Abstract
Human noroviruses (NoVs) of genogroup II, genotype 4 (GII.4) are the most common strains detected in outbreaks of acute gastroenteritis worldwide. To gain insight into the epidemiology and genetic variation of GII.4 strains, we analyzed 773 NoV outbreaks reported to the CDC from 1994 to 2006. Of these NoV outbreaks, 629 (81.4%) were caused by GII viruses and 342 (44.2%) were caused by GII.4 strains. The proportion of GII.4 outbreaks increased from 5% in 1994 to 85% in 2006, but distinct annual differences were noted, including sharp increases in 1996, 2003, and 2006 each associated with newly emerging GII.4 strains. Sequence analysis of the full-length VP1 gene of GII.4 strains identified in this study and from GenBank segregated these viruses into at least 9 distinct subclusters which had 1.3 to 3.2% amino acid variation between strains in different subclusters. We propose that GII.4 subclusters be defined as having >5% sequence variation between strains. Our data confirm other studies on the rapid emergence and displacement of highly virulent GII.4 strains.
Noroviruses (NoVs), members of the genus Norovirus of the family Caliciviridae (13), are the leading cause of acute gastroenteritis (AGE) worldwide. In the United States, 81 to 96% of the nonbacterial AGE outbreaks with fecal specimens sent to the Viral Gastroenteritis Unit at the Centers for Disease Control and Prevention (CDC) were attributable to NoVs (4, 11, 12). In Europe, surveillance data from 10 countries demonstrated that >85% of all AGE outbreaks were associated with NoVs (22), and in Japan, NoV outbreaks were responsible for 93 to 97% of all viral gastroenteritis outbreaks (Infectious Disease Surveillance Center [IDSC], National Institute of Infectious Diseases [NIID] website http://idsc.nih.go.jp/iasr/iasrcnt-e.html#srsv-e).
In the United States, NoVs account for an estimated 30 to 50% of all food-borne outbreaks (7a, 47). Norovirus outbreaks occur in a wide variety of settings year-round but are particularly common and protracted in the winter months in closed settings (e.g., hospitals and long-term care facilities [LTCFs]) where transmission is predominantly from person to person. Immunity to NoVs is poorly understood due to the lack of a cell culture system to measure neutralizing antibodies (9). Data from volunteer challenge studies demonstrate that immunity to NoVs seems to be short-term, and reinfections with heterogeneous and homologous strains have been documented (13). However, these conclusions are suspect because the challenge dose of virus used may have been 4 to 6 log10 units higher than the infectious dose (39).
NoVs have a 7.5- to 7.7-kb single-stranded genome of positive-sense RNA which contains three open reading frames (ORFs). ORF2 codes for the major capsid protein (VP1). NoVs are classified into five distinct genogroups (genogroup I [GI] to genogroup V [GV]) on the basis of VP1 sequencing analysis. These genogroups are further subdivided into at least 32 genetic clusters (13, 24, 44, 48). Of these, GI, GII, and GIV strains have been detected in humans, and viruses belonging to GII, genetic cluster or genotype 4 (GII.4) have emerged as the predominant strain over the last decade (4, 5, 10, 14, 15, 21, 23, 28, 33, 43, 46). The global distribution of a distinct GII.4 strain was first recognized in 1995 and 1996 (28). Between 1997 and 2002, NoV activity was moderate to low with different strains cocirculating without a distinct epidemic strain (6, 22). In 2002, 2004, and 2006, NoV outbreaks increased sharply and were associated with the emergence of new GII.4 strains (5, 7, 21, 46).
To understand the epidemiologic patterns of GII.4 outbreaks in the United States and to determine the genetic variation of GII.4 NoV strains over time, we systematically analyzed NoV outbreaks and specimens reported to the CDC from 1994 to 2006.
(This work was presented in part at the Third International Calicivirus Meeting, 10 to 13 November 2007, Cancun, Mexico, and at the 26th Annual Meeting of the American Society for Virology, 14 to 18 July 2007, Corvallis, OR.)
MATERIALS AND METHODS
Outbreak data.
We analyzed epidemiologic data (setting, transmission route, and onset date) for all confirmed NoV outbreaks for which specimens were sent to the CDC from 1994 through 2006. A confirmed NoV outbreak was defined as one from which at least 2 stools tested positive for NoV by reverse transcription-PCR (RT-PCR) (2, 4, 11, 27) or real-time TaqMan RT-PCR (40). Epidemiologic data of NoV-positive outbreaks were analyzed for statistical significance by using the chi-square test (SAS, version 9.1).
Laboratory data.
All NoV-positive specimens had previously been typed into genogroup or genotype as described previously (4, 11, 12, 27, 28). Strains that had been genotyped as GII.4 by sequence analyses of partial regions of the polymerase gene or VP1 gene (2, 4, 42) were included in the study for further characterization.
RT-PCR and sequencing.
We retested the selected specimens by previously described standard methods. In brief, viral RNA was extracted from a 10% stool suspension using the NucliSens automated nucleic acid extraction system (BioMérieux) (4). A 322-nucleotide (nt) region of the 5′ end of the VP1 gene of all strains was amplified with primer set Mon381-Mon383 (4, 11, 27). The full-length VP1 gene of 11 strains each representing a different GII.4 subcluster (Table 1 ) was also amplified using the Qiagen One-Step RT-PCR kit (Qiagen Inc., Valencia, CA) with forward primer DPZ-F291 (5′-ACT CAG ACA AAT GTA TTG GAC-3′) and reverse primer DPZ-R2457 (5′-ACC CTC TAG GAG CAT CGC CTG-3′). RT-PCR products were purified from 2% agarose gels and sequenced on an automated DNA sequencer (model 3130xl; Applied Biosystems, Foster City, CA) using the BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems).
TABLE 1.
Classification of 198 unique GII.4 sequence variants and proposed GII.4 subclusters based on full-length VP1 sequences
| GII.4 subclustera | GII.4 strains identified in this study |
GII.4 strains in GenBank |
||||||
|---|---|---|---|---|---|---|---|---|
| Representative strainb (GenBank accession no.) | No. of unique sequence variantsc | Yr(s) strains circulated | No. of GenBank hitsd | Yr(s) strains circulated | Range of amino acid variation (%)e (VP1) | Prototype strainf (GenBank accession no.) | Other strain(s) (reference) | |
| Bristol | 5 | 1987-1994 | 0.2-3.3 | Bristol-GBR1993 (X76716) | Lordsdale (23), Camberwell (20), and MD145-12 (14) | |||
| Richmond | Richmond-USA1994 (EU078406) | 2 | 1994 | 0 | N/A | |||
| 95/96-US | Houston-USA1995 (EU078407) | 40 | 1995-2002 | 43 | 1995-2002 | 0.2-3.3 | Grimsby-GBR1995 (AJ004864) | Grimsby (23) |
| Wellington-USA1995 (FJ411169) | 20 | 1995-1999 | 1996 variant (33) | |||||
| Erfurt | 2000 | N/A | Erfurt007-ESP01 (X76716) | |||||
| Henry | Henry-USA2000 (FJ411170) | 14 | 2000-2005 | 3 | 2000-2002 | 0.7-3.2 | AST-ESP2001 (AJ583672) | |
| Lonaconing-USA2001 (EU078408) | 2 | 2001 | ||||||
| Farmington Hills | Warren-USA2002 (EU078409) | 83 | 2002-2005 | 37 | 2002-2004 | 0.2-4.1 | Langen-DEU2002 (AY485642) | 2002 variant (33) |
| Hunter | Cumberland-USA2004 (EU078414) | 12 | 2003-2006 | 14 | 2004-2006 | 0.2-2.0 | NL317-NLD2004 (AY883096) | 2004 variant (33) |
| Chiba | QM2CS-USA2004 (EU078411) | 2 | 2004-2005 | 18 | 2004-2006 | 0.2-2.2 | Chiba04-JPN2004 (AB220925) | 2003 Asia (32) |
| Sakai (19) | ||||||||
| Yerseke | CLCS-USA2006 (EU078419) | 11 | 2005-2006 | 5 | 2006→ | 0.2-1.9 | Yerseke38-NLD2006 (EF126963) | Laurens (7), 2006a variant (33) |
| Osaka | SSCS-USA2005 (FJ411171) | 2 | 2005-2006 | 1 | 2005-2007 | 1.3 | Osaka07-JPN2007 (AB434770) | |
| Den Haag | OSDCS-USA2006 (EU078417) | 10 | 2006 to present | 15 | 2006→ | 0.2-1.7 | DenHaag89-NLD2006 (EF126965) | Minerva (7), 2006b variant (33) |
| All | 11 | 198 | 1994-2006 | 141 | 1987-2007 | 10.6 | ||
Subclusters were defined as strains having >5% amino acid variation in VP1. The names of subclusters were adopted either using the well-accepted names in publications or as the first publicly available sequence (see footnote f below).
Strains identified and sequenced in this study (Fig. 5).
Sequence variants were defined as strains having >1-nucleotide (nt) difference in a 277-nt region of VP1 (4).
GenBank BLAST search was done on 25 July 2008 using full-length VP1 sequences of each 11 strains identified in this study. Sequences which aligned with no gaps and that were unique per country per year were selected for subcluster variation and phylogenetic analysis. The numbers of hits do not include the strains detected in this study.
Sequences used to calculate the amino acid variation for each subcluster included the 11 US strains but excluded any identical sequences. Tentative subclusters Richmond and Erfurt include only one strain, and thus, amino acid variation is not applicable (N/A).
Prototype strains were defined as strains of which the VP1 sequences were the first publicly available.
Sequence and pairwise distance analysis.
All sequences generated in this study were edited with Sequencher 4.8 (Gene Codes Corporation, Ann Arbor, MI) and analyzed with several GCG programs (Wisconsin Package, version 11.1.2; Accelrys Inc.). The FastA program was used to search for the closest sequence match to identify unique sequences, the Distances program was used to calculate the uncorrected pairwise distances and to draw the phylogenetic trees, and the PAUP Search was used for bootstrapping the consensus tree.
Nucleotide sequences that were not identical to any other sequences in our database were assigned a unique identification number (sequence identification [SeqID]). Strains that had more than 85% sequence similarity to GII.4 reference strains (i.e., Bristol or Farmington Hills) were initially classified as GII.4 (28, 48) and included for further analysis. Phylogenetic trees were displayed with TreeView software (30).
GenBank BLAST search for additional NoV GII.4 sequences.
To compare GII.4 sequences from our study with strains that have been detected globally, a GenBank BLAST search was conducted using the 11 full-length GII.4 VP1 sequences that were generated in this study.
Nucleotide sequence accession numbers.
The VP1 sequences identified in this study were deposited in GenBank and have been assigned accession numbers as shown in Table 1.
RESULTS
NoV outbreaks.
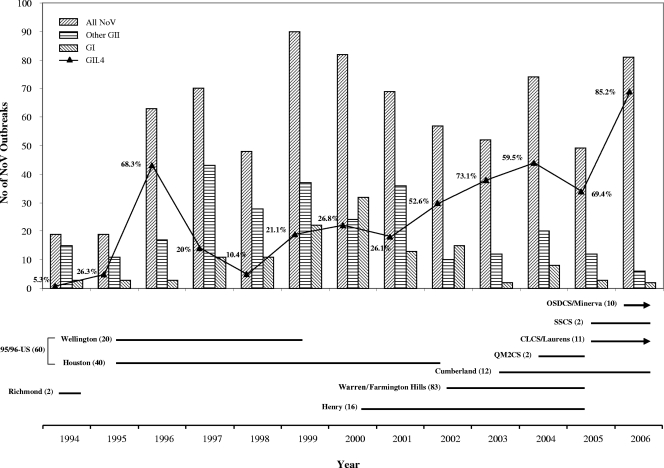
Between 1994 and 2006, CDC received fecal specimens from 997 outbreaks of acute gastroenteritis that had been submitted by state and local health departments for NoV testing and genotyping. Of these, 773 (77.5%) outbreaks were confirmed as NoV positive by conventional or real-time RT-PCR with at least two positive specimens per outbreak. The number of NoV-positive outbreaks varied from 19 to 90 each year with the proportion of GI outbreaks ranging between 2 and 15 per year except for 1999 (n = 22; 24.4%) and 2000 (n = 32; 39.0%) when a significant increase of the number of GI outbreaks was reported. The number of GII outbreaks ranged from 16 to 77 per year and accounted for 58 to 96% of all NoV outbreaks. Of the 773 NoV outbreaks, only two outbreaks (one in 1999 and one in 2000) were typed as GIV. Among the GII outbreaks, the number of GII.4 outbreaks ranged from 1 to 69 by year with 3 remarkable seasonal increases in 1995 to 1996 (5 to 43), 2002 to 2003 (18 to 44), and 2005 to 2006 (34 to 69). Since 2002, the proportion of GII.4 outbreaks never dropped below 52% (2005) and steadily increased to 85.2% in 2006 (Fig. 1).
FIG. 1.
Distribution of NoV outbreaks reported to the CDC between 1994 and 2006. (Top) Outbreaks (n = 773) were grouped by year and divided into “all NoV outbreaks,” “GI,” “GII.4,” and “other GII.” The solid black line connects the percentages of GII.4 outbreaks for the years shown. (Bottom) The horizontal lines refer to the years that the different GII.4 subclusters were detected. Lines with an arrowhead indicate that viruses belonging to that GII.4 subcluster have been circulating beyond 2006. The name chosen for each subcluster refers to the earliest strain detected in the United States, and the number (in parentheses) refers to how many sequence variants have been detected.
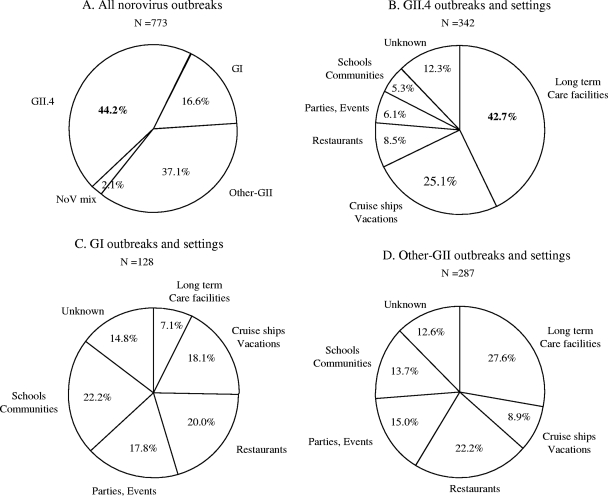
Overall, 342 (44.2%) of the 773 NoV outbreaks were typed as GII.4, 287 (37.1%) were typed as other GII strains, 128 (16.6%) were typed as GI, and 16 (2.1%) had mixed genotypes (Fig. 2A).
FIG. 2.
Epidemiologic characteristics of 773 NoV outbreaks in the United States between 1994 and 2006. Distribution by genogroup and genetic cluster (A) and by setting of GII.4 outbreaks (n = 342) (B), GI outbreaks (n = 128) (C), and other GII outbreaks (n = 287) (D).
Epidemiologic characteristics of GII.4 outbreaks.
GII.4 outbreaks occurred more frequently in long-term care facilities (LTCFs) (e.g., nursing homes) (42.7%) and cruise ships (25.1%) (P < 0.001) than in other settings, whereas GI and other GII viruses were more often associated with outbreaks in restaurants and parties (37.8 and 37.2%, respectively) than in LTCFs (7.1 and 27.6%, respectively) (Fig. 2B, C, and D) (P < 0.001).
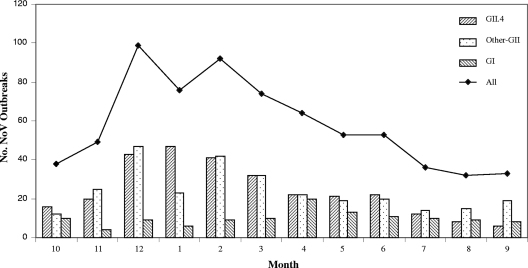
GII outbreaks (including GII.4) demonstrated a marked seasonal peak during the cooler months (December to March), whereas the GI outbreaks occurred year-round and did not exhibit any apparent seasonality (Fig. 3).
FIG. 3.
Seasonal distribution of NoV outbreaks by month (all [n = 708], GI [n = 119], other GII [n = 299], and GII.4 [n = 290]) reported to the CDC from 1994 to 2006. The months are indicated by the numbers 1 to 12 below the graph (e.g., 1 for January, 2 for February, etc.).
Prevalence and frequency of GII.4 subclusters in the United States from 1994 to 2006.
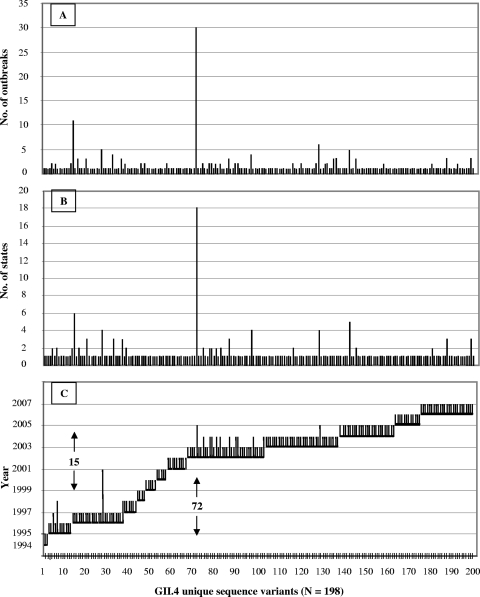
A total of 726 sequences were obtained from 342 GII.4 outbreaks. Among these, 198 unique sequences (differing by at least 1 nt) were identified, and these sequences were plotted by frequency, geographic distribution, and year (Fig. 4A to C).
FIG. 4.
Dynamic activities of GII.4 sequence variants in the United States from 1994 to 2006. Each of the GII.4 sequence variants (n = 198) was plotted against the number of outbreaks (A), number of states (B), and years (C). Each vertical line on the x axis represents one sequence variant. The numbers 15 and 72 refer to sequence variants 15 and 72 on the x axis.
Overall, 66 (33.3%) unique GII.4 sequences were detected from 1994 to 2001 and 132 (66.7%) unique GII.4 sequences were detected from 2002 to 2006 (Fig. 4C). The largest number of unique GII.4 sequences was detected in 1996 (n = 24), 2002 and 2003 (n = 95), and 2006 (n = 25). In contrast, during 1997 to 2001 when >50% of the NoV outbreaks were associated with non-GII.4 viruses, the number of unique GII.4 sequences ranged from 4 to 9 per year and they did not cause many outbreaks or spread widely.
The majority of the unique GII.4 sequences were associated with only one outbreak (80%) (Fig. 4A), in one state (88%) (Fig. 4B), and in one year (92%) (Fig. 4C). Some of the GII.4 sequences caused more outbreaks, had a larger geographic distribution, and circulated for a longer period of time. For example, GII.4 sequence 15 was associated with 11 outbreaks in 6 states but circulated only in 1996, whereas sequence 72, which first emerged in 2002, was detected in 30 outbreaks in 18 states during 2002 to 2005 (Fig. 4A to C).
Strain diversity and distribution of NoV GII.4 subclusters in the United States.
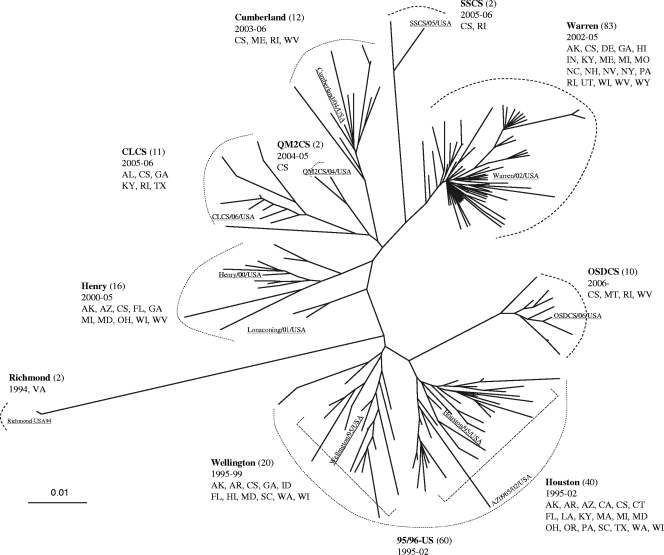
An unrooted phylogenetic distance tree was generated based on analysis of 198 unique partial ORF2 sequences from GII.4 strains collected during 1994 to 2006 in the United States (Fig. 5). These sequences fell into 9 subclusters of which 3 subclusters (Richmond, QM2CS, and SSCS) consisted of only 2 strains. Viruses of most subclusters circulated for 2 to 3 years before becoming extinct except for viruses in the subcluster 95/96-US which were detected for up to 7 years from 1995 to 2002 (28). The 60 unique GII.4 sequence variants formed 2 separate groups (Houston and Lonaconing) with <5% sequence diversity (Fig. 5 and Table 1). The GII.4 sequence variants detected after 2001 (n = 136) grouped into 7 subclusters. Viruses from subcluster Warren/Farmington Hills, which were detected from 2002 to 2005, demonstrated the greatest number of unique sequences (n = 83) and caused a remarkable increase of GII.4 outbreaks in 2002 (Fig. 1) (46). Viruses from subclusters Henry and Cumberland included 16 and 12 unique sequences, respectively, which were detected in 22 GII.4 outbreaks in 2000 to 2005 (Henry) and 13 GII.4 outbreaks in 2003 to 2006 (Cumberland). Viruses belonging to subclusters CLCS/Laurens and OSDCS/Minerva were responsible for a significant increase in the number of GII.4 outbreaks in 2006 (Fig. 1) (7). Unique GII.4 sequences that grouped into subclusters QM2CS and SSCS circulated for only 1 to 2 years in 2001 and 2005 to 2006, respectively, and did not cause a large number of outbreaks.
FIG. 5.
Genetic diversity of GII.4 sequence variants causing outbreaks in the United States from 1994 to 2006. An unrooted tree was generated on the basis of 198 unique GII.4 sequence variants (of a 277-nt region of VP1). Each of the subclusters was assigned a name of the representative strain that was first detected and selected for complete VP1 sequencing (underlined). The number of sequence variants is listed in parentheses; the years the viruses circulated and the states where the strains were detected are also given. The scale bar labeled 0.01 indicates the average distance by nucleotide differences (as a percentage). The states are indicated by the two-letter state codes as follows: Alabama (AL), Alaska (AK), Arkansas (AR), Arizona (AZ), California (CA), Colorado (CO), Connecticut (CT), Delaware (DE), Florida (FL), Georgia (GA), Illinois (IL), Indiana (IN), Kentucky (KY), Louisiana (LA), Idaho (ID), Massachusetts (MA), Maryland (MD), Maine (ME), Missouri (MO), Michigan (MI), Minnesota (MN), Montana (MT), North Carolina (NC), New Hampshire (NH), Nevada (NV), New Mexico (NM), New York (NY), Ohio (OH), Oregon (OR), Pennsylvania (PA), Rhode Island (RI), South Carolina (SC), Texas (TX), Tennessee (TN), Utah (UT), Virginia (VA), Washington (WA), Washington DC (DC), Wisconsin (WI), West Virginia (WV), Wyoming (WY). CS, cruise ship.
Phylogenetic relationship among GII.4 strains.
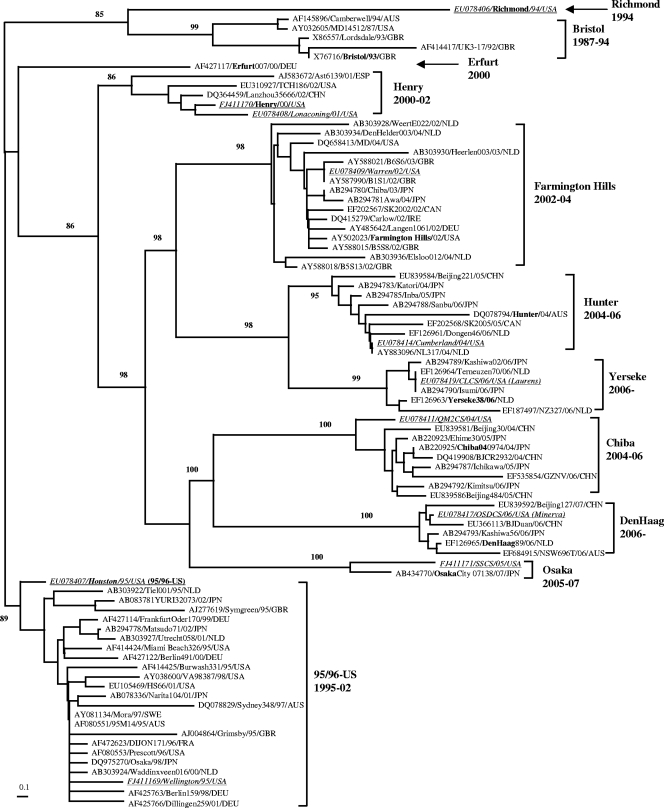
A BLAST search of the GenBank database using the 11 strains representing different GII.4 subclusters yielded 1,043 sequences of which 141 were unique at the nucleotide level. Of these, 73 strains differed at the amino acid level and were used to assess the phylogenetic relationship of the GII.4 strains detected in this study with the strains circulating in other parts of the world. Strains in each subcluster had a maximum amino acid variation of 1.3 to 4.1% (Table 1). The strain diversity among all GII.4 VP1 sequences was 10.6%, which is <15%, the cutoff for viruses classified into a cluster (48). Thus, all strains belong to the GII.4 cluster and can be grouped into 9 distinct subclusters with robust bootstrap values (85 to 100%), including Bristol, US95/96, Henry, Farmington Hills, Hunter, Chiba, Yerseke, Osaka, and Den Haag (Fig. 6). Two strains (Richmond and Erfurt007) could not be grouped in any of the 9 subclusters and tentatively represent additional subclusters. Viruses in subclusters US95/96 and Farmington Hills were associated with sequence variants 15 and 72, respectively (Fig. 4), which caused pandemics in 1996 and 2002 (21, 28, 46); viruses in Hunter and Chiba subclusters caused outbreaks during 2004 to 2006 with Hunter viruses detected around the globe (5), whereas Chiba viruses were mainly detected in Asia. Viruses in subclusters Yerseke and Den Haag both had a global distribution in 2006 (7, 31, 41). In contrast, viruses in subclusters Henry and Osaka were only found in 3 (China, Spain, and United States) and 2 (Japan, United States) countries, respectively. Overall, GII.4 viruses of a particular subcluster circulated between 2 and 6 years.
FIG. 6.
Phylogenetic relationships of GII.4 strains and proposed GII.4 subcluster nomenclature. The phylogenetic tree was based on 84 full-length VP1 amino acid sequences, including 11 representative sequences (underlined) characterized in this study and 73 sequences from GenBank. Subclusters are named after the first strain for which the VP1 sequence was publicly available in GenBank. The time span (in years) that strains in each subcluster circulated is listed. Sequence name was formatted as follows: GenBank accession number/strain name/year/country. Countries are shown as three-letter country codes as follows: Australia (AUS), China (CHN), Canada (CAN), Germany (DEU), France (FRA), Great Britain (GBR), Ireland (IRE), Japan (JPN), The Netherlands (NLD), Sweden (SWE), Spain (ESP), and the United States (USA).
DISCUSSION
We systematically analyzed the epidemiology and strain diversity of GII.4 NoV outbreaks reported to the CDC from 1994 to 2006. Overall, the proportion of GII.4 outbreaks increased from 5.3% in 1994 to 85.2% in 2006, but distinct annual differences with sharp increases in pandemic years were noted. The GII.4 outbreaks were most often identified in LTCFs (43%) and cruise ships (25%) and had distinct winter seasonality, whereas GI viruses caused outbreaks throughout the year, which also has been reported in Europe (19).
Comparison of GII.4 sequences circulating in the United States with sequences submitted to GenBank demonstrated that most GII.4 strains have a worldwide distribution. Phylogenetic analysis of full-length VP1 sequences demonstrated that all known GII.4 viruses can be grouped into at least 9 distinct subclusters (Fig. 6) with tentatively 2 additional subclusters (Richmond and Erfurt) represented by only one full-length VP1 sequence. On the basis of the maximum diversity within each subcluster (1.3 to 4.1%) (Table 1), we propose that 5% of amino acid variation in the full-length VP1 be the cutoff for the classification of GII.4 subclusters. For naming of the subclusters, we employed the same approach as has been used for naming of genotypes, i.e., the first publicly available VP1 protein sequence (48), which was adopted by participants of the Third International Calicivirus Meeting, Cancun, Mexico, 10 to 13 November 2007.
We detected 198 unique GII.4 sequence variants, the majority of which were associated with only one outbreak. A few sequence variants were associated with the emergence of new GII.4 subclusters (US95/96 in 1996, Farmington Hills in 2002, Hunter in 2004, and Yerseke and Den Haag in 2006) that gradually displaced previous GII.4 viruses in the population (20, 31, 34), an evolutionary pattern similar to the pattern described for influenza virus (17). An important feature of RNA virus replication is the lack of the proofreading function of its RNA polymerase (8). Due to the error-prone nature of RNA polymerases, the mutation rate for NoVs can be expected to be in the same range as estimated for RNA viruses, such as poliovirus and hepatitis C virus, with 1.44 × 10−3 to 3 × 10−3 base substitutions per site per year (29, 45). This could ultimately lead to the emergence of new GII.4 strains. Rapid spread of these newly emerging strains in an immunologically naïve population could result in large-scale outbreaks leading to a pandemic within a short amount of time (5, 7, 21, 23, 26, 28, 32, 41, 43, 46).
Recent studies have demonstrated that susceptibility to NoV infection, including GII.4 viruses, is associated with of the presence of certain histo-blood group antigens (HBGA) on host cells (36, 37). Moreover, the surface-exposed carbohydrate ligand-binding domain in the NoV capsid is under heavy immune selection and likely evolves by antigenic drift. Since no simple cell culture or small animal model is available to study NoVs, the exact mechanism behind the evolution and subsequent selection of a particular strain and its transmissibility remains unknown. Several research groups have used three-dimensional (3D) homology models to study the interactions between HBGA receptors and the protruding (P) domain of the NoV capsid monomer, the putative receptor-binding domain, to assess the host susceptibility to viral infections, which might help to understand the mechanism of GII.4 predominance (1, 35-38).
Although the CDC started to document outbreaks of acute viral gastroenteritis in 1982, the true burden of NoV outbreaks in the United States is unknown and is likely substantially underreported due to the passive surveillance system (4). In this study, we analyzed only outbreaks for which samples were submitted to the CDC. Prior to 1995, most outbreak samples were tested for NoV by electron microscopy or serology (3, 25); with the introduction of RT-PCR and real-time RT-PCR methods, the NoV detection rate has increased greatly (3, 25, 40).
To strengthen the current NoV outbreak surveillance and to obtain a better assessment of the true burden of NoV illness as well as the early detection of newly emerging GII.4 strains, real-time RT-PCR assays have been implemented in most laboratories of state health departments. In addition, several electronic surveillance systems for NoV outbreaks have recently been initiated. CaliciNet, a network of state and local public health and food regulatory agency laboratories in the United States is coordinated by the CDC and allows us to rapidly detect and link epidemiologic and NoV sequence data to identify multistate and international food-borne outbreaks. A global norovirus surveillance network, NoroNet, which includes data from CaliciNet as well as data from other surveillance networks (18, 26) will allow more timely identification of increased norovirus activity and emergence of novel pandemic strains.
The findings of this study highlight the importance of continued NoV outbreak surveillance as well as molecular characterization of strains to better understand the mechanism(s) driving the rapid evolving GII.4 viruses, to determine why they have an advantage over other NoV genotypes, and to assess whether other genotypes follow similar evolutionary patterns (16).
Acknowledgments
We thank all state and local health departments for providing us with epidemiologic data and outbreak specimens; Lenee H. Blanton, R. Suzanne Beard, and Belinda Vermeulen for their assistance with outbreak and laboratory investigations; and Larry Anderson, Jon Gentsch, and Olen Kew for their constructive comments.
The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention. This article did receive clearance through the appropriate channels at the CDC prior to submission.
Footnotes
Published ahead of print on 28 October 2009.
REFERENCES
- 1.Allen, D. J., J. J. Gray, C. I. Gallimore, J. Xerry, and M. Iturriza-Gomara. 2008. Analysis of amino acid variation in the P2 domain of the GII-4 norovirus VP1 protein reveals putative variant-specific epitopes. PLoS One 3:e1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ando, T., S. S. Monroe, J. R. Gentsch, Q. Jin, D. C. Lewis, and R. I. Glass. 1995. Detection and differentiation of antigenically distinct small round-structured viruses (Norwalk-like viruses) by reverse transcription-PCR and Southern hybridization. J. Clin. Microbiol. 33:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ando, T., M. N. Mulders, D. C. Lewis, M. K. Estes, S. S. Monroe, and R. I. Glass. 1994. Comparison of the polymerase region of small round structured virus strains previously classified in three antigenic types by solid-phase immune electron microscopy. Arch. Virol. 135:217-226. [DOI] [PubMed] [Google Scholar]
- 4.Blanton, L. H., S. M. Adams, R. S. Beard, G. Wei, S. N. Bulens, M. A. Widdowson, R. I. Glass, and S. S. Monroe. 2006. Molecular and epidemiologic trends of caliciviruses associated with outbreaks of acute gastroenteritis in the United States, 2000-2004. J. Infect. Dis. 193:413-421. [DOI] [PubMed] [Google Scholar]
- 5.Bull, R. A., E. T. Tu, C. J. McIver, W. D. Rawlinson, and P. A. White. 2006. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. J. Clin. Microbiol. 44:327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon, J. L., L. C. Lindesmith, E. F. Donaldson, L. Saxe, R. S. Baric, and J. Vinjé. 2009. Herd immunity to GII.4 noroviruses is supported by outbreak patient sera. J. Virol. 83:5363-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2007. Norovirus activity-United States, 2006-2007. MMWR Morb. Mortal. Wkly. Rep. 56:842-846. [PubMed] [Google Scholar]
- 7a.Centers for Disease Control and Prevention. 2009. Surveillance for foodborne disease outbreaks—United States, 2006. MMWR Morb. Mortal. Wkly. Rep. 58:609-615. [PubMed] [Google Scholar]
- 8.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 9.Duizer, E., K. J. Schwab, F. H. Neill, R. L. Atmar, M. P. Koopmans, and M. K. Estes. 2004. Laboratory efforts to cultivate noroviruses. J. Gen. Virol. 85:79-87. [DOI] [PubMed] [Google Scholar]
- 10.Estes, M. K., B. V. Prasad, and R. L. Atmar. 2006. Noroviruses everywhere: has something changed? Curr. Opin. Infect. Dis. 19:467-474. [DOI] [PubMed] [Google Scholar]
- 11.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 12.Fankhauser, R. L., J. S. Noel, S. S. Monroe, T. Ando, and R. I. Glass. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 178:1571-1578. [DOI] [PubMed] [Google Scholar]
- 13.Green, K. Y. 2007. Caliciviridae: the noroviruses, p. 949-979. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 14.Green, K. Y., G. Belliot, J. L. Taylor, J. Valdesuso, J. F. Lew, A. Z. Kapikian, and F. Y. Lin. 2002. A predominant role for Norwalk-like viruses as agents of epidemic gastroenteritis in Maryland nursing homes for the elderly. J. Infect. Dis. 185:133-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green, S. M., K. E. Dingle, P. R. Lambden, E. O. Caul, C. R. Ashley, and I. N. Clarke. 1994. Human enteric Caliciviridae: a new prevalent small round-structured virus group defined by RNA-dependent RNA polymerase and capsid diversity. J. Gen. Virol. 75:1883-1888. [DOI] [PubMed] [Google Scholar]
- 16.Iritani, N., H. Vennema, J. J. Siebenga, R. J. Siezen, B. Renckens, Y. Seto, A. Kaida, and M. Koopmans. 2008. Genetic analysis of the capsid gene of genotype GII.2 noroviruses. J. Virol. 82:7336-7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koelle, K., S. Cobey, B. Grenfell, and M. Pascual. 2006. Epochal evolution shapes the phylodynamics of interpandemic influenza A (H3N2) in humans. Science 314:1898-1903. [DOI] [PubMed] [Google Scholar]
- 18.Koopmans, M., H. Vennema, H. Heersma, E. van Strien, Y. van Duynhoven, D. Brown, M. Reacher, and B. Lopman. 2003. Early identification of common-source foodborne virus outbreaks in Europe. Emerg. Infect. Dis. 9:1136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroneman, A., L. Verhoef, J. Harris, H. Vennema, E. Duizer, Y. van Duynhoven, J. Gray, M. Iturriza, B. Bottiger, G. Falkenhorst, C. Johnsen, C. H. von Bonsdorff, L. Maunula, M. Kuusi, P. Pothier, A. Gallay, E. Schreier, M. Hohne, J. Koch, G. Szucs, G. Reuter, K. Krisztalovics, M. Lynch, P. McKeown, B. Foley, S. Coughlan, F. M. Ruggeri, I. Di Bartolo, K. Vainio, E. Isakbaeva, M. Poljsak-Prijatelj, A. H. Grom, J. Z. Mijovski, A. Bosch, J. Buesa, A. S. Fauquier, G. Hernandez-Pezzi, K. O. Hedlund, and M. Koopmans. 2008. Analysis of integrated virological and epidemiological reports of norovirus outbreaks collected within the foodborne viruses in Europe Network from 1 July 2001 to 30 June 2006. J. Clin. Microbiol. 46:2959-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindesmith, L. C., E. F. Donaldson, A. D. Lobue, J. L. Cannon, D. P. Zheng, J. Vinjé, and R. S. Baric. 2008. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med. 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopman, B., H. Vennema, E. Kohli, P. Pothier, A. Sanchez, A. Negredo, J. Buesa, E. Schreier, M. Reacher, D. Brown, J. Gray, M. Iturriza, C. Gallimore, B. Bottiger, K. O. Hedlund, M. Torven, C. H. von Bonsdorff, L. Maunula, M. Poljsak-Prijatelj, J. Zimsek, G. Reuter, G. Szucs, B. Melegh, L. Svennson, Y. van Duijnhoven, and M. Koopmans. 2004. Increase in viral gastroenteritis outbreaks in Europe and epidemic spread of new norovirus variant. Lancet 363:682-688. [DOI] [PubMed] [Google Scholar]
- 22.Lopman, B. A., M. H. Reacher, Y. Van Duijnhoven, F. X. Hanon, D. Brown, and M. Koopmans. 2003. Viral gastroenteritis outbreaks in Europe, 1995-2000. Emerg. Infect. Dis. 9:90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maguire, A. J., J. Green, D. W. Brown, U. Desselberger, and J. J. Gray. 1999. Molecular epidemiology of outbreaks of gastroenteritis associated with small round-structured viruses in East Anglia, United Kingdom, during the 1996-1997 season. J. Clin. Microbiol. 37:81-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martella, V., M. Campolo, E. Lorusso, P. Cavicchio, M. Camero, A. L. Bellacicco, N. Decaro, G. Elia, G. Greco, M. Corrente, C. Desario, S. Arista, K. Banyai, M. Koopmans, and C. Buonavoglia. 2007. Norovirus in captive lion cub (Panthera leo). Emerg. Infect. Dis. 13:1071-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monroe, S. S., R. I. Glass, N. Noah, T. H. Flewett, E. O. Caul, C. I. Ashton, A. Curry, A. M. Field, R. Madeley, and P. J. Pead. 1991. Electron microscopic reporting of gastrointestinal viruses in the United Kingdom, 1985-1987. J. Med. Virol. 33:193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Motomura, K., T. Oka, M. Yokoyama, H. Nakamura, H. Mori, H. Ode, G. S. Hansman, K. Katayama, T. Kanda, T. Tanaka, N. Takeda, and H. Sato. 2008. Identification of monomorphic and divergent haplotypes in the 2006-2007 norovirus GII/4 epidemic population by genomewide tracing of evolutionary history. J. Virol. 82:11247-11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noel, J. S., T. Ando, J. P. Leite, K. Y. Green, K. E. Dingle, M. K. Estes, Y. Seto, S. S. Monroe, and R. I. Glass. 1997. Correlation of patient immune responses with genetically characterized small round-structured viruses involved in outbreaks of nonbacterial acute gastroenteritis in the United States, 1990 to 1995. J. Med. Virol. 53:372-383. [DOI] [PubMed] [Google Scholar]
- 28.Noel, J. S., R. L. Fankhauser, T. Ando, S. S. Monroe, and R. I. Glass. 1999. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J. Infect. Dis. 179:1334-1344. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto, H., M. Kojima, S. Okada, H. Yoshizawa, H. Iizuka, T. Tanaka, E. E. Muchmore, D. A. Peterson, Y. Ito, and S. Mishiro. 1992. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology 190:894-899. [DOI] [PubMed] [Google Scholar]
- 30.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 31.Reuter, G., P. Pankovics, and G. Szucs. 2008. Genetic drift of norovirus genotype GII-4 in seven consecutive epidemic seasons in Hungary. J. Clin. Virol. 42:135-140. [DOI] [PubMed] [Google Scholar]
- 32.Siebenga, J., H. Vennema, D. P. Zheng, J. Vinjé, B. Lee, X. L. Pang, E. Ho, W. Lim, A. Choudekar, S. Broor, T. Halperin, N. Rasool, J. Hewitt, G. Greening, M. Jin, Z. J. Duan, Y. Lucero, M. O'Ryan, M. Hoehne, E. Schreier, R. M. Ratcliff, P. White, N. Iritani, G. Reuter, and M. Koopmans. 2009. Norovirus illness is a global problem; emergence and spread of norovirus GII.4 variants, 2001-2007. J. Infect. Dis. 200:802-812. [DOI] [PubMed] [Google Scholar]
- 33.Siebenga, J. J., H. Vennema, E. Duizer, and M. P. Koopmans. 2007. Gastroenteritis caused by norovirus GGII.4, The Netherlands, 1994-2005. Emerg. Infect. Dis. 13:144-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siebenga, J. J., H. Vennema, B. Renckens, E. de Bruin, B. van der Veer, R. J. Siezen, and M. Koopmans. 2007. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. J. Virol. 81:9932-9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan, M., P. Fang, T. Chachiyo, M. Xia, P. Huang, Z. Fang, W. Jiang, and X. Jiang. 2008. Noroviral P particle: structure, function and applications in virus-host interaction. Virology 382:115-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan, M., P. Huang, J. Meller, W. Zhong, T. Farkas, and X. Jiang. 2003. Mutations within the P2 domain of norovirus capsid affect binding to human histo-blood group antigens: evidence for a binding pocket. J. Virol. 77:12562-12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan, M., and X. Jiang. 2008. Association of histo-blood group antigens with susceptibility to norovirus infection may be strain-specific rather than genogroup dependent. J. Infect. Dis. 198:940-943. [DOI] [PubMed] [Google Scholar]
- 38.Tan, M., M. Jin, H. Xie, Z. Duan, X. Jiang, and Z. Fang. 2008. Outbreak studies of a GII-3 and a GII-4 norovirus revealed an association between HBGA phenotypes and viral infection. J. Med. Virol. 80:1296-1301. [DOI] [PubMed] [Google Scholar]
- 39.Teunis, P. F., C. L. Moe, P. Liu, S. E. Miller, L. Lindesmith, R. S. Baric, J. Le Pendu, and R. L. Calderon. 2008. Norwalk virus: how infectious is it? J. Med. Virol. 80:1468-1476. [DOI] [PubMed] [Google Scholar]
- 40.Trujillo, A. A., K. A. McCaustland, D. P. Zheng, L. A. Hadley, G. Vaughn, S. M. Adams, T. Ando, R. I. Glass, and S. S. Monroe. 2006. Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of norovirus. J. Clin. Microbiol. 44:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tu, E. T., R. A. Bull, G. E. Greening, J. Hewitt, M. J. Lyon, J. A. Marshall, C. J. McIver, W. D. Rawlinson, and P. A. White. 2008. Epidemics of gastroenteritis during 2006 were associated with the spread of norovirus GII.4 variants 2006a and 2006b. Clin. Infect. Dis. 46:413-420. [DOI] [PubMed] [Google Scholar]
- 42.Vinjé, J., R. A. Hamidjaja, and M. D. Sobsey. 2004. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J. Virol. Methods 116:109-117. [DOI] [PubMed] [Google Scholar]
- 43.Vipond, I. B., E. O. Caul, D. Hirst, B. Carmen, A. Curry, B. A. Lopman, P. Pead, M. A. Pickett, P. R. Lambden, and I. N. Clarke. 2004. National epidemic of Lordsdale Norovirus in the UK. J. Clin. Virol. 30:243-247. [DOI] [PubMed] [Google Scholar]
- 44.Wang, Q. H., M. G. Han, S. Cheetham, M. Souza, J. A. Funk, and L. J. Saif. 2005. Porcine noroviruses related to human noroviruses. Emerg. Infect. Dis. 11:1874-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward, C. D., M. A. Stokes, and J. B. Flanegan. 1988. Direct measurement of the poliovirus RNA polymerase error frequency in vitro. J. Virol. 62:558-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Widdowson, M. A., E. H. Cramer, L. Hadley, J. S. Bresee, R. S. Beard, S. N. Bulens, M. Charles, W. Chege, E. Isakbaeva, J. G. Wright, E. Mintz, D. Forney, J. Massey, R. I. Glass, and S. S. Monroe. 2004. Outbreaks of acute gastroenteritis on cruise ships and on land: identification of a predominant circulating strain of norovirus-United States, 2002. J. Infect. Dis. 190:27-36. [DOI] [PubMed] [Google Scholar]
- 47.Widdowson, M. A., A. Sulka, S. N. Bulens, R. S. Beard, S. S. Chaves, R. Hammond, E. D. Salehi, E. Swanson, J. Totaro, R. Woron, P. S. Mead, J. S. Bresee, S. S. Monroe, and R. I. Glass. 2005. Norovirus and foodborne disease, United States, 1991-2000. Emerg. Infect. Dis. 11:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312-323. [DOI] [PubMed] [Google Scholar]