Abstract
Vpx and Vpr are related lentiviral accessory proteins that enhance virus replication in macrophages and dendritic cells. Both proteins are packaged into virions and mediate their effects in the target cell through an interaction with an E3 ubiquitin ligase that contains DCAF1 and DDB1. When introduced into primary macrophages and dendritic cells in viruslike particles, Vpx can enhance the efficiency of a subsequent infection. Here, we confirm the ability of Vpx to enhance simian immunodeficiency virus (SIV) and human immunodeficiency virus type 1 (HIV-1) infection of macrophages up to 100-fold by using single-cycle reporter viruses and by pretreatment of the cells with Vpx-containing viruslike particles. Vpx was also active in differentiated THP-1 cells but not in other cell lines. Induction of an antiviral state in macrophages with type I interferon significantly magnified the effect of Vpx on HIV-1 infection, suggesting that Vpx helps the virus to overcome an inducible intracellular restriction. Quantitative PCR quantitation of SIV and HIV-1 reverse transcripts in newly infected macrophages showed that the block was at an early step in reverse transcription. In spite of its structural similarity, Vpr was inactive. This difference allowed us to map the functional domains of Vpx with a panel of Vpr/Vpx chimeras. Analysis of the chimeras demonstrated that the amino-terminal domain of Vpx is important for the enhancement of infection. Fine mapping of the region indicated that amino acids at positions 9, 12, and 15 to 17 were required. Although the mutants failed to enhance infection, they retained their ability to interact with DCAF1. These findings suggest that the Vpx amino terminus contains an activation domain that serves as the binding site for a cellular restriction factor.
The Vpx and Vpr lentiviral accessory proteins enhance virus replication in macrophages and dendritic cells (DC) but have little effect on replication in activated CD4+ T cells (2, 10, 16, 18, 44). The proteins are ca. 50% identical in amino acid sequence, suggesting that they arose by gene duplication (39). Most lentiviruses encode Vpr, whereas a Vpx gene is present in human immunodeficiency virus type 2 (HIV-2), simian immunodeficiency virus of sooty mangabey (SIVsm), and SIV of rhesus macaque (SIVmac) but absent from HIV-1. Although Vpr and Vpx are dispensable in vitro, studies in nonhuman primate models have demonstrated their importance in vivo (15, 19). Rhesus macaques infected with Δvpx SIV have reduced virus loads and progress more slowly to AIDS, while animals infected with a Δvpx/Δvpr SIVmac double mutant showed little evidence of disease progression and maintained low virus loads.
Vpr and Vpx are specifically packaged into the virion, a feature that is unique among the accessory proteins. Both proteins are packaged during virus assembly through an interaction with amino acid motifs in the p6 region of the Gag polyprotein (1, 30). The presence of Vpr and Vpx in the virion suggests that they play a role early in virus replication. Both proteins localize to the nucleus of the infected cell and have been proposed to facilitate the nuclear import of the preintegration complex (5, 12, 13). However, a report that Δvpr HIV-1 maintains its ability to infect nondividing cells argues against a role in nuclear import (42). In addition, recent studies have indicated a role for Vpx in reverse transcription, a process that mainly occurs in the cytoplasm prior to nuclear import (14, 18, 35).
Goujon et al. have found that Vpx increases the infectibility of monocyte-derived macrophages (MDM) and DC when introduced into the cell in trans in viruslike particles (VLP) (17). Vpx-containing VLP were also found to promote the transduction of DC with episomal lentiviral vectors by increasing the number of cells infected and the level of transgene expression (9). The effect of Vpx could be partially mimicked by treating the DC with the proteasome inhibitor MG132, suggesting that Vpx allows the virus to escape a degradative pathway operative in these cells (18).
Lentiviral accessory and regulatory proteins either co-opt specific cellular proteins to facilitate virus replication or interfere with the antiviral activity of intracellular restriction factors. In the infected cell, Vpr and Vpx both form complexes with an E3 ubiquitin ligase that is composed of Cullin4a, DDB1, Rbx1, and DCAF1 (6, 20, 24, 31, 38). In the complexes, Vpr appears to directly bind DCAF1. Vpx also binds to DCAF1 but with reduced affinity (24). Modeling of Vpx on the NMR structure solved for Vpr suggests that Vpx consists of three central α-helices that form a hydrophobic core flanked by flexible amino-terminal and carboxy-terminal regions (22, 25). Le Rouzic et al. have found that Vpx and Vpr bind DCAF1 via conserved amino acids in α-helix 3 (24). The interaction of Vpr and Vpx with the E3 ubiquitin ligase is required for biological function. Vpr point mutants that are defective for DCAF1 binding do not arrest the cell cycle or induce apoptosis (24), and Vpx point mutants that are defective for DCAF1 binding do not facilitate MDM and DC infection (8, 14, 35). By analogy with HIV-1 Vif, the involvement of an E3 ubiquitin ligase suggests a role in inducing the degradation of a host protein (26, 34, 36, 43). Support for such a role was reported by Sharova et al. (32), who fused macrophages to COS cells. The resulting heterokaryons were resistant to Δvpx SIV, supporting a role for Vpx in counteracting a dominant inhibitor.
To investigate the role of Vpx and Vpr on MDM infection, we used single-cycle SIV reporter viruses and Vpx-containing SIV VLP that were generated by vectors that are identical except for mutations in vpr or vpx. In single-cycle infections, Vpx enhanced the infection of MDM by more than 100-fold. Pretreatment of MDM with Vpx-containing VLP also dramatically enhanced the SIV and HIV-1 infection of MDM, definitively mapping the effect to Vpx and confirming earlier reports (17). The VLP were active, albeit to a lesser extent, in phorbol ester differentiated THP-1. The enhancement required the interaction of Vpx with DCAF1 and served to relieve a block early in reverse transcription. Pretreatment of the MDM with type I interferon (IFN) magnified the effect of Vpx-containing VLP, suggesting a role in overcoming an IFN-inducible intracellular restriction. Vpr had little activity in either assay, a finding that we exploited to map functional domains of the protein with a panel of Vpr/Vpx chimeras. Analysis of the activity of the chimeras demonstrated a critical role for the amino-terminal domain. Analysis of point mutants in this region mapped the activity to single amino acids. Several mutants failed to enhance infection but retained the ability to bind DCAF1, suggesting that the amino-terminal domain may be a binding site for a cellular restriction factor.
MATERIALS AND METHODS
Cells and cell culture.
293T and HeLa cells were cultured in Dulbecco modified Eagle medium (DMEM)-10% fetal bovine serum. THP-1 and SUPT1 were cultured in RPMI 1640-10% fetal bovine serum. Where indicated, THP-1 cells were differentiated for 3 days in medium containing 50 nM phorbol myristic acid (PMA). Monocytes were purified from healthy donor peripheral blood mononuclear cells (PBMC) by positive selection on anti-CD14-coated magnetic beads (Miltenyi Biotech, Inc.) and were typically >98% CD14+. Monocytes were differentiated to MDM by culturing for 4 to 6 days in medium containing 50 ng of granulocyte-macrophage colony-stimulating factor (Invitrogen)/ml.
Plasmids.
The SIV-derived proviral plasmids, termed pSIVmac, are based on SIVmac239 with the exception of pSIV3+, provided by A. Cimarelli (Ecole Normale Supérieure de Lyon), which is based on SIVmac251 (17). SIVmac Vpx and Vpr expression vectors were codon optimized by overlap extension PCR and cloned into the EcoRI and XhoI sites of pcDNA6/myc-His (Invitrogen), placing the epitope tag at the carboxy terminus. Vpx point mutants and Vpr/Vpx chimeras were generated by PCR mutagenesis. pNL-luc-E− and pSIVmac-luc-E− plasmids have been previously described (10, 27). The Δvpx and Δvpr variants—pSIV3+R−, pSIV3+X−, and pSIV3+X−R−—were generated by PCR mutagenesis. To generate pSIV3+X−, a vpx/vpr-containing fragment of SIVmac251 (accession no. M19499) from the BstBI site (nucleotide [nt] 5852) to the AfeI site (nt 6329) was amplified by using the primers SIV3+stopvpx.BstBI.sense (5′-GAGGCCTTCGAATAGCTAAACAGAACAG) and SIV3+AfeI.antisense (5′-GAAGAGCGCTCGTTGG) that introduced a stop codon at amino acid 24 of vpx (nt 5858). pSIV3+R− was generated by overlapping PCR. The 5′ fragment was amplified with the primers SIV3+BstBI.sense (5′-GAGGCCTTCGAATGGCTAAACAGAACAG) and SIV3+stop.vpr.overlap.antisense (5′-CTGGAGGTCTTTCTTACATTTATGCTAG), and the 3′ fragment was amplified with the primers SIV3+stop.vpr.overlap.sense (5′-CTAGCATAAATGTAAGAAAGACCTCCAG) and SIV3+AfeI.antisense (5′-GAAGAGCGCTCGTTGG). The fragments were mixed and amplified with the outer primers. The resulting amplicon was digested with BstBI and AfeI and cloned back into pSIV3+, resulting in a stop codon in vpr at amino acid 2 (nt 6130). pSIV3+X−R− was constructed by overlapping PCR with SIV3+stopvpx.BstBI.sense as the outer primer, introducing a stop codon in vpx (nt 5858). The PCR fragments were cloned into the BstBI and AfeI sites of pSIV3+. PCR-amplified sequences were confirmed by nucleotide sequence analysis.
Virus preparation and infections.
HIV-1 and SIVmac luciferase reporter viruses were generated as described previously (10, 27). Briefly, 293T cells were cotransfected by using Lipofectamine 2000 (Invitrogen) with reporter virus plasmid and vesicular stomatitis virus glycoprotein (VSV-G) expression plasmid, pcVSV-G. To produce VLP, 293T cells were cotransfected with pSIV3+, R−, X−, or R−X− and pcVSV-G at a mass ratio of 2:1. To produce trans-complemented reporter viruses, 293T cells were cotransfected with pSIVmac-luc-E−X−R−, pcVSV-G, and either pcVpr, pcVpx, or pcDNA at a ratio of 2:1:1. Supernatants were harvested 48 h posttransfection, passed through 0.4-μm-pore-size filters, divided into aliquots, and frozen at −80°C. Luciferase reporter viruses were normalized on 293T cells by luciferase assay. For this, 1 × 104 293T cells were infected with 50 μl of luciferase reporter virus resulting in 1 × 106 to 3 × 106 luciferase counts per second (cps) on average. VLP-containing supernatants were normalized for p27 measured by enzyme-linked immunosorbent assay (ELISA) (Zeptometrix Corp.).
MDM (1 × 105) and THP-1 (5 × 104) were seeded in a 96-well plate and spin infected with a normalized volume of reporter virus for 2 h at 500 × g. Viral supernatant used for infection was equivalent to 3 × 105 cps for MDM or 1 × 105 for THP-1 cells. Where indicated, cells were preincubated with VLP for 2 h and then infected with reporter virus at a 10:1 ratio of VLP to virus p27 or p24. To induce with type I IFN, MDM were treated 14 h prior to infection with 1 to 100 U of universal type I IFN (PBL Biomedical Laboratories)/ml. The medium was replaced 6 h postinfection, and the luciferase activity was measured 3 days (THP-1, HeLa, and SUPT1) or 4 days (MDM) postinfection using commercial reagents (Promega). The data are presented as the average cps of triplicate infections, with error bars to indicate the standard deviation.
Quantitative real-time PCR (qPCR).
MDM were infected at a multiplicity of infection (MOI) of 1 with virus stocks that had been treated for 1 h with 50 U of Benzonase (Invitrogen)/ml to remove the plasmid DNA. To control for residual plasmid DNA, 25 μM zidovudine (AZT) was added to one control well 14 h prior to infection. DNA was isolated 6, 24, and 48 h postinfection by using a DNAeasy kit (Qiagen). Reverse transcripts in 250 ng of DNA were quantitated by qPCR with an ABI Prism 7300 (Applied Biosystems) and SYBR green reagent (Applied Biosystems). The primer pairs used were mac239.early.sense (5′-TGGGTGTTCCCTGCTAGACTC) and mac239.early.antisense (5′-CAAGCGTGGAGTCACTCTGC) for early products, mac239.late.sense (5′-TTGGGAAACCGAAGCAGG) and mac239.late.antisense (5′-TCTCTCACTCTCCTTCAAGTCCC) for late products, and SIV.2-LTR.reverse.U3 (5′-CTTGCACTGTAATAAATCCC) and SIV.2-LTR-forward.U5 (5′-CCTTTCTGCTTTGGGAAACCG) for two-LTR circles. Standard curves were generated by amplification of serially diluted proviral and two-LTR plasmids.
Immunoblot and coimmunoprecipitation.
Virions were harvested from culture supernatants 2 days posttransfection, filtered, and pelleted through 20% sucrose at 100,000 × g for 20 min at 4°C before lysis. The cells were lysed in buffer containing 1% NP-40. Cell and virus proteins were analyzed on immunoblots as previously described. The filters were probed with anti-myc monoclonal antibody (MAb) 9E10 (Covance), anti-p27 MAb 55-2F12 (AIDS Research and Reference Reagent Program, NIH), anti-VprBP MAb (Shanghai Genomics), and anti-α-tubulin (Sigma). Bound antibodies were detected with biotinylated goat anti-rabbit or goat anti-mouse immunoglobulin, followed by streptavidin DyeLight 800 conjugate (Pierce) and imaged on an Odyssey infrared imaging system (LiCOR) at 800 nm. To coimmunoprecipitate Vpx/DCAF1 complexes, 293T cells were transfected with pcVpx-myc or empty vector. After 2 days, cell lysates were prepared and precleared for 30 min at 4°C with 70 μl of protein G-Sepharose (GE Healthcare). Vpx was then immunoprecipitated for 2 h with 3 μl of anti-myc MAb, followed by 1 h with 80 μl of protein G-Sepharose. The beads were washed four times with lysis buffer, resuspended in reducing sample buffer (Invitrogen), and heated to 95°C. The immunoprecipitates were then analyzed on an immunoblot.
siRNA knockdown in MDM.
MDM (7 × 105) were nucleofected (Amaxa) with 3 μg of siRNA targeting DCAF1 (5′-GGAGGGAAUUGUCGAGAAUUU) or with nonspecific control small interfering RNA (siRNA). After 24 h, the MDM were removed from the dish by using phosphate-buffered saline containing 5 mM EDTA and plated in a 96-well dish at 3 × 104 cells per well. The cells were infected 24 h later with luciferase reporter virus and, after 4 days, the luciferase was measured.
RESULTS
Vpx promotes SIV infection of MDM and differentiated THP-1 cells.
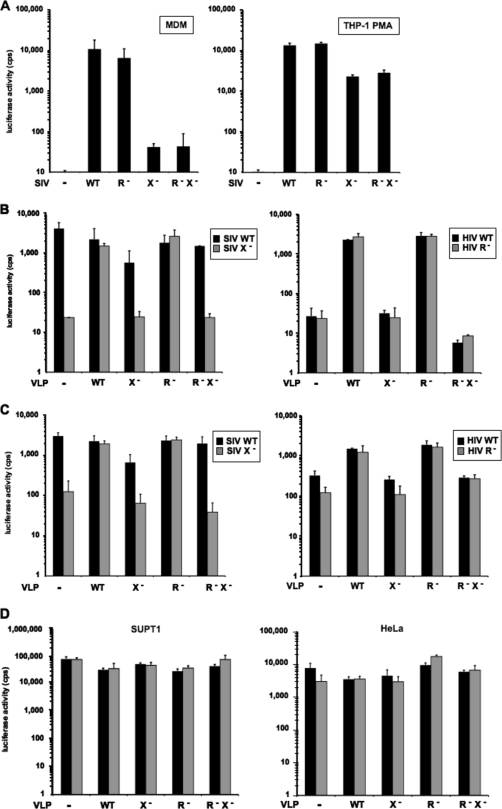
To test the role of Vpx in SIV replication, we generated Δvpr, Δvpx, and Δvpr/Δvpx single-cycle luciferase reporter virus plasmids that contained termination codons near the 5′ ends of vpr and vpx in pSIVmac-luc-E−. Virus stocks were produced in transfected 293T cells as VSV-G pseudotypes and then normalized on the basis of the infectious titer on 293T cells, a cell line in which Vpr and Vpx have no significant effect on infectivity (see Fig. S1 in the supplemental material). To determine the effect of the accessory proteins on early virus replication, we compared the infectivity of the viruses on primary MDM and on the myeloid cell lines U937 and THP-1. U937 and THP-1 can be differentiated into macrophagelike cells with the phorbol ester PMA (7). Consistent with previous reports (16, 32, 35), Vpx showed a pronounced phenotype in MDM (Fig. 1A). In analyses of MDM derived from multiple donors, the wild-type virus was up to 100-fold more infectious than Δvpx SIV. Vpx was not active in infection of U937 or undifferentiated THP-1 (data not shown) but enhanced the infection of PMA-differentiated THP-1 by 10-fold (Fig. 1A). Deletion of Vpr had only a small effect. It reduced SIV infectivity by <2-fold and did not synergize with Vpx.
FIG. 1.
Vpx facilitates SIVmac and HIV-1 infection of MDM and PMA-differentiated THP-1 cells. (A) MDM and differentiated THP-1 cells were infected with VSV-G pseudotyped SIVmac luciferase reporter viruses normalized for infectivity. (B) Enhancement of SIV and HIV-1 infection of MDM by Vpx-containing VLP. Cells were preincubated with VLP for 2 h and then infected with SIV (left) or HIV-1 (right) reporter virus at a 10:1 ratio of VLP to virus. (C) Effect of VLP on SIV and HIV-1 infection of THP-1. THP-1 cells were preincubated with VLP and infected with reporter virus. (D) Effect of Vpx-containing VLP on HIV-1 infection of SUPT1 and HeLa cells. The cells were preincubated for 2 h with VLP and infected with HIV-1 reporter virus. The luciferase activity was measured 3 days postinfection for THP-1 or 4 days postinfection for MDM. The results shown are representative of results obtained with three independent experiments with different donor PBMC.
Vpx-containing VLP promote SIVmac and HIV-1 infection in trans.
In the studies of Goujon et al. that showed that preincubation of DC and MDM with Vpx-containing SIV VLP enhanced HIV-1 infection and compensated for Vpx in Δvpx SIV infection (17), the VLP were produced by using an SIVmac251-based plasmid, pSIV3+, in which the cytomegalovirus promoter drives the transcription of a SIVmac genome deleted for the packaging signal, nef, and env (17, 28). A combination of vectors that lacked an intact vpx gene was used to deduce the requirement for packaged Vpx. To simplify this system such that the VLP were produced from vectors that differed only by whether they contained Vpr and/or Vpx, we modified pSIV3+ by introducing termination codons into vpr and vpx. The resulting vectors were isogenic except for the Δvpx, Δvpr, and Δvpr/Δvpx mutations. We used these to generate VLP that differed only by the presence or absence of packaged Vpr or Vpx.
To test the effect of Vpx-containing VLP on SIV infection, we generated VSV-G-pseudotyped VLP in 293T cells transfected with wild-type or mutant pSIV3+ vector. We normalized the VLP for p27 content and added them to cultures of normal donor MDM. After 2 h, we infected the cells with wild-type, Δvpr, or Δvpx SIV luciferase reporter virus and 4 days later, determined the efficiency of infection by measuring the intracellular luciferase activity. We found that VLP that contained Vpr and Vpx (termed “wild-type VLP”) rescued Δvpx SIV, increasing its infectivity close to that of wild-type SIVmac (Fig. 1B). VLP that lacked Vpr were as effective as wild type. In contrast, VLP that lacked Vpx had no effect on Δvpx SIV infection. In these experiments, the VLP had a small (twofold) inhibitory effect on wild-type SIV, perhaps caused by competition of the VLP for a positively acting cellular factor or by induction of an antiviral response. This effect did not alter the outcome of the experiment. The Vpx-containing VLP also enhanced the infection of MDM by HIV-1, resulting in a nearly 100-fold increase in infectibility of cells treated with wild-type VLP compared to Δvpx VLP, which is consistent with the findings of Goujon et al. (Fig. 1B) (17). THP-1 cells serve as a useful model for myeloid cells since they have the ability to differentiate from a monocytic to an MDM-like phenotype upon differentiation with phorbol ester (7). Incubation of phorbol ester-differentiated THP-1 cells with Vpx-containing VLP resulted in a 10-fold enhancement of infection compared to cells treated with Δvpx VLP (Fig. 1C). Vpx-containing VLP had no effect on HIV-1 infection of HeLa and SUPT1 cells, confirming the cell type specificity of Vpx (Fig. 1D).
The restriction to Δvpx SIV and HIV-1 infection of MDM is early in reverse transcription.
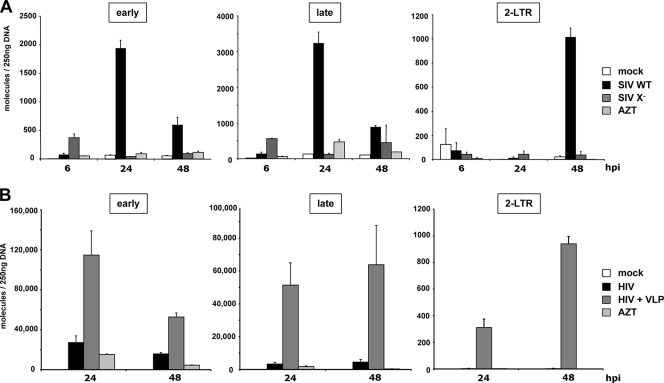
To determine the step in virus replication that is enhanced by Vpx in MDM, we infected MDM with wild-type or Δvpx SIV and determined the number of reverse transcribed viral DNA molecules present. We isolated the DNA of the infected MDM after 6, 24, and 48 h and determined the viral DNA copy number by qPCR using primer pairs specific to the early, late, and two-LTR circular DNA. We found that in cells infected with wild-type SIV, the early and late reverse transcripts peaked at 24 h and diminished 48 h postinfection (Fig. 2A). In contrast, in cells infected with Δvpx virus, few early, late, or two-LTR forms were present. In cells infected with Δvpx virus, a small peak of early cDNA appeared 6 h postinfection and then disappeared by 24 h. These may be partial reverse transcripts that had been aborted during synthesis or degraded following synthesis. An AZT control was included in each set of infections. These contained few DNA molecules, demonstrating the absence of input plasmid DNA. These findings suggest that Vpx acts at or shortly after the initiation of reverse transcription.
FIG. 2.
Δvpx SIVmac and HIV-1 is blocked at reverse transcription in MDM. (A) MDM were either not infected (mock), infected with wild-type SIV (SIV WT), or with Δvpx SIV (SIV X−). Cellular DNA was purified 6, 24, and 48 h postinfection (hpi). The DNA (250 ng) was used as a template in qPCR to amplify reverse transcriptase products or two-LTR circles using primer pairs specific for minus-strand strong-stop (early), second strand initiation (late), or two-LTR circle formation (2-LTR). (B) MDM were incubated with VLP or medium and, after 2 h, were either not infected (mock) or infected with HIV-1. Cellular DNA was purified 24 and 48 h postinfection (hpi) and 250 ng of DNA was used as a template in qPCR to amplify early and late reverse transcriptase products and 2-LTR circles. To control for plasmid contamination, 25 μM AZT was added to one well 14 h prior to infection with either SIVmac (A) or HIV-1 (B) (AZT). The data are presented as the average of triplicate reactions, with error bars indicating the standard deviation. The results of one of two representative experiments are shown.
We next tested whether Vpx acts on HIV-1 infection at a similar step in virus replication. For this, we preincubated MDM with Vpx-containing VLP and, after 2 h, infected the cells with HIV-1. We found that 24 and 48 h postinfection the Vpx-containing VLP caused a dramatic increase in the number of early, late, and two-LTR circle cDNA compared to control infections. This result suggests that Vpx enhances HIV-1 and SIV by a similar mechanism.
Interaction with DCAF1 is required for Vpx-mediated enhancement of SIV and HIV-1 infection.
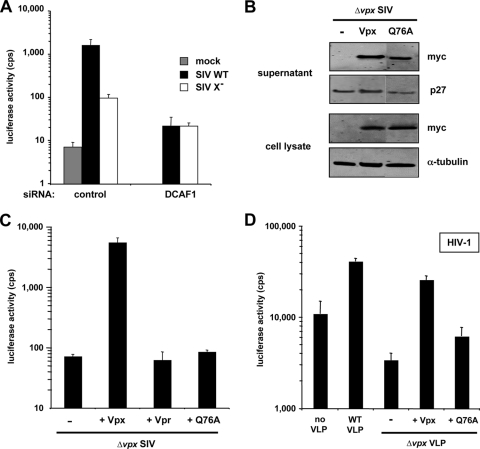
Vpx, like Vpr, interacts with the DCAF1/DDB1-containing E3 ubiquitin ligase (24, 31). To determine whether the interaction of Vpx with the DCAF1/DDB1-containing E3 ubiquitin ligase is required for Vpx to enhance SIV infection of MDM, we knocked down DCAF1 with siRNA and then infected the cells with wild-type or Δvpx SIV. To knock down DCAF1, we transfected MDM by nucleofection with DCAF1-specific siRNA or control siRNA. In MDM transfected with control siRNA, Vpx enhanced wild-type SIV infection 20-fold compared to Δvpx SIV (Fig. 3A). In contrast, in MDM in which DCAF1 had been knocked down, wild-type virus was no more infectious than the Δvpx virus.
FIG. 3.
SIV Vpx interacts with DCAF1 to facilitate SIVmac and HIV-1 infection of MDM. (A) DCAF1 knockdown in MDM. MDM were transfected by nucleofection with 2 pmol of siRNA specific for DCAF1 or control siRNA and, after 2 days, were infected with wild-type or Δvpx SIV luciferase reporter viruses. (B) Expression and packaging of Vpx Q76A. 293T cells were cotransfected with the Δvpx SIVmac plasmid and pcVpx or empty vector. After 2 days, cell lysates were prepared and analyzed on an immunoblot probed with anti-p27 MAb, anti-myc MAb, or anti-α-tubulin MAb. (C) Infection of MDM with Q76A-containing SIVmac. SIVmac Δvpx/Δvpr viruses were produced in the absence (pcDNA) or the presence of the indicated Vpx/Vpr expression plasmid. MDM were infected with the complemented reporter viruses. The luciferase activity was measured after 4 days. (D) Effect of Q76A-containing VLP on HIV-1 infection of MDM. Δvpx VLP were produced in the absence (pcDNA) or presence of the indicated Vpx protein and normalized on p27 amount. Cells were preincubated for 2 h with the different VLP before infection with HIV-1 reporter virus. Similar results were obtained with cells from two other donors.
To further test the importance of DCAF1, we generated Δvpx/Δvpr SIV virions that were trans complemented with Vpr, Vpx, or Q76A Vpx, a Vpx mutant that does not bind DCAF1 (24), and determined their infectivity on MDM. Virus that had packaged wild-type Vpx enhanced infection ∼100-fold compared to virus that lacked Vpx. Q76A failed to enhance infection (Fig. 3C), although it was efficiently packaged into virions (Fig. 3B). Taken together, these findings suggest that Vpx acts through DCAF1 to enhance infection of MDM, a conclusion that is consistent with the findings of Le Rouzic et al. (24).
To determine whether the effect of Vpx on HIV-1 infection is also mediated by the interaction with DCAF1, we generated VLP containing wild-type or Q76A Vpx. The VLP were added to MDM, and 2 h later, the cells were infected with HIV-1 luciferase reporter virus. The results showed that the Q76A mutant was deficient, although it was not completely inactive (Fig. 3D). The low level activity may be explained by the weak binding of the mutant to DCAF1, as shown below (see Fig. 6). Δvpx VLP reduced HIV-1 infectivity slightly, probably due to competition for a limiting cellular factor or induction of an antiviral response in the MDM. Taken together, these results strongly suggest that Vpx enhances SIV and HIV-1 infection through its interaction with DCAF1.
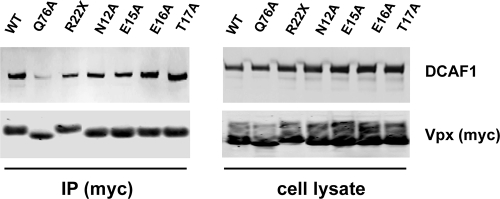
FIG. 6.
Coimmunoprecipitation of Vpx with DCAF1. 293T cells were transfected with expression plasmids encoding the indicated Vpx mutants. Cells were lysed after 2 days, and Vpx proteins were immunoprecipitated from the lysate via their carboxy-terminal myc tag using anti-myc MAb. Cell lysate and immunoprecipitate (IP) were analyzed on an immunoblot probed with anti-myc MAb for the myc-tagged Vpx proteins and with anti-DCAF1 MAb for endogenous DCAF1. One representative experiment of three is shown.
The amino terminus of Vpx is required to enhance infection but not for DCAF1 binding.
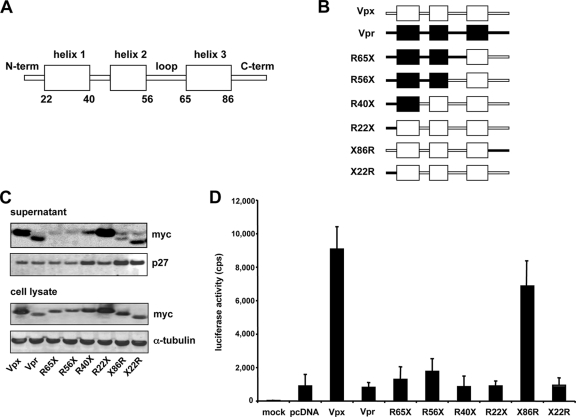
Based on the nuclear magnetic resonance structure of Escherichia coli-produced Vpr, Vpx is thought to consist of three central α-helices connected by flexible loops and unstructured amino and carboxy termini (Fig. 4A) (22, 25). An analysis of SIVmac Vpx with the 3D-JIGSAW structure prediction software further supports the positioning of the proposed α-helices (Fig. 4A) (3, 4, 11). A binding site for DCAF1 is proposed to lie in helix 3 (24). To identify functional domains of Vpx, we took advantage of the structural similarity of Vpr and Vpx to construct the myc-tagged Vpx/Vpr chimeras R22X, R40X, R56X, R65X, and X22R, in which the Vpx/Vpr junctions fall at the beginning or end of each α-helix (Fig. 4B). To test the chimeras, we packaged them in Δvpx/Δvpr SIV virions and analyzed them on an immunoblot. The chimeras were stably expressed in the cell and were packaged into virions (Fig. 4C). Chimeras R56X and R65X, which have junctions in the flexible loop between helix 2 and 3, were packaged with reduced efficiency, perhaps as a result of improper folding. To test the biological activity of the chimeras, we measured the infectivity of the complemented viruses on MDM. All of the chimeras that contained the amino terminus of Vpr failed to enhance MDM infection (Fig. 4D). Only X86R, in which the carboxy terminus of Vpx was replaced by Vpr, was active. Thus, the Vpx carboxy terminus, with its prominent string of prolines, is not required for function. The prolines may be required for stability of the protein, as the chimera was expressed at slightly lower levels than wild type. Chimera X22R, in which the amino terminus of Vpr was replaced by Vpx, failed to rescue Δvpx SIV, indicating that the amino terminus was necessary but not sufficient for biological activity. Taken together, these results point to the importance of the amino terminus of Vpx for the infection of MDM.
FIG. 4.
Analysis of SIVmac Vpx/Vpr chimeric proteins shows that the amino terminus of Vpx is required for function in MDM. (A) Schematic representation of Vpx. The numbers indicate the amino acid positions at the start and the end of the α-helices. (B) Chimeras between SIVmac Vpx (X, white) and Vpr (R, black). Numbers indicate the Vpx/Vpr junctions. (C) Expression and packaging of the Vpx/Vpr chimeras. 293T cells were cotransfected with the proviral plasmid encoding Δvpr/Δvpx SIVmac and the expression plasmid for the indicated chimera. After 2 days, lysate and supernatant were analyzed on an immunoblot probed with anti-p27 MAb and anti-myc MAb. The results shown are representative of three independent experiments. (D) Effect of Vpr/Vpx chimeras on SIV infection of MDM. Δvpx/Δvpr SIVmac reporter viruses were complemented with Vpx, Vpr, or the indicated chimeras by cotransfecting the respective expression plasmid or empty vector (pcDNA), together with the proviral plasmid and the Env expression vector into 293T cells. MDM were infected with the complemented viruses. The luciferase activity was measured after 4 days. The results shown are representative of three independent experiments using cells obtained from three different donors.
The amino-terminal mutants retain the ability to interact with DCAF1.
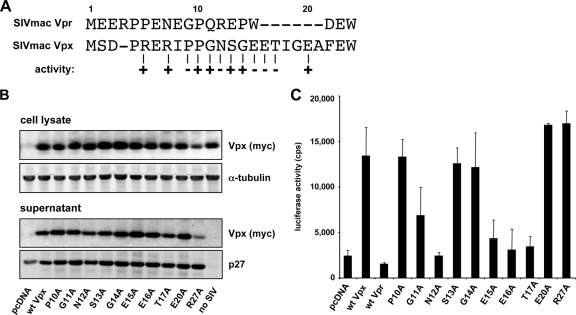
To further dissect the role of the amino terminus of Vpx, we generated point mutants in residues 1 to 22 (Fig. 5A). The sequence of Vpx and Vpr are similar at amino acids 1 to 10 but differ at amino acids 11 to 22 with additional amino acids in vpx. To analyze residues 1 to 10, we exchanged amino acids 6, 8, and 9 of Vpx with those of Vpr and tested the mutated proteins in the complemented luciferase reporter virus assay. Exchange of the amino acids at positions 6 and 8 had no effect, while the exchange of proline to glycine at position 9 inactivated Vpx (data not shown). To test amino acids 10 to 22, we mutated amino acids at positions 10 to 17, 20, and 27 to alanine. The mutants were stably expressed and efficiently packaged into virions (Fig. 5B). Four of the mutants (N12A, E15A, E16A, and T17A) were inactive (Fig. 5C). Thus, specific residues within the amino-terminal region are required for function, which is consistent with a role as an interaction site for another protein.
FIG. 5.
Analysis of the Vpx amino terminus. (A) An alignment of the SIVmac Vpx and Vpr amino terminus is shown. Numbers refer to the amino acid position in Vpx. The ability of each Vpx mutant to enhance infection of MDM is indicated as (+) or (−). The arginines at position 5 and 7 and the proline at position 9 were mutated to the corresponding Vpr amino acid (data not shown). Amino acids 10 to 17, 20, and 27 were changed to alanine. (B) Expression and packaging of the amino-terminal mutants. 293T cells were cotransfected with the proviral plasmid pSIVmac Δvpx and an expression plasmid for Vpx, mutated Vpx, or empty vector (pcDNA). Cell lysate and supernatant were analyzed on an immunoblot probed with anti-p27 MAb and anti-myc MAb. No SIV, transfection of Vpx plasmid without the proviral plasmid. (C) Effect of the amino-terminal mutants on SIV infection of MDM. Δvpx SIVmac reporter viruses were complemented with Vpx mutant or empty vector (pcDNA). MDM were infected with the complemented reporter viruses, and luciferase activity was measured after 4 days. The results shown are representative of three independent experiments using MDM prepared from PBMC of three different donors.
In light of these findings, we tested whether the amino-terminal domain might be involved in the interaction with DCAF1. To determine whether this was the case, we tested whether the mutated Vpx proteins would coimmunoprecipitate with DCAF1. We transfected 293T cells with expression vectors for wild-type Vpx, the point mutants, or the R22X chimera. After 2 days, the cells were lysed and the Vpx proteins were immunoprecipitated with anti-myc MAb and coimmunoprecipitated DCAF-1 was detected on an immunoblot probed with anti-DCAF1 antibody. We found that the point mutants and the R22X chimera retained the ability to bind DCAF1 (Fig. 6). The negative control point mutant Q76A, which has been previously found to be defective for the interaction with DCAF1, was included and found to inefficiently coimmunoprecipitate (24, 35). These findings suggest that the failure of these mutant proteins to enhance infection is not caused by a failure to interact with DCAF1 but rather that the amino-terminal domain has another role.
Vpx allows HIV-1 to circumvent the type I IFN-induced antiviral state.
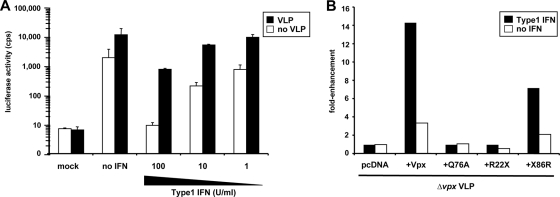
Type I IFN induces an array of cellular genes, resulting in an antiviral state that is effective against a wide range of viruses, including retroviruses. BST2/tetherin, a retroviral restriction factor, is strongly induced by type I IFN (29, 41). Because of the potential role of Vpx in counteracting an intracellular restriction, we sought to determine whether the activity of the putative factor might be IFN inducible in MDM. To test this, we determined the effect of Vpx-containing VLP on HIV-1 infection of type I IFN-induced MDM. We induced MDM for 14 h with increasing concentrations of type I IFN and then added Vpx-containing VLP. After 2 h, we infected the cells with HIV-1 reporter virus. Treatment of the MDM with type I IFN caused them to become resistant to HIV-1, with 100 U of type I IFN/ml reducing the infection to a background level (Fig. 7A). Vpx-containing VLP enhanced the infection of treated and untreated MDM, but the effect was significantly more pronounced in the type I IFN-treated MDM. Without type I IFN, the Vpx-containing VLP enhanced HIV-1 infection by 10-fold. In MDM induced with 100 U of type I IFN/ml, the VLP were 10-fold more effective, providing an overall 100-fold increase in infection. The VLP did not fully restore the infection, probably because multiple antiviral pathways are induced, not all of which are counteracted by Vpx. The effect of type I IFN was not modeled in differentiated THP-1. In these cells, type I IFN restricted HIV-1 infection, but the enhancing effect of Vpx was not magnified in the presence of IFN (see Fig. S3 in the supplemental material). We also tested whether Vpx helped SIVmac to counteract the type I IFN-induced antiviral state. We found that type I IFN-treated MDM were relatively resistant to SIV and that wild-type SIV was more infectious than Δvpx at lower type I IFN concentrations. However, the magnitude of the Vpx effect was not increased by type I IFN (see Fig. S2 in the supplemental material).
FIG. 7.
Vpx allows HIV-1 to overcome the type I IFN-induced antiviral state. (A) MDM were induced for 14 h with increasing concentrations of type I IFN, incubated for 2 h with wild-type VLP or medium, and then infected with HIV-1 luciferase reporter virus. One set of wells was left uninfected (mock) to control for background signal. Luciferase activity was measured 4 days postinfection. The results are representative of two independent experiments with MDM from different donors. (B) MDM were induced with 100 U of type I IFN/ml overnight. The cells were then incubated with Δvpx VLP complemented with empty vector (pcDNA), wild-type or Q76A Vpx, or the chimeras R22X or X86R and after 2 h, infected with HIV-1 reporter virus. Luciferase activity was measured 4 days postinfection. The data are presented as the fold enhancement of HIV-1 infection relative to cells incubated with Δvpx VLP. The results are representative of three independent experiments using MDM prepared from PBMC of three different donors.
To determine the mechanism by which the VLP enhanced the infection of type I IFN-induced MDM, we compared the effect of wild-type, Δvpx, Q76A-containing, and R22X chimera-containing VLP on HIV-1 infection of type I IFN-induced MDM (Fig. 7B). Δvpx VLP, Q76A, and the R22X chimera were inactive, whereas wild-type Vpx and the X86R chimera enhanced the infection. Thus, the activity of the VLP was Vpx dependent and mediated by the interaction with DCAF1.
DISCUSSION
An obstacle to understanding the roles of the Vpr and Vpx lentiviral accessory proteins has been their relatively modest phenotype in vitro. Although Vpr remains enigmatic, recent findings from several laboratories have established a clear, cell-type-specific phenotype for Vpx (8, 14, 18, 32, 35). Here, we show that in both single-cycle reporter virus assays and in trans by preincubation with VLP, Vpx can provide a 100-fold boost to the infection of MDM. Some donor variability was apparent in independent repetitions of the experiments, but in cells from all donors tested, Vpx provided a significant boost to infection. The VLP were produced from vectors that differed only by the presence or absence of Vpx. In addition, in the single-cycle assays, Δvpx SIV was complemented with a Vpx expression vector, definitively assigning the effects on infection to virion-packaged Vpx and ruling out confounding effects, such as differences in genomic viral RNA structure. Vpr and Vpx are related by amino acid sequence, have a similar cellular localization, are packaged into virions through an association with Gag p6, and associate with the same E3 ubiquitin ligase through an interaction with DCAF1 (1, 12, 13, 24, 40). Yet, for reasons that are not clear, Vpr is nearly inactive in both assays used in the present study.
The similarity in predicted structures of Vpr and Vpx provided an opportunity to map the functional domains of Vpx by the analysis of a panel of Vpx/Vpr chimeras. Determination of the function of the chimeras in the single-cycle complementation assay mapped the amino terminus of Vpx as critical. Dissection of this region by the analysis of point mutants in this domain identified amino acids within positions 9 to 22 that were required for Vpx to enhance HIV-1 and SIV infection of MDM. Biochemical analysis of the mutant proteins demonstrated that they retained the ability to bind DCAF1 despite their failure to enhance infection. This finding is consistent with localization of a DCAF1 binding site more toward the carboxy terminus in helix 3, where Q76 is located (24). Vpx contains a prominent polyproline stretch at the carboxy terminus, but this was not required for biological function. The finding that the point mutants retain the ability to bind DCAF1 suggests that the amino-terminal domain serves another role, potentially serving as a binding site for the proposed cellular restriction factor.
Support for the hypothesis that Vpx serves to counteract a cellular restriction to lentivirus replication in MDM and DC was provided by Sharova et al. (32), who showed that heterokaryons formed between MDM and COS were resistant to Δvpx but not wild-type SIV. This finding, together with the association of Vpx with an E3 ubiquitin ligase (8, 32, 35), is consistent with a role for Vpx in inducing the ubiquitination and degradation of a host restriction factor. Such a role would be analogous to that of Vif, which associates with an E3 ubiquitin ligase to induce the degradation of APOBEC3 proteins (26, 33, 36, 43). The finding that amino-terminal mutants of Vpx retain the ability to associate with DCAF1 and to package into virions, yet fail to enhance infection, is consistent with this hypothesis.
The pattern of permissivity for Δvpx virus does not coincide with that for Δvif virus, arguing against a role for APOBEC3G as the Vpx target (16, 23). APOBEC3A, which has been proposed to act on in-coming virions, is theoretically a target; however, we did not detect an interaction between these two proteins by coimmunoprecipitation (data not shown). THP-1 was the only cell line we found that models the Vpx phenotype. This cell line becomes nonpermissive upon differentiation into macrophage-like cells with PMA (16), which is consistent with the induction of a developmentally controlled restriction factor. The ability of Vpx to function when introduced in trans by VLP suggests that it does not need to associate with the virion that it protects (17). It is unlikely that Vpx molecules released from the VLP would associate with a separate incoming virion, since the affinity of Vpx for Gag p6 is relatively low (21).
Type I IFN induces a panoply of antiviral cellular factors that restrict the replication of viruses, including lentiviruses. Although the Vpx phenotype was present in uninduced MDM, induction with type I IFN increased the magnitude of the effect. This finding suggests that the Vpx target is type I IFN-inducible and that Vpx plays a role in allowing the virus to escape this arm of the innate immune system. Precedent for such a role has been provided by Vpu, which counteracts the type I IFN inducible restriction factor, tetherin/BST2 (29, 41). For SIVmac infection, the Vpx effect was only marginally increased in MDM induced with type I IFN. We cannot explain this difference between HIV-1 and SIVmac, but it is possible that MDM express multiple restriction factors to which the two viruses differ in susceptibility. SIV could be sensitive to a restriction factor that is induced by type I IFN but not targeted by Vpx, accounting for why Vpx countered the restriction to HIV-1 but not to SIV in the type I IFN-induced MDM.
Quantitation of the viral cDNA in newly infected MDM demonstrated that Vpx acts early in reverse transcription of SIVmac and HIV-1; this is consistent with the findings of Srivastava et al. (35). A role early in reverse transcription argues against a role in nuclear import, a process that occurs only after reverse transcription is largely completed. In this analysis, Δvpx SIVmac generated a small peak of early reverse transcripts that were rapidly degraded. This could have been caused by abortive reverse transcription due to a block to uncoating or decreased reverse transcriptase processivity. TRIM5α acts at a similar stage in the virus replication cycle (37). However, the block to Δvpx virus is not consistent with restriction by TRIM5α, which is abrogated by preincubation of target cells with VLP, regardless of whether they contain Vpx.
It is paradoxical that Vpx provides a dramatic enhancement to HIV-1 infection and yet does not itself encode this protein. Instead, it encodes Vpr, which in the in vitro assays, had only a modest effect on MDM infection (a <2-fold boost to Δvpx SIV infection of MDM on average). It remains possible that Vpr plays an important and similar role for HIV-1 replication but that it counteracts a restriction factor that is not highly expressed under the conditions in which the MDM were cultured in these studies. Further advances in understanding these questions will be facilitated by the identification of the cellular targets of these proteins, the existence of which are further supported by these studies.
Supplementary Material
Acknowledgments
This study was supported by grants from the NIH (AI067059 and AI058864). T.G. is the recipient of a Research Fellowship of the Deutsche Forschungsgemeinschaft (GR3355/1-1), and N.R.L. is an Elizabeth Glaser fellow of the Pediatric AIDS Foundation.
We thank Andrea Cimarelli for pSIV3+.
Footnotes
Published ahead of print on 18 November 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Accola, M. A., A. A. Bukovsky, M. S. Jones, and H. G. Gottlinger. 1999. A conserved dileucine-containing motif in p6(gag) governs the particle association of Vpx and Vpr of simian immunodeficiency viruses SIV(mac) and SIV(agm). J. Virol. 73:9992-9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balliet, J. W., D. L. Kolson, G. Eiger, F. M. Kim, K. A. McGann, A. Srinivasan, and R. Collman. 1994. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: mutational analysis of a primary HIV-1 isolate. Virology 200:623-631. [DOI] [PubMed] [Google Scholar]
- 3.Bates, P. A., L. A. Kelley, R. M. MacCallum, and M. J. Sternberg. 2001. Enhancement of protein modeling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins 2001(Suppl. 5):39-46. [DOI] [PubMed] [Google Scholar]
- 4.Bates, P. A., and M. J. Sternberg. 1999. Model building by comparison at CASP3: using expert knowledge and computer automation. Proteins 1999(Suppl. 3):47-54. [DOI] [PubMed] [Google Scholar]
- 5.Belshan, M., L. A. Mahnke, and L. Ratner. 2006. Conserved amino acids of the human immunodeficiency virus type 2 Vpx nuclear localization signal are critical for nuclear targeting of the viral preintegration complex in non-dividing cells. Virology 346:118-126. [DOI] [PubMed] [Google Scholar]
- 6.Belzile, J. P., G. Duisit, N. Rougeau, J. Mercier, A. Finzi, and E. A. Cohen. 2007. HIV-1 Vpr-mediated G2 arrest involves the DDB1-CUL4AVPRBP E3 ubiquitin ligase. PLoS Pathog. 3:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bender, A. T., C. L. Ostenson, D. Giordano, and J. A. Beavo. 2004. Differentiation of human monocytes in vitro with granulocyte-macrophage colony-stimulating factor and macrophage colony-stimulating factor produces distinct changes in cGMP phosphodiesterase expression. Cell Signal. 16:365-374. [DOI] [PubMed] [Google Scholar]
- 8.Bergamaschi, A., D. Ayinde, A. David, E. Le Rouzic, M. Morel, G. Collin, D. Descamps, F. Damond, F. Brun-Vezinet, S. Nisole, F. Margottin-Goguet, G. Pancino, and C. Transy. 2009. The human immunodeficiency virus type 2 Vpx protein usurps the CUL4A-DDB1 DCAF1 ubiquitin ligase to overcome a postentry block in macrophage infection. J. Virol. 83:4854-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berger, G., C. Goujon, J. L. Darlix, and A. Cimarelli. 2009. SIVMAC Vpx improves the transduction of dendritic cells with nonintegrative HIV-1-derived vectors. Gene Ther. 16:159-163. [DOI] [PubMed] [Google Scholar]
- 10.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 11.Contreras-Moreira, B., and P. A. Bates. 2002. Domain fishing: a first step in protein comparative modeling. Bioinformatics 18:1141-1142. [DOI] [PubMed] [Google Scholar]
- 12.Di Marzio, P., S. Choe, M. Ebright, R. Knoblauch, and N. R. Landau. 1995. Mutational analysis of cell cycle arrest, nuclear localization and virion packaging of human immunodeficiency virus type 1 Vpr. J. Virol. 69:7909-7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fletcher, T. M., III, B. Brichacek, N. Sharova, M. A. Newman, G. Stivahtis, P. M. Sharp, M. Emerman, B. H. Hahn, and M. Stevenson. 1996. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIV(SM). EMBO J. 15:6155-6165. [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita, M., M. Otsuka, M. Miyoshi, B. Khamsri, M. Nomaguchi, and A. Adachi. 2008. Vpx is critical for reverse transcription of the human immunodeficiency virus type 2 genome in macrophages. J. Virol. 82:7752-7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbs, J. S., A. A. Lackner, S. M. Lang, M. A. Simon, P. K. Sehgal, M. D. Daniel, and R. C. Desrosiers. 1995. Progression to AIDS in the absence of a gene for vpr or vpx. J. Virol. 69:2378-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goujon, C., V. Arfi, T. Pertel, J. Luban, J. Lienard, D. Rigal, J. L. Darlix, and A. Cimarelli. 2008. Characterization of simian immunodeficiency virus SIVSM/human immunodeficiency virus type 2 Vpx function in human myeloid cells. J. Virol. 82:12335-12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goujon, C., L. Jarrosson-Wuilleme, J. Bernaud, D. Rigal, J. L. Darlix, and A. Cimarelli. 2006. With a little help from a friend: increasing HIV transduction of monocyte-derived dendritic cells with virion-like particles of SIV(MAC). Gene Ther. 13:991-994. [DOI] [PubMed] [Google Scholar]
- 18.Goujon, C., L. Riviere, L. Jarrosson-Wuilleme, J. Bernaud, D. Rigal, J. L. Darlix, and A. Cimarelli. 2007. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirsch, V. M., M. E. Sharkey, C. R. Brown, B. Brichacek, S. Goldstein, J. Wakefield, R. Byrum, W. R. Elkins, B. H. Hahn, J. D. Lifson, and M. Stevenson. 1998. Vpx is required for dissemination and pathogenesis of SIV(SM) PBj: evidence of macrophage-dependent viral amplification. Nat. Med. 4:1401-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hrecka, K., M. Gierszewska, S. Srivastava, L. Kozaczkiewicz, S. K. Swanson, L. Florens, M. P. Washburn, and J. Skowronski. 2007. Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl. Acad. Sci. U. S. A. 104:11778-11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenkins, Y., O. Pornillos, R. L. Rich, D. G. Myszka, W. I. Sundquist, and M. H. Malim. 2001. Biochemical analyses of the interactions between human immunodeficiency virus type 1 Vpr and p6(Gag). J. Virol. 75:10537-10542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khamsri, B., F. Murao, A. Yoshida, A. Sakurai, T. Uchiyama, H. Shirai, Y. Matsuo, M. Fujita, and A. Adachi. 2006. Comparative study on the structure and cytopathogenic activity of HIV Vpr/Vpx proteins. Microbes Infect. 8:10-15. [DOI] [PubMed] [Google Scholar]
- 23.Koning, F. A., E. N. Newman, E. Y. Kim, K. J. Kunstman, S. M. Wolinsky, and M. H. Malim. 2009. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 83:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Rouzic, E., N. Belaidouni, E. Estrabaud, M. Morel, J. C. Rain, C. Transy, and F. Margottin-Goguet. 2007. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 6:182-188. [DOI] [PubMed] [Google Scholar]
- 25.Mahnke, L. A., M. Belshan, and L. Ratner. 2006. Analysis of HIV-2 Vpx by modeling and insertional mutagenesis. Virology 348:165-174. [DOI] [PubMed] [Google Scholar]
- 26.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 27.Munk, C., S. M. Brandt, G. Lucero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. U. S. A. 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Negre, D., P. E. Mangeot, G. Duisit, S. Blanchard, P. O. Vidalain, P. Leissner, A. J. Winter, C. Rabourdin-Combe, M. Mehtali, P. Moullier, J. L. Darlix, and F. L. Cosset. 2000. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther. 7:1613-1623. [DOI] [PubMed] [Google Scholar]
- 29.Neil, S. J., T. Zang, and P. D. Bieniasz. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425-430. [DOI] [PubMed] [Google Scholar]
- 30.Paxton, W., R. I. Connor, and N. R. Landau. 1993. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J. Virol. 67:7229-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schrofelbauer, B., Y. Hakata, and N. R. Landau. 2007. HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proc. Natl. Acad. Sci. U. S. A. 104:4130-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharova, N., Y. Wu, X. Zhu, R. Stranska, R. Kaushik, M. Sharkey, and M. Stevenson. 2008. Primate lentiviral Vpx commandeers DDB1 to counteract a macrophage restriction. PLoS Pathog. 4:e1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 34.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 35.Srivastava, S., S. K. Swanson, N. Manel, L. Florens, M. P. Washburn, and J. Skowronski. 2008. Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog. 4:e1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 37.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 38.Tan, L., E. Ehrlich, and X. F. Yu. 2007. DDB1 and Cul4A are required for human immunodeficiency virus type 1 Vpr-induced G2 arrest. J. Virol. 81:10822-10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tristem, M., C. Marshall, A. Karpas, and F. Hill. 1992. Evolution of the primate lentiviruses: evidence from vpx and vpr. EMBO J. 11:3405-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tristem, M., A. Purvis, and D. L. Quicke. 1998. Complex evolutionary history of primate lentiviral vpr genes. Virology 240:232-237. [DOI] [PubMed] [Google Scholar]
- 41.Van Damme, N., D. Goff, C. Katsura, R. L. Jorgenson, R. Mitchell, M. C. Johnson, E. B. Stephens, and J. Guatelli. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita, M., and M. Emerman. 2005. The cell cycle independence of HIV infections is not determined by known karyophilic viral elements. PLoS Pathog. 1:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 44.Yu, X. F., Q. C. Yu, M. Essex, and T. H. Lee. 1991. The vpx gene of simian immunodeficiency virus facilitates efficient viral replication in fresh lymphocytes and macrophage. J. Virol. 65:5088-5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.