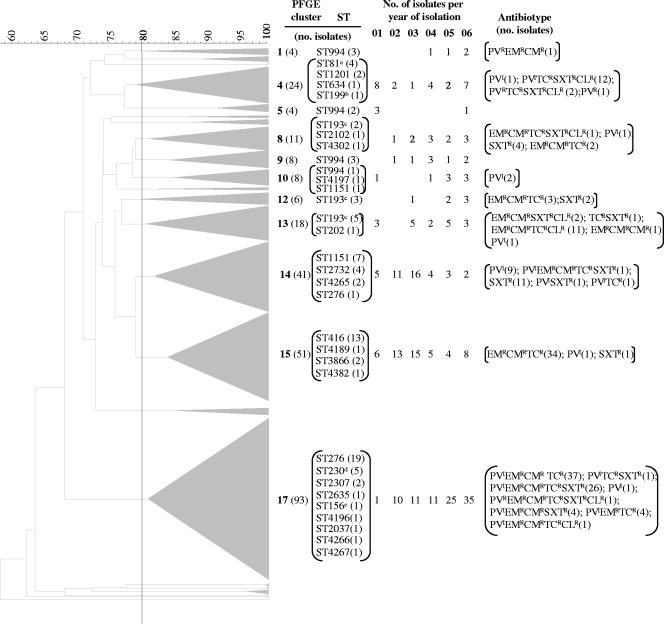
The three populations of serotype 19A were compared genotypically by a combination of PFGE and MLST. All isolates were analyzed by PFGE, and representatives of the PFGE clusters were characterized by MLST (n = 95). Overall, serotype 19A strains were distributed throughout 23 different PFGE clusters (Fig. 1), and 26 distinct STs were detected. The PFGE cluster and the ST demonstrated a good correlation. In fact, the Wallace value between the ST and the PFGE cluster was 0.772 (95% CI, 0.678 to 0.865), indicating that there is a high probability that two strains that have the same ST will also belong to the same PFGE cluster. This value is even higher if one considers the clonal complexes defined by goeBURST (Wallace value = 0.833 and 95% CI = 0.744 to 0.922; i.e., four of five pairs of isolates belonging to the same clonal complex will also be classified in the same PFGE cluster). Despite these high Wallace values, a few STs were detected in several PFGE groups, such as ST193 and ST994. Nevertheless, the Wallace values indicate that most isolates belonging to the same genetic lineage, as defined by goeBURST analysis, would be classified in the same PFGE cluster, allowing us to compare our data based on the results of PFGE analysis with data in other reports in which only MLST data are available.
FIG. 1.
Macrorestriction dendrograms and ST information for serotype 19A isolates. Dice coefficient values (percentages) are indicated in the scale above the dendrogram. Whenever two or more isolates had a macrorestriction pattern with a Dice coefficient of ≥80%, a triangle proportional to the number of isolates is indicated in the dendrogram. The number in boldface type beside each triangle indicates the PFGE cluster number, and the number in parentheses indicates the number of isolates grouped in that cluster. All STs determined for isolates of each PFGE cluster are indicated, and the number in parentheses indicates the number of isolates exhibiting that ST. The distribution of the isolates found in each PFGE cluster (n ≥ 4) over the years of the study is presented. The superscript letters identify STs identifying clones spread internationally, as follows: a, Spain23F-ST81; b, Netherlands15B-ST199; c, Greece21-ST193; d, Denmark14-ST230; e, Spain9V-ST156. Nonsusceptibility to various antimicrobial agents is indicated by superscripts, as follows: R, resistant; I, intermediate; CL, chloramphenicol; CM, clindamycin; EM, erythromycin; PV, penicillin; SXT, co-trimoxazole; TC, tetracycline. The number of isolates sharing the same antimicrobial resistance profile is indicated in parentheses.
The number of invasive isolates of serotype 19A recovered from children (age, <6 years; n = 45) was less than the number of isolates recovered from individuals in other age groups (n = 76). This reduced number of isolates could compromise the statistical efficiency of the tests comparing invasive isolates to isolates from other sources. However, if it could be demonstrated that the distribution of clones between children (age, <6 years) and older individuals was indistinguishable, all invasive isolates could be regarded as representing a single population, greatly enhancing the power of the comparisons. In fact, we could not show a difference in the distribution of the PFGE clusters between the two age groups (Fisher's exact test, P = 0.775), indicating that all serotype 19A invasive isolates constitute a homogeneous population independent of the age group considered. We therefore used the entire collection of invasive isolates in the comparisons with the isolates from other sources.
Isolates from all three sources presented a high degree of genetic diversity, as determined by PFGE cluster analysis, with high and undistinguishable SID values (Table 2). Analysis of the distribution of isolates between PFGE clusters showed that most clusters included isolates from the three distinct sources. This is highlighted by an adjusted Rand value of 0.044, which indicates a low overall level of congruence between the PFGE cluster and the isolate source. Despite the absence of a strong correlation between the isolate source and the PFGE cluster, some PFGE clusters were not uniformly distributed among the three populations (Table 2).
TABLE 2.
Diversity and major PFGE clusters of the three serotype 19A populations analyzed
| Population | % SID (95% CI) | % of isolates in the following PFGE clustera: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 4 | 8 | 9 | 10 | 13 | 14 | 15 | 17 | ||
| Carriage isolates (n = 110) | 78.8 (74.5-83.0) | 6 | 1 | 1 | 2 | 2 | 21 | 34 | 24 |
| Invasive isolates (n = 121) | 82.2 (76.8-87.5) | 12 | 5 | 5 | 1 | 12 | 7 | 7 | 37 |
| Noninvasive isolates (n = 57) | 81.4 (73.3-89.6) | 4 | 7 | 2 | 9 | 4 | 16 | 9 | 39 |
PFGE clusters that included at least 5% of any of the three populations analyzed are represented. When the proportion of the population grouped into one of the PFGE clusterswas ≥10%, the value is highlighted in boldface.
To identify the serotype 19A PFGE clusters associated with carriage, the ORs for the main clusters were determined on the basis of the number of isolates of that particular cluster found to be carriage and invasive disease isolates with reference to all other isolates from these two sources. This approach identified two PFGE clusters associated with carriage: PFGE cluster 14 (OR = 0.30 [95% CI = 0.12 to 0.73]) and PFGE cluster 15 (OR = 0.16 [95% CI = 0.06 to 0.36]), both of which are significant after correction for FDR (P = 0.030 and P < 10−4, respectively). A cluster with enhanced invasive disease potential, PFGE cluster 13 (OR = 7.02 [95% CI = 1.55 to 65.18], P = 0.030), was also identified. Since the MLST analysis of PFGE cluster 13 revealed isolates mostly representing ST193, we repeated the analysis by grouping together all PFGE clusters in which ST193 was found (PFGE clusters 8, 12, and 13), and the result was also significant (OR = 8.76 [95% CI = 2.54 to 46.80], P < 10−4).
Temporal fluctuations in PFGE clusters.
To probe for temporal variations in the proportions of serotype 19A isolates in each PFGE cluster, the Cochran-Armitage test was used. This analysis was performed for the major PFGE clusters (n ≥ 10) found among isolates causing infection and also by considering as a single genetic lineage all PFGE clusters associated with ST994 (n = 24) and as another single genetic lineage all those associated with ST193 (n = 35). The test revealed that among the serotype 19A isolates responsible for infections, the incidence of isolates in PFGE cluster 14 decreased during the study period (P < 10−4), while the incidence of isolates in PFGE cluster 17 increased (P = 0.002).
PFGE cluster and antibiotic resistance.
The relationships between PFGE cluster and antimicrobial resistance were examined by determination of the odds ratios for each antimicrobial, and the significant values are presented in Table 3. PFGE cluster 17 stood out in the analysis, since it was associated with resistance to all antimicrobials and antimicrobial classes with the exception of chloramphenicol.
TABLE 3.
Significant antimicrobial resistance associations with PFGE clones of serotype 19A strains
| PFGE cluster (ST in PFGE cluster)a | Penicillin |
Erythromycin |
Chloramphenicol |
Tetracycline |
Co-trimoxazole |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| ORb (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |
| 4 (ST1201, [ST81, ST634], ST199) | 0 (0.00 to 0.13) | <10−4 | 18.32 (7.09 to 47.31) | <10−4 | 3.47 (1.48 to 8.13) | 0.003 | ||||
| 9 (ST994)c | 0 (0.00-0.44) | 0.002 | 0 (0.00-0.44) | 0.001 | ||||||
| 10 (ST994, ST4197, ST1151)c | 0 (0.00-0.44) | 0.002 | 0 (0.00-0.44) | 0.001 | ||||||
| 13 (ST193, ST202)d | 0.07 (0.01-0.55) | 0.001 | 4.85 (1.37-17.15) | 0.007 | 29.76 (9.94-89.07) | <10−4 | ||||
| 14 ([ST1151, ST2732, ST9001], ST276) | 0.03 (0.01-0.14) | <10−4 | 0 (0.00-0.74) | 0.021 | 0.06 (0.02-0.18) | <10−4 | ||||
| 15 ([ST416, ST4189, ST3866], ST9004) | 0 (0.00-0.07) | <10−4 | 0 (0.00-0.56) | 0.008 | 0.04 (0.01-0.29) | <10−4 | ||||
| 17e ([ST276, ST230, ST2037, ST2307, ST4196, ST9002, ST9003], [ST156, ST2635]) | 83.34 (31.46-220.74) | <10−4 | 24.01 (10.52-54.82) | <10−4 | 0.14 (0.03-0.59) | 0.002 | 9.70 (4.96-18.96) | <10−4 | 2.34 (1.37-3.99) | 0.002 |
Only PFGE clusters with a significant association with at least one antimicrobial are indicated. Brackets indicate STs that belong to the same eBURST group.
Only significant values are shown. An OR of >1 (in boldface) indicates a significant association with resistance, whereas an OR of <1 indicates a significant association with susceptibility.
If all PFGE clusters exclusively representing ST994 (PFGE clusters 1, 5, 9, and 10) are analyzed together, a significant association with susceptibility to co-trimoxazole (OR = 0 [95% CI = 0.00 to 0.40]; P = 0.002) emerges, in addition to those already identified for PFGE clusters 9 and 10.
If all PFGE clusters exclusively representing ST193 (PFGE clusters 12 and 13) are analyzed together, no qualitative changes occurred in relation to those identified for PFGE cluster 13.
PFGE cluster 17 was the only one presenting significant associations with both multidrug resistance and simultaneous resistance to penicillin and erythromycin.
DISCUSSION
Several studies have demonstrated that after the introduction of a conjugate vaccine, significant changes in the distribution of the serotypes responsible for invasive infections occur. These changes are characterized by a sharp decrease in the number of infections caused by vaccine types and an increase in the number of infections caused by nonvaccine serotypes, even if they are only incremental (2, 4, 18, 27, 39). The pneumococcal serotypes not represented in vaccine formulations are generally considered to be less virulent (38), yet the lower incidence of invasive infections caused by nonvaccine serotypes could be due not to reduced virulence but to a lower transmissibility or to a weaker competitive capacity for nasopharyngeal colonization. If the latter was the case, then the removal by vaccination of the serotypes commonly found in the nasopharynx would allow the establishment of nonvaccine serotypes that could then emerge as important causes of invasive infections (21). In this scenario, all other things being equal, one could expect that the more virulent clones found among nonvaccine serotypes would increase in prevalence as a cause of invasive infections. Another hypothesis for vaccine selection of nonvaccine serotypes would be the emergence of capsular transformants (30). These would retain the successful genotypes previously found among vaccine serotypes but would express a capsular polysaccharide not targeted by the vaccine (7).
In line with the findings of other studies, an increased proportion of infections caused by serotype 19A isolates has been noted in Portugal, despite nonuniversal vaccination (2; this study). If occupation of a vacant colonization niche was the sole driver behind the increase in the incidence of serotype 19A infection, then an increase in the incidence of 19A colonization would be expected. Although we did observe an increase in the incidence of serotype 19A isolates among carriage isolates, it was not statistically significant and may therefore not offer a full explanation for the rise in the incidence of serotype 19A infections. A possible explanation for the more modest increase in the incidence of serotype 19A carriage isolates may be that isolates of the other serotypes may be equally adapted at exploiting the vacant niche left by the reduction in the incidence of isolates of the vaccine serotypes as colonizers.
Each of the three serotype 19A populations analyzed was highly diverse, and several genetic lineages were identified, most of which were found among all three populations. This finding is in contrast to the findings of other studies that included isolates expressing all serotypes, which found a genetically more diverse population among carriage isolates (17, 32). A higher degree of diversity among carriage isolates is frequently interpreted as supporting the hypothesis that only a few lineages are capable of causing invasive infections. However, this conclusion may be conditioned by the larger number of serotypes found among carriage isolates. Studies of other pneumococcal populations homogeneous for their serotype are needed to clarify if the similar genetic diversity found among serotype 19A isolates causing infections and those carried asymptomatically is an unusual characteristic of serotype 19A or a more general property.
Previous studies have suggested that the capsular serotype may be more important than the genotype in determining the ability of pneumococci to cause invasive disease (6). Of interest, among all serotype 19A lineages identified, two groups established opposing relationships with the human host: one was associated with asymptomatic carriage (PFGE clusters 14 and 15, representing ST1151 and ST416 and closely related STs, respectively), while the other was associated with invasive disease (PFGE cluster 13, mostly representing ST193). The existence of several lineages within serotype 19A with opposing properties and their different prevalences in various geographic regions may explain why some studies identified isolates of this serotype as having enhanced invasive disease potential (32) and others did not (6).
Neither the lineage defined by ST193 (including all PFGE clusters presenting this ST) nor the one defined by ST416 and its single-locus variants increased among the infection isolates over time. Instead, two other genetic lineages did show temporal variations. The incidence of the lineage identified by PFGE cluster 14 (mostly representing isolates of ST1151 and ST2732 that are single-locus variants of each other and STs apparently only detected in Portugal), which was significantly associated with carriage and susceptibility to most antimicrobials, declined as a cause of infection during the study period. On the other hand, the incidence of the lineage characterized by PFGE cluster 17, associated with resistance to most antimicrobials, including ß-lactams and macrolides (the antibiotics of choice for the treatment of pneumococcal infections), as a cause of infection increased significantly over time. In fact, this lineage was the most abundant in all collections. A comparison based on the PFGE profiles identified members of cluster 17 as being closely related to clone Denmark14-ST230 (24). MLST analysis of representative isolates of this PFGE cluster identified ST276 (a single-locus variant of ST230) as the dominant ST and found that most isolates (30 of 32) belonged to CC230, corroborating the findings that isolates in this PFGE cluster mostly represent the Denmark14-ST230 clone.
Studies characterizing invasive isolates in both France (22) and Spain (4) have also identified representatives of clone Denmark14-ST230, in particular, ST276, as major causes of serotype 19A invasive disease in both children and adults. A study from Israel also identified ST276 as an important cause of acute otitis media in recent years (11). The latter is a particularly interesting study, since it associated the presence of ST276 with high levels of antibiotic consumption, a situation that may be mirrored in Portugal.
Studies from two other non-European countries, South Korea and the United States, have also analyzed in great detail the genetic diversity of the pneumococcal population expressing serotype 19A (19, 26). The study from South Korea revealed a serotype 19A population with limited diversity (only 4 STs), while that from the United States revealed a much more diverse population (73 STs); but the major clonal complex found in South Korea (n = 53/58), CC320, was also a major lineage in the United States (n = 111/528). In contrast, in Portugal, ST320 was not detected, and a single isolate recovered in Portugal was a member of the most frequent lineage in the United States (ST199) and was in a medium-sized PFGE cluster (PFGE cluster 4). The most frequent lineage in our study, PFGE cluster 17, representing the Denmark14-ST230 clone (n = 93/288, or 32%), was a minor lineage in the United States (accounting for 3% of the serotype 19A isolates) and was not found in South Korea. These data highlight the genetic diversity and geographic differences behind the increase in the incidence of serotype 19A isolates and suggest that the locally circulating clones, as well as other selective pressures, such as antibiotic consumption and vaccination, may play equally important roles in the emergence of serotype 19A as a major cause of pneumococcal infections.
Although this study has limitations, we consider that these do not affect our conclusions. First, isolates causing infections were recovered from centers geographically dispersed in Portugal, whereas pneumococcal carriage isolates were collected only in the Lisbon area. Our previous studies with isolates causing invasive infections did not identify any significant regional asymmetries (2, 33), so we do not expect these to exist in colonization. Second, the three major clones found to be colonizers, which together accounted for close to 80% of the colonization isolates, were each found in six DCCs, indicating that these clones are widely disseminated and do not correspond to local clusters. Third, no colonization isolates were available from 2004 and 2005 because no sampling was performed in those years. This did not allow the analysis of the temporal variations in the clonal composition of the colonization population, but a qualitative evaluation indicated that there was a strong parallel with the changes detected in the infection isolates. The major clones (n ≥ 10) found among the carriage isolates were present in all years studied, indicating no sudden variations in the clonal distribution. These two observations reassured us that we were effectively sampling the carriage population. Fourth, we included in our comparison isolates recovered from both adults and children with invasive infections, whereas the carriage isolates were recovered only from children. Our choice was guided by the smaller number of isolates that would have been analyzed if we had excluded those from adults, which would have resulted in a much lower statistical efficiency, and by the observation that there were no differences in the clonal compositions among invasive isolates when they were stratified by age group. In fact, the rate of asymptomatic carriage in adults is low and children are considered the major reservoir of pneumococci (16). Further confirmation of the pivotal role of children in adult infections was obtained from the recent observation that the vaccination of children had a major impact on the incidence of pneumococcal infections in adults (2, 4, 9).
In summary, we have shown that different genetic lineages expressing serotype 19A preferentially establish colonization or cause infections in the human host. The lineage currently expanding as a cause of infection in Portugal and other southern European countries was not identified as being particularly virulent. The availability of an enlarged nasopharyngeal niche seems to have allowed the expansion of a clone competent at colonization and infection, which drove the rise in the incidence of serotype 19A infections. Similar to what was found elsewhere, this dominant serotype 19A lineage was resistant to most antimicrobials. Together with the decline in the incidence of a successful local clone that was mostly susceptible, this observation suggests that antibiotic use may also be an important factor shaping the pneumococcal population in Portugal and may have been decisive as a cause for the clonal fluctuations among serotype 19A isolates causing infections observed in Portugal and other Mediterranean countries. Nevertheless, the reasons why the successful multiresistant ST320 lineage is not found in Portugal or why the equally resistant ST199 lineage did not expand to dominate the serotype 19A population, as it did in the United States, remain unclear and point to the importance of circulating clones and other local selective forces in driving the expansion of the most successful serotype 19A clones. These data reinforce the importance of continuous surveillance to understand changes in the incidences of particular serotypes in the era of pneumococcal conjugate vaccines and highlight the important differences in the incidences of particular serotypes between Europe and the United States. Such knowledge may allow us to understand the emergence of nonvaccine serotypes and guide the design and use of future pneumococcal vaccines.
Acknowledgments
Members of the Portuguese Surveillance Group for the Study of Respiratory Pathogens are gratefully thanked for their valuable collaboration in this study.
S. I. Aguiar was supported by grant SFRH/BD/27518/2006 and S. Nunes was supported by grant SFRH/BD/40706/2007 from the Fundação para a Ciência e Tecnologia, Portugal. Partial support for this work was provided by EURIS (contract QKL2-CT-2000-01020) and PREVIS (contract LSHM-CT-2003-503413) from the European Community, the Fundação para a Ciência e Tecnologia (grants PDTC/SAU-ESA/6888/2006, PTDC/SAU-ESA/65048/2006, and PIC/IC/83065/2007), and the Fundação Calouste Gulbenkian.
Footnotes
Published ahead of print on 28 October 2009.
REFERENCES
- 1.Agresti, A. 1990. Categorical data analysis. John Wiley & Sons, Inc., New York, NY.
- 2.Aguiar, S. I., I. Serrano, F. R. Pinto, J. Melo-Cristino, and M. Ramirez. 2008. Changes in Streptococcus pneumoniae serotypes causing invasive disease with non-universal vaccination coverage of the seven-valent conjugate vaccine. Clin. Microbiol. Infect. 14:835-843. [DOI] [PubMed] [Google Scholar]
- 3.Altman, D. G. 1999. Practical statistics for medical research. Chapman & Hall/CRC, Boca Raton, FL.
- 4.Ardanuy, C., F. Tubau, R. Pallares, L. Calatayud, M. A. Dominguez, D. Rolo, I. Grau, R. Martin, and J. Linares. 2009. Epidemiology of invasive pneumococcal disease among adult patients in Barcelona before and after pediatric 7-valent pneumococcal conjugate vaccine introduction, 1997-2007. Clin. Infect. Dis. 48:57-64. [DOI] [PubMed] [Google Scholar]
- 5.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57:289-300. [Google Scholar]
- 6.Brueggemann, A. B., D. T. Griffiths, E. Meats, T. Peto, D. W. Crook, and B. G. Spratt. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424-1432. [DOI] [PubMed] [Google Scholar]
- 7.Brueggemann, A. B., R. Pai, D. W. Crook, and B. Beall. 2007. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 3:e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carriço, J. A., C. Silva-Costa, J. Melo-Cristino, F. R. Pinto, H. de Lencastre, J. S. Almeida, and M. Ramirez. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 44:2524-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2005. Direct and indirect effects of routine vaccination of children with 7-valent pneumococcal conjugate vaccine on incidence of invasive pneumococcal disease—United States, 1998-2003. MMWR Morb. Mortal. Wkly. Rep. 54:893-897. [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing, 16th informational supplement, CLSI document M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 11.Dagan, R., N. Givon-Lavi, E. Leibovitz, D. Greenberg, and N. Porat. 2009. Introduction and proliferation of multidrug-resistant Streptococcus pneumoniae serotype 19A clones that cause acute otitis media in an unvaccinated population. J. Infect. Dis. 199:776-785. [DOI] [PubMed] [Google Scholar]
- 12.Dagan, R., N. Givon-Lavi, O. Zamir, M. Sikuler-Cohen, L. Guy, J. Janco, P. Yagupsky, and D. Fraser. 2002. Reduction of nasopharyngeal carriage of Streptococcus pneumoniae after administration of a 9-valent pneumococcal conjugate vaccine to toddlers attending day care centers. J. Infect. Dis. 185:927-936. [DOI] [PubMed] [Google Scholar]
- 13.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 14.Eskola, J., T. Kilpi, A. Palmu, J. Jokinen, J. Haapakoski, E. Herva, A. Takala, H. Kayhty, P. Karma, R. Kohberger, G. Siber, and P. H. Makela. 2001. Efficacy of a pneumococcal conjugate vaccine against acute otitis media. N. Engl. J. Med. 344:403-409. [DOI] [PubMed] [Google Scholar]
- 15.Francisco, A. P., M. Bugalho, M. Ramirez, and J. A. Carriço. 2009. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinform. 10:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frazão, N., A. Brito-Avô, C. Simas, J. Saldanha, R. Mato, S. Nunes, N. G. Sousa, J. A. Carriço, J. S. Almeida, I. Santos-Sanches, and H. de Lencastre. 2005. Effect of the seven-valent conjugate pneumococcal vaccine on carriage and drug resistance of Streptococcus pneumoniae in healthy children attending day-care centers in Lisbon. Pediatr. Infect. Dis. J. 24:243-252. [DOI] [PubMed] [Google Scholar]
- 17.Hanage, W. P., T. H. Kaijalainen, R. K. Syrjanen, K. Auranen, M. Leinonen, P. H. Makela, and B. G. Spratt. 2005. Invasiveness of serotypes and clones of Streptococcus pneumoniae among children in Finland. Infect. Immun. 73:431-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hicks, L. A., L. H. Harrison, B. Flannery, J. L. Hadler, W. Schaffner, A. S. Craig, D. Jackson, A. Thomas, B. Beall, R. Lynfield, A. Reingold, M. M. Farley, and C. G. Whitney. 2007. Incidence of pneumococcal disease due to non-pneumococcal conjugate vaccine (PCV7) serotypes in the United States during the era of widespread PCV7 vaccination, 1998-2004. J. Infect. Dis. 196:1346-1354. [DOI] [PubMed] [Google Scholar]
- 19.Hwa Choi, E., S. Hee Kim, B. Wook Eun, S. Jung Kim, N. Hee Kim, J. Lee, and H. Jong Lee. 2008. Streptococcus pneumoniae serotype 19A in children, South Korea. Emerg. Infect. Dis. 14:275-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kyaw, M. H., R. Lynfield, W. Schaffner, A. S. Craig, J. Hadler, A. Reingold, A. R. Thomas, L. H. Harrison, N. M. Bennett, M. M. Farley, R. R. Facklam, J. H. Jorgensen, J. Besser, E. R. Zell, A. Schuchat, and C. G. Whitney. 2006. Effect of introduction of the pneumococcal conjugate vaccine on drug-resistant Streptococcus pneumoniae. N. Engl. J. Med. 354:1455-1463. [DOI] [PubMed] [Google Scholar]
- 21.Lipsitch, M. 1997. Vaccination against colonizing bacteria with multiple serotypes. Proc. Natl. Acad. Sci. U. S. A. 94:6571-6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahjoub-Messai, F., C. Doit, J. L. Koeck, T. Billard, B. Evrard, P. Bidet, C. Hubans, J. Raymond, C. Levy, R. Cohen, and E. Bingen. 2009. Population snapshot of Streptococcus pneumoniae serotype 19A isolates before and after introduction of seven-valent pneumococcal vaccination for French children. J. Clin. Microbiol. 47:837-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mato, R., I. S. Sanches, C. Simas, S. Nunes, J. A. Carriço, N. G. Sousa, N. Frazão, J. Saldanha, A. Brito-Avô, J. S. Almeida, and H. de Lencastre. 2005. Natural history of drug-resistant clones of Streptococcus pneumoniae colonizing healthy children in Portugal. Microb. Drug Resist. 11:309-322. [DOI] [PubMed] [Google Scholar]
- 24.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefevre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the Pneumococcal Molecular Epidemiology Network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melo-Cristino, J., M. L. Fernandes, N. Serrano, and the Portuguese Surveillance Group for the Study of Respiratory Pathogens. 2001. A multicenter study of the antimicrobial susceptibility of Haemophilus influenzae, Streptococcus pneumoniae, and Moraxella catarrhalis isolated from patients with community-acquired lower respiratory tract infections in 1999 in Portugal. Microb. Drug Resist. 7:33-38. [DOI] [PubMed] [Google Scholar]
- 26.Moore, M. R., R. E. Gertz, Jr., R. L. Woodbury, G. A. Barkocy-Gallagher, W. Schaffner, C. Lexau, K. Gershman, A. Reingold, M. Farley, L. H. Harrison, J. L. Hadler, N. M. Bennett, A. R. Thomas, L. McGee, T. Pilishvili, A. B. Brueggemann, C. G. Whitney, J. H. Jorgensen, and B. Beall. 2008. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J. Infect. Dis. 197:1016-1027. [DOI] [PubMed] [Google Scholar]
- 27.Muñoz-Almagro, C., I. Jordan, A. Gene, C. Latorre, J. J. Garcia-Garcia, and R. Pallares. 2008. Emergence of invasive pneumococcal disease caused by nonvaccine serotypes in the era of 7-valent conjugate vaccine. Clin. Infect. Dis. 46:174-182. [DOI] [PubMed] [Google Scholar]
- 28.Pai, R., M. R. Moore, T. Pilishvili, R. E. Gertz, C. G. Whitney, and B. Beall. 2005. Postvaccine genetic structure of Streptococcus pneumoniae serotype 19A from children in the United States. J. Infect. Dis. 192:1988-1995. [DOI] [PubMed] [Google Scholar]
- 29.Pinto, F. R., J. Melo-Cristino, and M. Ramirez. 2008. A confidence interval for the Wallace coefficient of concordance and its application to microbial typing methods. PLoS One 3:e3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramirez, M., and A. Tomasz. 1999. Acquisition of new capsular genes among clinical isolates of antibiotic-resistant Streptococcus pneumoniae. Microb. Drug Resist. 5:241-246. [DOI] [PubMed] [Google Scholar]
- 31.Sá-Leão, R., S. Nunes, A. Brito-Avô, N. Frazão, A. S. Simões, M. I. Crisostomo, A. C. Paulo, J. Saldanha, I. Santos-Sanches, and H. de Lencastre. 2009. Changes in pneumococcal serotypes and antibiotypes carried by vaccinated and unvaccinated day-care centre attendees in Portugal, a country with widespread use of the seven-valent pneumococcal conjugate vaccine. Clin. Microbiol. Infect. 15:1002-1007. [DOI] [PubMed] [Google Scholar]
- 32.Sandgren, A., K. Sjostrom, B. Olsson-Liljequist, B. Christensson, A. Samuelsson, G. Kronvall, and B. Henriques Normark. 2004. Effect of clonal and serotype-specific properties on the invasive capacity of Streptococcus pneumoniae. J. Infect. Dis. 189:785-796. [DOI] [PubMed] [Google Scholar]
- 33.Serrano, I., J. Melo-Cristino, J. A. Carriço, and M. Ramirez. 2005. Characterization of the genetic lineages responsible for pneumococcal invasive disease in Portugal. J. Clin. Microbiol. 43:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serrano, I., M. Ramirez, the Portuguese Surveillance Group for the Study of Respiratory Pathogens, and J. Melo-Cristino. 2004. Invasive Streptococcus pneumoniae from Portugal: implications for vaccination and antimicrobial therapy. Clin. Microbiol. Infect. 10:652-656. [DOI] [PubMed] [Google Scholar]
- 35.Singleton, R. J., T. W. Hennessy, L. R. Bulkow, L. L. Hammitt, T. Zulz, D. A. Hurlburt, J. C. Butler, K. Rudolph, and A. Parkinson. 2007. Invasive pneumococcal disease caused by nonvaccine serotypes among Alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. JAMA 297:1784-1792. [DOI] [PubMed] [Google Scholar]
- 36.Sorensen, U. B. 1993. Typing of pneumococci by using 12 pooled antisera. J. Clin. Microbiol. 31:2097-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sousa, N. G., R. Sá-Leão, M. I. Crisóstomo, C. Simas, S. Nunes, N. Frazão, J. A. Carriço, R. Mato, I. Santos-Sanches, and H. de Lencastre. 2005. Properties of novel international drug-resistant pneumococcal clones identified in day-care centers of Lisbon, Portugal. J. Clin. Microbiol. 43:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spratt, B. G., and B. M. Greenwood. 2000. Prevention of pneumococcal disease by vaccination: does serotype replacement matter? Lancet 356:1210-1211. [DOI] [PubMed] [Google Scholar]
- 39.Whitney, C. G., M. M. Farley, J. Hadler, L. H. Harrison, N. M. Bennett, R. Lynfield, A. Reingold, P. R. Cieslak, T. Pilishvili, D. Jackson, R. R. Facklam, J. H. Jorgensen, and A. Schuchat. 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348:1737-1746. [DOI] [PubMed] [Google Scholar]