Abstract
Sepsis is caused by a heterogeneous group of infectious etiologies. Early diagnosis and the provision of appropriate antimicrobial therapy correlate with positive clinical outcomes. Current microbiological techniques are limited in their diagnostic capacities and timeliness. Multiplex PCR has the potential to rapidly identify bloodstream infections and fill this diagnostic gap. We identified patients from two large academic hospital emergency departments with suspected sepsis. The results of a multiplex PCR that could detect 25 bacterial and fungal pathogens were compared to those of blood culture. The results were analyzed with respect to the likelihood of infection, sepsis severity, the site of infection, and the effect of prior antibiotic therapy. We enrolled 306 subjects with suspected sepsis. Of these, 43 were later determined not to have infectious etiologies. Of the remaining 263 subjects, 70% had sepsis, 16% had severe sepsis, and 14% had septic shock. The majority had a definite infection (41.5%) or a probable infection (30.7%). Blood culture and PCR performed similarly with samples from patients with clinically defined infections (areas under the receiver operating characteristic curves, 0.64 and 0.60, respectively). However, blood culture identified more cases of septicemia than PCR among patients with an identified infectious etiology (66 and 46, respectively; P = 0.0004). The two tests performed similarly when the results were stratified by sepsis severity or infection site. Blood culture tended to detect infections more frequently among patients who had previously received antibiotics (P = 0.06). Conversely, PCR identified an additional 24 organisms that blood culture failed to detect. Real-time multiplex PCR has the potential to serve as an adjunct to conventional blood culture, adding diagnostic yield and shortening the time to pathogen identification.
The emergency department (ED) often serves as the front line for the initial evaluation and management of patients with infections, with estimates indicating that there are more than 3.5 million visits to EDs annually (26). Community-acquired sepsis carries a large health care burden and accounts for a substantial proportion of these visits. Furthermore, recent studies suggest a rising burden of sepsis, with the annualized increase being about 8.7% (23). Many of these patients, estimated at 3 per 1,000 population (1), progress to severe sepsis. Despite improvements in the management of severe sepsis (15, 27), the case fatality rate ranges from 10% in children to 38.4% in the elderly (1). However, sepsis case fatality rates can be reduced by 33% to 77% with the early administration of adequate antibiotics (4, 10, 17, 18, 34). The antibiotics initially chosen are often broad and empirical, with modifications being made after microbiological diagnostic information becomes available. Blood culture remains the “gold standard” for the identification of bacterial and fungal bloodstream infections, although it is limited by high volume requirements to maximize its sensitivity and often prolonged incubation times. In an effort to address some of these limitations, many advances have been developed to improve the sensitivity and decrease the time to identification of the cause of bloodstream infections.
Molecular amplification techniques have been developed to replace the incubation step in blood culture. Conserved regions of microbial genomes serve as targets for amplification, such as the rRNA genes and the 16S-23S interspace region (13, 25). The identification of this species-specific amplified DNA is coupled with one of a variety of detection schemes for the rapid real-time identification of pathogen DNA in the bloodstream. Many protocols that target individual species (8) or multiple pathogens within a particular clade have been developed, such as panbacterial and panfungal assays (6, 12, 29). Within the past year, the simultaneous detection of up to 25 common bacterial and fungal pathogens by the use of a multiplex real-time PCR format has been reported (19, 21, 22, 30, 32, 33). However, many of those studies defined operating characteristics on the basis of repeated sampling of a small but highly morbid patient population, which can lead to spectrum and context biases. We hypothesized that PCR and blood culture can identify a partially overlapping group of bloodstream infections, making PCR a useful adjunct to blood culture. This study aims to define the operating characteristics of the SeptiFast multiplex real-time PCR assay compared to those of conventional blood culture for the diagnosis of bloodstream infections in patients with suspected community-acquired sepsis presenting to the ED.
MATERIALS AND METHODS
Study site and patients.
Subjects were recruited from the Duke University Medical Center ED (DUMC Level 1 Trauma Center, which has an annual census of 65,000) and the Veterans Affairs Medical Center in Durham, NC (DVAMC; which has an annual ED census of 40,000). This study was approved by the institutional review board at each institution, and written, informed consent was obtained from all study participants or their legal representatives. Subjects were screened between July 2003 and February 2009. Patients were considered for inclusion in the study if they had a known or suspected infection on the basis of clinical data at the time of screening and if they exhibited two or more signs of the systemic inflammatory response syndrome (SIRS) within a 24-h period (3). Patients were excluded if they were <18 years old, if they had an imminently terminal comorbid condition, or if they were participating in an ongoing clinical trial. Only subjects admitted to the hospital and for whom blood culture results were available were included in this analysis.
Data collection.
Following the provision of informed consent, the patient or the patient's representative completed a standardized questionnaire that included questions about demographic factors, recent exposures, and symptoms. We obtained samples of blood for culture from all patients, but the samples for culture from other anatomical sites were ordered by the treating physician, as clinically indicated. Trained study coordinators at each site reviewed and abstracted vital signs and the microbiology, laboratory, and imaging results from the initial ED encounter and at 24-h intervals for up to 72 h or until death. Following discharge from the hospital, research personnel abstracted the duration of hospitalization, the length of stay in an intensive care unit (ICU), the in-hospital mortality rate, the timing and appropriateness of antimicrobial administration, and microbiological culture results from the medical record.
Adjudication of infections and patient status.
Research personnel blinded to the hypothesis adjudicated the patient records at least 30 days after hospital discharge in order to determine the discharge status, including the likelihood of infection; the site of infection, if one was present; the causative organisms; and the patient outcome. One of two study physicians with board certification in emergency medicine (S.W.G.) or internal medicine (E.L.T.) determined the infection status after a review of all study data and the complete patient medical record, including ED dictations, hospital admission and discharge summaries, and any other relevant information (e.g., consultant reports). The likelihood of infection was categorized according to a five-point scale (11). Category 1 (confirmed) was defined as having clinical evidence of infection without evidence of a noninfectious process and an identified etiologic agent. Category 2 (probable) was the same as category 1 but in the absence of an identified etiologic agent. Category 3 (possible) was reserved for indeterminate cases in which infection could be neither confirmed nor excluded. Category 4 (negative) was defined as no evidence of infection and no evidence of a noninfectious process. Finally, category 5 (negative) was no evidence of infection and the identification of a noninfectious etiology, which was required.
For patients who could not be definitively placed in an infection category by the primary adjudicator (S.W.G.), a second individual with specialty training and board certification in infectious diseases (C.W.W.) reviewed the medical record and made the final determination. A third individual with specialty training in infectious diseases (E.L.T.) performed an independent adjudication of a 10% sample of the patient records. Agreement on the infection classification between this individual and the primary adjudicator was high (kappa statistic = 0.82; 95% confidence interval [CI] = 0.62 to 1.00). For determination of the infection classification and the etiologic agents involved, investigators were blinded to the multiplex PCR test results. The identification of blood culture contamination was based on previously published criteria and included the probability that the organism was a skin contaminant, the number of independent positive and negative cultures, other concurrent microbiology results, and clinical compatibility (9, 31).
Study definitions.
The severity of sepsis was determined on the basis of the patient's status at the time of study enrollment. Patients later determined not to have infection (infection categories 4 and 5) were labeled as noninfected and SIRS positive. Sepsis was defined as SIRS with evidence of infection but no evidence of end-organ damage. Severe sepsis occurred in the presence of end-organ damage, which included metabolic damage (a lactate concentration of >1.5 times the upper limit of normal or an arterial pH of <7.30), hematologic damage (platelet count of <80,000/high-power field), pulmonary damage (intubation or a partial arterial O2 concentration/fractional inspired O2 concentration ratio, <250), or renal damage (urine output, <0.5 ml/kg of body weight/h, despite adequate fluid resuscitation) (14, 24). Sepsis in the presence of hypotension (a systolic blood pressure of <90 mm Hg or a mean arterial pressure of <65 mm Hg), despite fluid challenge, or a blood lactate concentration of ≥4 mmol/liter was defined as septic shock (14, 24).
Sample processing.
Blood was collected for culture by the use of sterile technique. The volume inoculated was not monitored and was subject to user variability. At the Veterans Affairs Medical Center, the BacT/Alert system (bioMérieux, Marcy l'Etiole, France) was used. At the Duke University Medical Center, the BacT/Alert system was used along with the BD Bactec system (Becton Dickinson and Company, Franklin Lakes, NJ). DNA was extracted by use of a SeptiFast Lys kit on a MagNA Lyser platform (Roche Diagnostics), followed by silica matrix purification. We performed multiplex PCR using the LightCycler SeptiFast MGRADE test, version 2.0 (SeptiFast test; Roche, Basel, Switzerland), an in vitro nucleic acid amplification test for the detection and identification of DNA from bacterial and fungal microorganisms (see the SeptiFast master list in Table 1). Internal controls for the amplification step are included with the assay. Gram-positive and gram-negative organisms were targeted via the internal transcribed spacer between the 16S and 23S rRNA genes, whereas fungi were amplified by using the 18S and 5.8S rDNA sequence.
TABLE 1.
SeptiFast master list
| Organism group and organism |
|---|
| Gram-negative organisms |
| Escherichia coli |
| Klebsiella pneumoniae/Klebsiella oxytoca |
| Serratia marcescens |
| Enterobacter cloacae/Enterobacter aerogenes |
| Proteus mirabilis |
| Pseudomonas aeruginosa |
| Acinetobacter baumannii |
| Stenotrophomonas maltophilia |
| Gram-positive organisms |
| Staphylococcus aureus |
| Coagulase-negative Staphylococcus spp.a |
| Streptococcus pneumoniae |
| Streptococcus spp.b |
| Enterococcus faecalis |
| Enterococcus faecium |
| Fungi |
| Candida albicans |
| Candida glabrata |
| Candida krusei |
| Candida tropicalis |
| Candida parapsilosis |
| Aspergillus fumigatus |
Includes S. hominis subsp. novobiosepticus, S. pasteuri, S. warneri, S. cohnii subsp. urealyticum, S. hominis subsp. hominis, S. lugdunensis, S. cohnii subsp. cohnii, S. capitis subsp. ureolyticus, S. capitis subsp. capitis, S. caprae, S. saprophyticus, S. saprophyticus subsp. saprophyticus, S. xylosus, S. epidermidis, and S. haemolyticus.
Includes S. agalactiae, S. anginosus, S. bovis, S. constellatus, S. cristatus, S. gordonii, S. intermedius, S. milleri, S. mitis, S. mutans, S. oralis, S. parasanguinis, S. pneumoniae, S. pyogenes, S. salivarius, S. sanguinis, S. thermophilus, S. vestibularis, and S. viridans.
At the time of the initial ED visit, 1.5 ml of whole blood was drawn from each subject and stored at −70°C at Duke University. Blood for experimental analysis, including blood culture, was typically obtained within 120 to 180 min after the initial clinical assessment. The frozen samples were later thawed and processed according to the manufacturing guidelines and as published previously (19), and negative and reagent controls were included. SeptiFast identification software (SIS; version 1.1) was used to identify the organisms on the SeptiFast master list by melting curve analysis. All tests were performed and all results were interpreted by a single research laboratory technician (D.J.) blinded to the hypothesis. The processing time totaled about 6.5 h and consisted of 30 min for sample thawing, 90 min for DNA extraction, 45 min for PCR setup, and 3.5 h for the PCR itself, followed by 5 min for automated software analysis.
Statistical analysis.
Unless otherwise specified, the frequency (percentage) is reported for categorical variables, and mean (range) is presented for continuous variables. For the purposes of defining the operating characteristics of the PCR, we treated polymicrobial infections as a single event. In contrast, these polymicrobial isolates were counted as independent events and were analyzed qualitatively when the microbiology of PCR and blood culture were considered. The area under the receiver operating characteristic curve (AUC) was determined as previously recommended for binary test results (5). The time to blood culture positivity among those subjects with concordant versus discordant blood culture and PCR results was compared by using a two-sided t test. Comparison of the blood culture and PCR results with respect to the likelihood of infection, the severity of sepsis, the site of infection, and a history of antibiotic administration was assessed by McNemar's test. Statistical significance was set at a two-sided P value of <0.05. The kappa statistic was calculated to measure the level of agreement between blood culture and PCR results, where indicated. The likelihood of infection was recoded into a dichotomous outcome (infection present versus infection absent). “Infection present” comprised infection categories 1 (definite), 2 (probable), and 3 (possible). Infection categories 4 and 5 (no infection) defined the “infection absent” group. All analyses were performed with the SAS (Cary, NC) program (version 9.2).
RESULTS
Subject characteristics.
A total of 306 subjects were evaluated by both conventional blood culture techniques and the SeptiFast PCR. The demographic characteristics of the subject population are presented in Table 2. Infection categories 1 and 2 together included those with clinically determined infections. There were a combined 221 subjects in these two categories, whereas 42 were identified as having a possible or unknown infection (category 3) and 43 had no evidence of infection (categories 4 and 5) (Table 2). The most common sites of infection included the lung (n = 55), urinary tract (n = 46), and skin (n = 41), which together accounted for 66.0% of the identifiable infection sites. Twenty-two different etiologic agents were identified in 123 (40.2%) of the subjects, with the remainder having no identified etiologic agent. However, Staphylococcus aureus and Escherichia coli together accounted for the majority (56.9%) of the etiologic agents identified. The etiologic agents were identified by a variety of techniques, including culture of blood, urine, sputum, or other specimens; conventional PCR (e.g., for cytomegalovirus, Neisseria gonorrhoeae, Chlamydia trachomatis, etc.); antigen or serologic testing (e.g., for Streptococcus pneumoniae urinary antigen); and the use of special stains, among others, but without the use of multiplex PCR.
TABLE 2.
Baseline demographic information for the 306 enrolled subjects
| Characteristic | Value |
|---|---|
| Mean (range) age | 54.1 (18-97) |
| No. (%) of subjects | |
| Male | 168 (54.9) |
| Race | |
| Caucasian | 165 (53.9) |
| African American | 126 (41.2) |
| Unknown/other | 15 (4.9) |
| Mortality | 8 (2.6) |
| Length of stay (days) | |
| Mean (range) | 6.3 (1-111) |
| Median (IQRa) | 4 (2-7) |
| No. (%) of subjects with the following: | |
| Noninfected SIRS positive | 43 (14.1) |
| Sepsis | 184 (60.1) |
| Severe sepsis | 42 (13.7) |
| Septic shock | 37 (12.1) |
| No. (%) of subjects in the following infection category: | |
| 1 (confirmed) | 127 (41.5) |
| 2 (probable) | 94 (30.7) |
| 3 (possible) | 42 (13.7) |
| 4 (negative) | 4 (1.3) |
| 5 (negative) | 39 (12.7) |
| No. (%) of subjects with infection at the following site: | |
| Lung | 55 (18.0) |
| Urinary tract | 46 (15.0) |
| Skin | 41 (13.4) |
| Intra-abdominal | 25 (8.2) |
| Intravascular catheter | 16 (5.2) |
| Otherb | 32 (10.5) |
| Unknownc | 91 (29.7) |
| No. (%) of subjects infected with the following etiologic agentd: | |
| Staphylococcus aureus | 42 (34.1) |
| Escherichia coli | 28 (22.8) |
| Klebsiella pneumoniae/Klebsiella oxytoca | 11 (8.9) |
| Streptococcus pneumoniae | 9 (7.3) |
| Streptococcus species | 6 (4.9) |
| Cryptococcus neoformans | 2 (1.6) |
| Proteus mirabilis | 2 (1.6) |
| Pseudomonas aeruginosa | 2 (1.6) |
| Other etiologic agentse | 21 (17.1) |
| Unidentified | 183 |
IQR, interquartile range.
Includes bone (n = 9), central nervous system (n = 8), ear-nose-throat (n = 7), gynecologic (n = 6), and cardiac (n = 2).
Subjects in infection categories 4 and 5 are considered to have an “unknown” site of infection.
The percentage is based on the total number of etiologies identified. Each etiologic agent was identified by conventional microbiological techniques but excluding multiplex PCR.
Other etiologic agents include one isolate each of Apophysomyces elegans, Campylobacter jejuni/Campylobacter coli, Candida glabrata, Chlamydia trachomatis, Clostridium difficile, cytomegalovirus, Enterobacter species, Enterococcus faecalis, Fusobacterium nucleatum, Haemophilus influenzae, herpes simplex virus, influenza virus, Listeria monocytogenes, Morganella morganii, Neisseria gonorrhoeae, Peptostreptococcus species, Pneumocystis jirovecii, Rickettsia rickettsii, Salmonella enterica serovar Paratyphi, Serratia marcescens, and Staphylococcus species (coagulase-negative staphylococcus).
Performance characteristics of PCR versus those of blood culture.
In order to define the performance characteristics of PCR compared to those of blood culture, the likelihood of infection was dichotomized. Infection categories 1 to 3 were redefined as infection present, whereas infection categories 4 and 5 were redefined as infection absent (Table 3). By using clinical criteria as the reference standard, blood culture had a sensitivity of 0.25, a negative predictive value (NPV) of 0.18, and an AUC of 0.63 (when the data for contaminants were excluded). PCR had a sensitivity of 0.20, an NPV of 0.17, and an AUC of 0.60. When the data for contaminants were included in the analysis, the operating characteristics of blood culture were essentially unchanged: a sensitivity of 0.32, a specificity of 0.95, an NPV of 0.19, a positive predictive value (PPV) of 0.98, and an AUC of 0.64. No contaminants were detected by PCR. When PCR and blood culture were used in combination, the sensitivity was 0.30, the NPV was 0.19, and the AUC was 0.65 (excluding the data for contaminants). When blood culture was used as the reference standard, PCR had a sensitivity of 0.61, a specificity of 0.95, an NPV of 0.90, a PPV of 0.76, and an AUC of 0.78 (on the basis of the data for noncontaminant blood culture isolates that were represented on the PCR list in Table 1). The kappa statistic for the agreement of the blood culture and PCR results for these subjects was 0.59 (95% CI, 0.48 to 0.69).
TABLE 3.
Assay performance as a function of infection status, excluding data for contaminantsa
| Assay result | No. of subjects in which infection was: |
No. of subjects with the following blood culture result: |
||
|---|---|---|---|---|
| Present | Absent | Positive | Negative | |
| Blood culture positive | 66 | 0 | ||
| Blood culture negative | 197 | 43 | ||
| PCR positive | 53 | 0 | 40 | 13 |
| PCR negative | 210 | 43 | 26 | 227 |
| Blood culture and/or PCR positive | 79 | 0 | ||
| Blood culture and PCR negative | 184 | 43 | ||
Blood culture and PCR results are presented separately, followed by performance of a combined blood culture and PCR strategy. The performance of PCR versus that of blood culture as the reference standard is also presented.
Microbiological concordance/discordance.
The distribution of organisms identified by blood culture, PCR, or both methods is presented in Table 4. Statistical comparisons of the blood culture and PCR results stratified by organism were precluded by the relatively small number of results for any given organism. Nevertheless, there are several noteworthy observations. Many more contaminants were obtained by blood culture (20 isolates for 19 subjects) than by PCR (no contaminants). Fourteen of the blood culture contaminants consisted of coagulase-negative staphylococci. The majority of the Staphylococcus aureus isolates (17 of 24) were detected by both blood culture and PCR, with only a few more isolates added by either assay alone. This is in contrast to the results for Streptococcus pneumoniae, for which the results of both assays overlapped in only one case. Blood culture identified five additional cases of S. pneumoniae bacteremia, whereas PCR alone identified no additional cases. In cases in which the blood culture result was positive but the PCR result was negative, we determined if the discordance was due to the failure of the PCR, such as the inability of the primers to hybridize to the target site. We therefore retrieved the isolates from the cultures of blood from those subjects with a corresponding negative PCR result. Addition of these isolates from the solid culture media to the PCR resulted in positive results for the specified organism in 18 of 19 cases, suggesting that the originally negative PCR result was not intrinsic to the reaction itself. Finally, the specific probes used in this multiplex PCR platform were designed to detect the most common pathogens leading to nosocomial bloodstream infections. There were 26 different pathogens identified as causing bloodstream infections in this cohort, of which 20 are on the list of SeptiFast PCR-detectable organisms (77%).
TABLE 4.
Microorganisms identified by PCR and/or blood culturea
| Microorganism | No. of isolates identified by: |
||
|---|---|---|---|
| Both methods (n = 40) | PCR only (n = 24) | BC only (n = 52) | |
| Bacterial panel | |||
| Staphylococcus aureus | 17 | 2 | 5 |
| Staphylococcus species (CoNS) | 0 | 1 | 1 |
| Streptococcus pneumoniae | 1 | 0 | 5 |
| Streptococcus species | 1 | 1 | 2 |
| Enterococcus faecalis | 0 | 0 | 1 |
| Enterococcus faecium | 0 | 0 | 0 |
| Acinetobacter baumannii | 0 | 0 | 0 |
| Enterobacter aerogenes/Enterobacter cloacae | 2 | 2 | 0 |
| Escherichia coli | 11 | 5 | 7 |
| Klebsiella pneumoniae/Klebsiella oxytoca | 6 | 6 | 3 |
| Proteus mirabilis | 0 | 0 | 1 |
| Pseudomonas aeruginosa | 1 | 2 | 0 |
| Serratia marcescens | 1 | 1 | 0 |
| Stenotrophomonas maltophilia | 0 | 1 | 0 |
| Fungal panel | |||
| Candida albicans | 0 | 0 | 0 |
| Candida glabrata | 0 | 1 | 1 |
| Candida krusei | 0 | 1 | 0 |
| Candida parapsilosis | 0 | 0 | 0 |
| Candida tropicalis | 0 | 0 | 0 |
| Aspergillus fumigatus | 0 | 1 | 0 |
| Contaminants | |||
| Staphylococcus species (CoNS) | 0 | 0 | 14 |
| Streptococcus species | 0 | 0 | 2 |
| Enterococcus faecalis | 0 | 0 | 2 |
| Bacillus (non-Bacillus anthracis) | — | — | 1 |
| Propionibacterium acnes | — | — | 1 |
| Not in PCR menu | |||
| Citrobacter amalonaticus | — | — | 1 |
| Fusobacterium species | — | — | 1 |
| Kluyvera species | — | — | 1 |
| Listeria monocytogenes | — | — | 1 |
| Peptostreptococcus species | — | — | 1 |
| Salmonella species | — | — | 1 |
BC, blood culture; CoNS, coagulase-negative Staphylococcus; —, organisms not on the SeptiFast PCR-detectable master list.
Of the 53 subjects with positive PCR results, 6 had polymicrobial infections, for the isolation of a total of 64 organisms (Table 4). Of the 66 subjects with positive blood culture results (excluding contaminants), 3 had polymicrobial infections, for a total of 72 organisms. The lack of independence in identifying polymicrobial infections precludes their inclusion in an analysis of the test characteristics, as discussed in Materials and Methods. However, by taking these factors into account and excluding those blood culture isolates that are not on the PCR list, the rate of discordance is equalized (24 blood culture-negative and PCR-positive results versus 26 blood culture-positive and PCR-negative results).
An overall comparison of the concordance between the two tests is presented in Fig. 1A. Of the 40 subjects with concordant positive results, blood culture and PCR identified the same etiologic organism in 38 cases. There were two cases in which both assays were concordant positive but were concordant for different organisms. In the first, Staphylococcus aureus was detected by blood culture and Klebsiella pneumoniae/Klebsiella oxytoca was identified by PCR. The patient involved in this case had an indwelling central venous catheter in the femoral vein as well as osteonecrosis of the femoral heads with a possible associated infection (but the infection was not confirmed). The other case was a patient with a left ventricular assist device who clinically had a drive-line infection. PCR detected coagulase-negative Staphylococcus, whereas blood culture and urine culture detected Proteus mirabilis, considered to represent a coincident case of urinary tract infection.
FIG. 1.
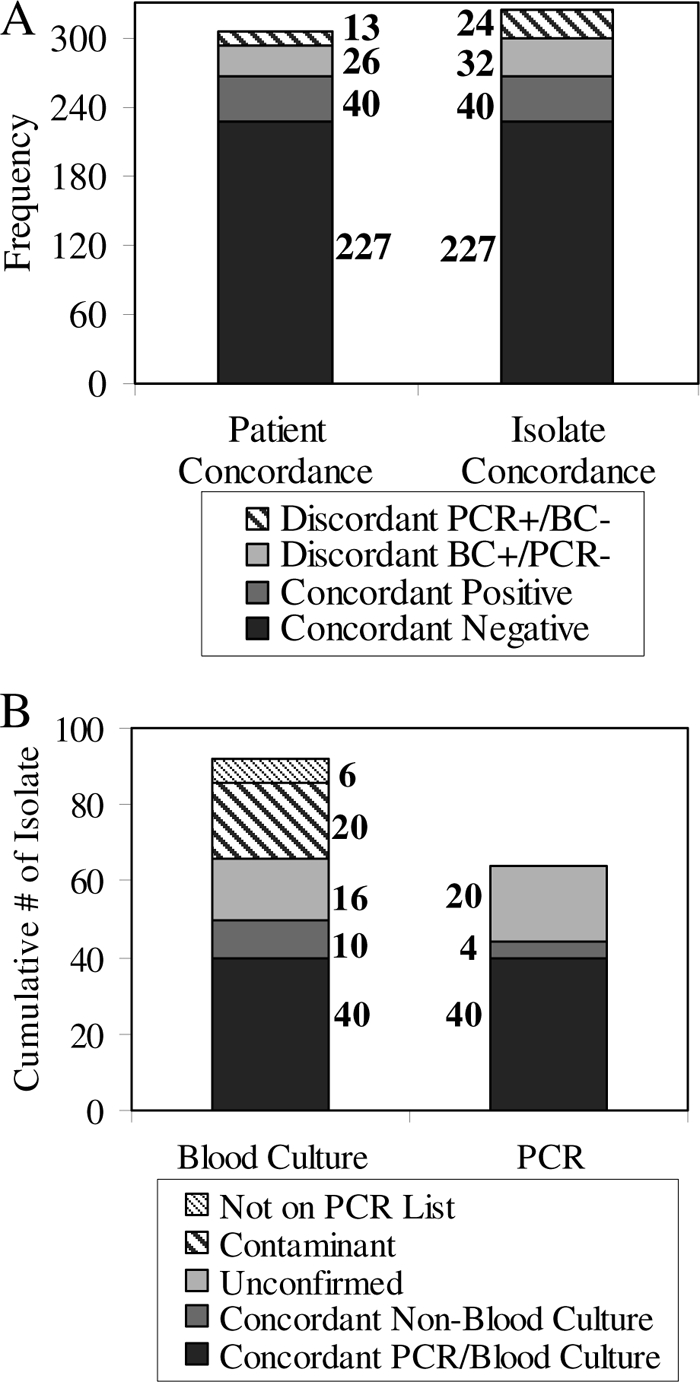
Positive and negative agreement of PCR and blood culture (BC) results. (A) “Patient concordance” defines concordant positive as a single subject with a positive blood culture result and a positive PCR result but not necessarily for the same organism or number of organisms. “Isolate concordance” defines concordant positive as a single instance in which the same organism was isolated by blood culture and PCR, which might occur more than once in a subject with a polymicrobial infection. Subjects with polymicrobial infections are counted only once under patient concordance, but each isolate is counted independently under isolate concordance, resulting in more isolates than the total number of subjects. (B) Results of blood culture and PCR reconciled with other available culture data. “Concordant non-blood culture” represents instances in which some other microbiological investigation (e.g., urine culture or antigen testing) identified the same infectious etiology that PCR or blood culture did.
The data in Fig. 1B expand on the issue of concordance by treating each isolate from polymicrobial infections as a separate event. Excluding contaminants and those organisms isolated by blood culture that are not on the SeptiFast master list, a total of 40 organisms were isolated by both assays. Blood culture added an additional 26 isolates, 10 of which could be confirmed by culture of other non-blood specimens (e.g., urine culture). PCR added an additional 24 isolates, 4 of which could be confirmed by culture of other non-blood specimens (termed “concordant non-blood culture” in Fig. 1B).
Time to positivity.
The time to blood culture positivity, defined as the time from the start of incubation to the time of the detection of growth in at least one blood culture, correlates with the severity of sepsis and the burden of infection (2, 16, 20). We assessed whether the time to blood culture positivity, as a proxy for the bacterial burden, correlated with the likelihood of blood culture and PCR concordance and found no significant difference. The mean time to positivity among those subjects with concordant blood culture and PCR results was 23 h 15 min, whereas it was 26 h 12 min for subjects with discordant blood culture and PCR results (a difference of 177 min; 95% CI, −583 to 936 min). Stratification by the Gram stain result did not reveal any significant differences between subjects with concordant and discordant results. The mean time to positivity among those with concordant results and gram-positive bacterial infections was 17 h 52 min, whereas it was 26 h 11 min for those with discordant results and gram-positive bacterial infections (a difference of 499 min; 95% CI, −516 to 1,514 min). The mean time to positivity for subjects with concordant blood culture and PCR results and gram-negative bacterial infections was 28 h 19 min, whereas it was 25 h 9 min for those with discordant results and gram-negative bacterial infections (a difference of 190 min; 95% CI, −1,484 to 1,104 min).
Correlation with infection and sepsis severity.
The results were stratified by the five-level infection category scheme defined in Materials and Methods and are presented in Table 5. Among those patients with a definite infection, which was determined on the basis of the blood culture data but blinded with respect to the PCR results, blood culture was superior to PCR (sensitivities, 0.52 and 0.36, respectively; P = 0.0004). Among the category 1 patients, the value of the kappa statistic was 0.50 (95% CI, 0.35 to 0.65), correlating to a moderate strength of agreement between blood culture and PCR. There were two category 3 infections (possible infection) in which PCR identified an organism. In one case, Serratia marcescens was identified by PCR in a 33-year-old woman who presented for the evaluation of fever, abdominal pain, and diarrhea after having an indwelling vascular catheter accessed. The etiology was never identified but was presumed to be gastroenteritis rather than a line infection. She was empirically treated with broad-spectrum antibiotics for the length of her 2-day hospitalization but was discharged without antibiotics. Her follow-up care did not reveal any evidence of a recurrence. The other case involved a 48-year-old man with increased drainage from a surgical drain 3 weeks following pancreatectomy, splenectomy, and cholecystectomy for chronic pancreatitis. PCR identified Staphylococcus aureus, but blood cultures and drain fluid cultures were negative.
TABLE 5.
Performance of blood culture versus performance of PCR stratified by infection category
| Infection category (no. of samples)a | Blood culture result | No. of samples with the following PCR result: |
% Concordance (kappa statistic value) | |
|---|---|---|---|---|
| Positive | Negative | |||
| Category 1, definite (127) | + | 40 | 26 | 74.8 (0.50) |
| − | 6 | 55 | ||
| Category 2, probable (94) | + | 0 | 0 | 94.7 (0) |
| − | 5 | 89 | ||
| Category 3, possible (42) | + | 0 | 0 | 95.2 (0) |
| − | 2 | 40 | ||
| Category 4, negative (4) | + | 0 | 0 | 100 (0) |
| − | 0 | 4 | ||
| Category 5, negative (39) | + | 0 | 0 | 100 (0) |
| − | 0 | 39 | ||
For category 1, P = 0.0004 by McNemar's test for agreement between blood culture and PCR stratified by infection category. McNemar's test could not be performed for categories 2 to 5 due to the absence of positive blood cultures.
The severity of sepsis was assessed for the subjects and was stratified by the assay results (Table 6). By definition, subjects in infection categories 4 and 5 did not have an infection and were therefore classified as noninfected SIRS positive. The majority of subjects with confirmed, probable, or possible infections (n = 263) met the criteria for sepsis (n = 184, or 70%). Forty-two (16%) subjects had severe sepsis, and 37 (14%) had septic shock. Blood culture and PCR performed similarly across sepsis severities. The value of the kappa statistic across all sepsis severities was 0.58 (95% CI, 0.46 to 0.70), which corresponds to a moderate strength of agreement between blood culture and PCR.
TABLE 6.
Performance of blood culture versus that of PCR stratified by severity of sepsis
| Sepsis severity (no. of samples)a | Blood culture result | No. of samples with the following PCR result: |
% Concordance (kappa statistic value) | |
|---|---|---|---|---|
| Positive | Negative | |||
| Noninfected SIRS positive (43) | + | 0 | 0 | 100 (0) |
| − | 0 | 43 | ||
| Sepsis (184) | + | 23 | 17 | 85.9 (0.55) |
| − | 9 | 135 | ||
| Severe sepsis (42) | + | 9 | 5 | 83.3 (0.60) |
| − | 2 | 26 | ||
| Septic shock (37) | + | 8 | 4 | 83.8 (0.61) |
| − | 2 | 23 | ||
For the sepsis group, P = 0.12; for the severe sepsis group, P = 0.26; for the septic shock group, P = 0.41. Statistical analysis for the noninfected SIRS-positive group is not available due to the absence of positive blood cultures.
Performance by site of infection.
The site of infection was determined for all subjects with definite, probable, or possible infections. An overall distribution of the sites involved is presented in Table 2. The performance of blood culture compared to that of PCR with respect to the adjudicated site of infection was not significantly different for any site. However, it is noteworthy that a majority of the subjects with infections of the urinary tract (25 of 46) or infections due to intravascular catheters (14 of 16) had a positive blood culture and/or a positive PCR result. A majority of the subjects in all other infection site categories had concordant negative blood culture and PCR results (126 of 153).
Prior antibiotic exposure.
Antibiotic administration can rapidly sterilize the bloodstream, whereas bacterial DNAemia may persist. We evaluated the effect of prior antibiotic administration on the performance characteristics of blood culture and PCR. There were 69 subjects (22.5% of 306) who received at least one dose of antibiotic before blood was collected for culture or PCR. Among those patients who did not previously receive a dose of antibiotics, the performance characteristics of blood culture and PCR were not statistically different. However, among the 69 patients who did previously receive antibiotics, blood culture alone was positive in 8 cases, PCR alone was positive in 2 cases, and both assays were concordant positive in 5 cases (P = 0.06).
DISCUSSION
Patients presenting to the ED with suspected sepsis can follow a spectrum of clinical courses, ranging from prompt resolution to substantial morbidity and death. Many factors that help determine which trajectory a patient takes have been identified. Vitally important to clinical recovery is the prompt administration of appropriate antibiotics (4, 10, 17, 18, 34). Currently, the initial choice of antibiotics is empirical and broad and covers the most likely etiologic agents. The major limitation to a more targeted antibiotic strategy is the lack of an available diagnostic method for the rapid and reliable identification of the etiologic agent. Blood culture is the most widely utilized technique for the diagnosis of bloodstream infections. However, despite its high degree of specificity, its sensitivity remains low and the mean time to positivity in our study was approximately 24 h and reached nearly 5 days in some cases. Antigen and serological testing is highly specific but has great variations in sensitivity, depending on the targeted organism. Furthermore, it is generally limited to a particular organism, and consequently, the suspected etiologic agent must be specified a priori. This study aimed to evaluate a real-time multiplex PCR assay as an adjunct to blood culture for the rapid identification of bloodstream infections.
The findings of this study suggest a high degree of concordance between the results of blood culture and PCR, with the two methods having similar operating characteristics (sensitivities, 0.25 and 0.20, respectively). This is most evident among those patients without infection and is manifested by high specificities: 95% for blood culture and 100% for PCR (when contaminants are included). Interestingly, of 127 subjects with definite infections, 55 had concordant negative results, whereas 40 had concordant positive results, for overall agreement in 74.8% of cases. The remaining 32 discordant cases are represented by 26 blood culture-positive and PCR-negative results and 6 blood culture-negative and PCR-positive results. Due to the significant number of infections that would be missed, it does not appear that this multiplex PCR assay could replace blood culture for the identification of bloodstream infections in patients presenting to the ED with suspected sepsis. However, the time to the identification of the etiologic agent is clearly much shorter with PCR than with blood culture. PCR also added diagnostic yield, with PCR detecting an additional 24 isolates that were missed by blood culture. The clinical relevance of such PCR-positive but blood culture-negative situations is an important question. Our results suggest that the organisms identified by PCR alone were clinically relevant on the basis of chart review and confirmation of the result by other microbiological diagnostic methods, such as urine culture and antigen testing. Potential reasons for the discordant results are numerous. We were unable to ensure that an adequate volume of blood for culture was collected, which could have contributed to the discordant blood culture-negative and PCR-positive results. Furthermore, the ability of PCR to detect DNA in sterilized blood, such as after antibiotic administration, could also have yielded these discordant results. However, an analysis of the PCR and blood culture results stratified by prior antibiotic use did not support this hypothesis. In contrast, discordant blood culture-positive and PCR-negative results could arise from the intrinsic variability of PCR, inhibiting factors present in the patient sample or other unidentified factors.
The cases in which PCR identified an infection are those in which the benefits of PCR can be most clearly realized. When there is concordance between PCR and blood culture results, the primary advantage to a PCR-based assay is the timeliness with which those results are available compared to the time to the availability of conventional blood culture results: 6 to 7 h and 24 to 72 h, respectively. This may allow the treating physician to narrow antibiotic coverage early in the course of treatment, potentially avoiding the toxicity and costs associated with the use of broad and empirical antimicrobial therapy. PCR can also identify an infection that was not being adequately treated. For example, empirical antibiotic choices typically do not treat fungal infections. However, the identification of fungemia by PCR can allow the physician to appropriately expand the antimicrobial regimen early in the course of disease. The potential clinical utility of the SeptiFast PCR to effect the clinical treatment and outcome was not addressed in this study, which used banked blood samples. Nevertheless, our results affirm the hypothesis that PCR and blood culture can identify a partially overlapping set of bloodstream infections, making PCR a useful adjunct to blood culture. Prospective clinical trials for the assessment of treatment and patient outcomes will be important to the further advancement of a multiplex PCR approach.
This study has several strengths in comparison to some previous evaluations of this multiplex PCR platform. First, patients were recruited from the ED on the basis of a suspicion of community-acquired sepsis. This relatively undifferentiated patient group is in contrast to the patient populations used in other studies, which enriched for patients with confirmed infection or hospital inpatients, who are at particularly high risk of developing bloodstream infections. Extrapolation of the findings from those studies could otherwise lead to context and spectrum biases. Furthermore, our use of blinded clinical adjudicators to determine infection status resulted in a better classification system. As a result, 14% of patients in this study were ultimately determined to have a noninfectious disease process, serving as a valuable internal control.
Second, another significant strength of this study is the way in which this new diagnostic tool was evaluated. Most published reports evaluating the SeptiFast PCR platform have tested multiple isolates from a small number of subjects, leading to spectrum bias, such that multiple samples are more likely to be drawn for sicker patients and these patients are subsequently more likely to have positive assay results (7, 22, 30, 32, 33). In contrast, our study is the largest to date in which subjects were identified prospectively and samples were collected from any given subject only once (21).
There are several potential limitations to this study. First, patients were recruited from the EDs of two tertiary-care centers, and consequently, the results may not be applicable to other clinical settings with different patient characteristics, resources, and laboratory procedures. A second potential limitation in interpreting the results of this study is the heterogeneity in the methods by which blood samples for culture were drawn. Although there are specific guidelines for how blood samples for culture should be drawn, including guidelines on skin cleansing, the volume per bottle, and the number of bottles, there is no way to ensure that all samples were collected according to those guidelines. This can have implications both for the sensitivity and for the specificity of blood culture, leading to an inadequate criterion bias, although it is more representative of actual clinical practice. Furthermore, although infection status is based on a multitude of factors, blood culture results are part of the criterion standard. As a result, incorporation bias likely plays a role in our observation that blood culture was statistically significantly better than PCR for those patients with definite infections. In addition, our clinical adjudication of infection includes viral, rickettsial, and other infectious etiologies that are expected to yield negative blood culture and PCR results, such as herpes simplex virus, influenza virus, Rickettsia rickettsii, and Clostridium difficile. This would tend to underestimate the performance characteristics of blood culture and PCR when the clinical infection criterion is used as the gold standard.
Current unanswered questions include the optimal assay conditions needed to minimize PCR inhibition by substances such as the heme in blood, the ethanol introduced during the extraction process, and other unidentified inhibitors found in blood (8, 28). In addition, the patient population most likely to benefit from this diagnostic tool needs to be better defined. Clinical utility may vary depending on the population prevalence of septicemia, with potentially lower levels being detected in the ED setting than in the ICU setting. Finally, the assays were performed with frozen samples and did not inform actual clinical practice. To that end, a recent study by Dierkes et al. reported on the effects of performing the SeptiFast-based PCR in parallel with blood culture with samples from a cohort of hospitalized patients (7). PCR was performed at the discretion of treating physicians without any subject enrollment or randomization, and the analysis was retrospective. Despite the biases that this might introduce, their findings are largely consistent with our own and serve as an important step toward assessing the directed clinical application of this technology.
In summary, PCR is a promising tool for the rapid identification of bloodstream infections as an adjunct to blood culture. Overall, the operating characteristics of PCR are similar to those of blood culture for the diagnosis of infection, but a significant number of additional isolates are identified by PCR. High-quality research into the real-clinical-time application of these results is a critical next step in the development of this technology.
Acknowledgments
We thank Debbie Freeman, Arturo Suarez, Christine Oien, and Sara Hoffman for their logistical support, without which this study could not have been carried out.
The cohort defined in this study draws from two other studies (ClinicalTrials.gov identifier NCT00258869 and grant/cooperative agreement number U38/CCU423095). This work was supported by NIH grant 5U01-AI066569-5 from the National Institute of Allergy and Infectious Diseases, as well as a research grant from Roche Molecular Sciences (to C.W.W.).
Lynette Waring was previously employed by Roche Molecular Sciences. Oliver Liesenfeld is employed by Roche Molecular Sciences. Charles B. Cairns is a consultant with a research contract with bioMérieux and is a consultant with Brahms. Christopher W. Woods is a consultant with a research contract with bioMérieux, a consultant with Cepheid Diagnostics, and a consultant with a research contract with Roche Molecular Diagnostics. None of the other authors has a potential conflict of interest to report.
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Angus, D. C., W. T. Linde-Zwirble, J. Lidicker, G. Clermont, J. Carcillo, and M. R. Pinsky. 2001. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit. Care Med. 29:1303-1310. [DOI] [PubMed] [Google Scholar]
- 2.Blot, F., E. Schmidt, G. Nitenberg, C. Tancrede, B. Leclercq, A. Laplanche, and A. Andremont. 1998. Earlier positivity of central-venous- versus peripheral-blood cultures is highly predictive of catheter-related sepsis. J. Clin. Microbiol. 36:105-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bone, R. C., R. A. Balk, F. B. Cerra, R. P. Dellinger, A. M. Fein, W. A. Knaus, R. M. Schein, and W. J. Sibbald. 1992. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee, American College of Chest Physicians/Society of Critical Care Medicine. Chest 101:1644-1655. [DOI] [PubMed] [Google Scholar]
- 4.Bryant, R. E., A. F. Hood, C. E. Hood, and M. G. Koenig. 1971. Factors affecting mortality of gram-negative rod bacteremia. Arch. Intern. Med. 127:120-128. [PubMed] [Google Scholar]
- 5.Cantor, S. B., and M. W. Kattan. 2000. Determining the area under the ROC curve for a binary diagnostic test. Med. Decis. Making 20:468-470. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, J. C., C. L. Huang, C. C. Lin, C. C. Chen, Y. C. Chang, S. S. Chang, and C. P. Tseng. 2006. Rapid detection and identification of clinically important bacteria by high-resolution melting analysis after broad-range ribosomal RNA real-time PCR. Clin. Chem. 52:1997-2004. [DOI] [PubMed] [Google Scholar]
- 7.Dierkes, C., B. Ehrenstein, S. Siebig, H. J. Linde, U. Reischl, and B. Salzberger. 2009. Clinical impact of a commercially available multiplex PCR system for rapid detection of pathogens in patients with presumed sepsis. BMC Infect. Dis. 9:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Espy, M. J., J. R. Uhl, L. M. Sloan, S. P. Buckwalter, M. F. Jones, E. A. Vetter, J. D. Yao, N. L. Wengenack, J. E. Rosenblatt, F. R. Cockerill III, and T. F. Smith. 2006. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin. Microbiol. Rev. 19:165-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everts, R. J., E. N. Vinson, P. O. Adholla, and L. B. Reller. 2001. Contamination of catheter-drawn blood cultures. J. Clin. Microbiol. 39:3393-3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freid, M. A., and K. L. Vosti. 1968. The importance of underlying disease in patients with gram-negative bacteremia. Arch. Intern. Med. 121:418-423. [PubMed] [Google Scholar]
- 11.Glickman, S. W., C. B. Cairns, R. M. Otero, C. W. Woods, E. L. Tsalik, R. J. Langley, J. C. van Velkinburgh, L. P. Park, L. T. Glickman, V. G. Fowler, S. F. Kingsmore, and E. P. Rivers. Disease progression in hemodynamically stable patients presenting to the emergency department with sepsis. Acad. Emerg. Med., in press. [DOI] [PMC free article] [PubMed]
- 12.Greisen, K., M. Loeffelholz, A. Purohit, and D. Leong. 1994. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J. Clin. Microbiol. 32:335-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurtler, V., and V. A. Stanisich. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142(Pt 1):3-16. [DOI] [PubMed] [Google Scholar]
- 14.Hollenberg, S. M., T. S. Ahrens, D. Annane, M. E. Astiz, D. B. Chalfin, J. F. Dasta, S. O. Heard, C. Martin, L. M. Napolitano, G. M. Susla, R. Totaro, J. L. Vincent, and S. Zanotti-Cavazzoni. 2004. Practice parameters for hemodynamic support of sepsis in adult patients: 2004 update. Crit. Care Med. 32:1928-1948. [DOI] [PubMed] [Google Scholar]
- 15.Jones, A. E., M. D. Brown, S. Trzeciak, N. I. Shapiro, J. S. Garrett, A. C. Heffner, and J. A. Kline. 2008. The effect of a quantitative resuscitation strategy on mortality in patients with sepsis: a meta-analysis. Crit. Care Med. 36:2734-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khatib, R., K. Riederer, S. Saeed, L. B. Johnson, M. G. Fakih, M. Sharma, M. S. Tabriz, and A. Khosrovaneh. 2005. Time to positivity in Staphylococcus aureus bacteremia: possible correlation with the source and outcome of infection. Clin. Infect. Dis. 41:594-598. [DOI] [PubMed] [Google Scholar]
- 17.Kollef, M. H., G. Sherman, S. Ward, and V. J. Fraser. 1999. Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients. Chest 115:462-474. [DOI] [PubMed] [Google Scholar]
- 18.Kreger, B. E., D. E. Craven, and W. R. McCabe. 1980. Gram-negative bacteremia. IV. Re-evaluation of clinical features and treatment in 612 patients. Am. J. Med. 68:344-355. [DOI] [PubMed] [Google Scholar]
- 19.Lehmann, L. E., K. P. Hunfeld, T. Emrich, G. Haberhausen, H. Wissing, A. Hoeft, and F. Stuber. 2008. A multiplex real-time PCR assay for rapid detection and differentiation of 25 bacterial and fungal pathogens from whole blood samples. Med. Microbiol. Immunol. 197:313-324. [DOI] [PubMed] [Google Scholar]
- 20.Liao, C. H., Y. T. Huang, F. Y. Chu, T. H. Lin, and P. R. Hsueh. 2008. Lack of increase in time to blood culture positivity in a patient with persistent methicillin-resistant Staphylococcus aureus bacteremia predicts failure of antimicrobial therapy. J. Microbiol. Immunol. Infect. 41:355-357. [PubMed] [Google Scholar]
- 21.Louie, R. F., Z. Tang, T. E. Albertson, S. Cohen, N. K. Tran, and G. J. Kost. 2008. Multiplex polymerase chain reaction detection enhancement of bacteremia and fungemia. Crit. Care Med. 36:1487-1492. [DOI] [PubMed] [Google Scholar]
- 22.Mancini, N., D. Clerici, R. Diotti, M. Perotti, N. Ghidoli, D. De Marco, B. Pizzorno, T. Emrich, R. Burioni, F. Ciceri, and M. Clementi. 2008. Molecular diagnosis of sepsis in neutropenic patients with haematological malignancies. J. Med. Microbiol. 57:601-604. [DOI] [PubMed] [Google Scholar]
- 23.Martin, G. S., D. M. Mannino, S. Eaton, and M. Moss. 2003. The epidemiology of sepsis in the United States from 1979 through 2000. N. Engl. J. Med. 348:1546-1554. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen, H. B., E. P. Rivers, F. M. Abrahamian, G. J. Moran, E. Abraham, S. Trzeciak, D. T. Huang, T. Osborn, D. Stevens, and D. A. Talan. 2006. Severe sepsis and septic shock: review of the literature and emergency department management guidelines. Ann. Emerg. Med. 48:28-54. [DOI] [PubMed] [Google Scholar]
- 25.Olsen, G. J., D. J. Lane, S. J. Giovannoni, N. R. Pace, and D. A. Stahl. 1986. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 40:337-365. [DOI] [PubMed] [Google Scholar]
- 26.Pitts, S., R. Niska, J. Xu, and C. Burt. 2008. National Hospital Ambulatory Medical Care Survey: 2006 emergency department summary, vol. 7. National Center for Health Statistics, Hyattsville, MD. [PubMed]
- 27.Rivers, E., B. Nguyen, S. Havstad, J. Ressler, A. Muzzin, B. Knoblich, E. Peterson, and M. Tomlanovich. 2001. Early goal-directed therapy in the treatment of severe sepsis and septic shock. N. Engl. J. Med. 345:1368-1377. [DOI] [PubMed] [Google Scholar]
- 28.Valentine-Thon, E. 2002. Quality control in nucleic acid testing—where do we stand? J. Clin. Virol. 25(Suppl. 3):S13-S21. [DOI] [PubMed] [Google Scholar]
- 29.Van Burik, J.-A., D. Myerson, R. W. Schreckhise, and R. A. Bowden. 1998. Panfungal PCR assay for detection of fungal infection in human blood specimens. J. Clin. Microbiol. 36:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Lilienfeld-Toal, M., L. E. Lehmann, A. D. Raadts, C. Hahn-Ast, K. S. Orlopp, G. Marklein, I. Purr, G. Cook, A. Hoeft, A. Glasmacher, and F. Stuber. 2009. Utility of a commercially available multiplex real-time PCR assay to detect bacterial and fungal pathogens in febrile neutropenia. J. Clin. Microbiol. 47:2405-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinstein, M. P., M. L. Towns, S. M. Quartey, S. Mirrett, L. G. Reimer, G. Parmigiani, and L. B. Reller. 1997. The clinical significance of positive blood cultures in the 1990s: a prospective comprehensive evaluation of the microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin. Infect. Dis. 24:584-602. [DOI] [PubMed] [Google Scholar]
- 32.Wellinghausen, N., A. J. Kochem, C. Disque, H. Muhl, S. Gebert, J. Winter, J. Matten, and S. G. Sakka. 2009. Diagnosis of bacteremia in whole-blood samples by a commercial universal 16S rRNA gene-based PCR and sequence analysis. J. Clin. Microbiol. 47:2759-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westh, H., G. Lisby, F. Breysse, B. Boddinghaus, M. Chomarat, V. Gant, A. Goglio, A. Raglio, H. Schuster, F. Stuber, H. Wissing, and A. Hoeft. 2009. Multiplex real-time PCR and blood culture for identification of bloodstream pathogens in patients with suspected sepsis. Clin. Microbiol. Infect. 15:544-551. [DOI] [PubMed] [Google Scholar]
- 34.Young, L. S., W. J. Martin, R. D. Meyer, R. J. Weinstein, and E. T. Anderson. 1977. Gram-negative rod bacteremia: microbiologic, immunologic, and therapeutic considerations. Ann. Intern. Med. 86:456-471. [DOI] [PubMed] [Google Scholar]


