Abstract
The 3′ termini of Alfalfa mosaic virus (AMV) RNAs adopt two mutually exclusive conformations, a coat protein binding (CPB) and a tRNA-like (TL) conformer, which consist of a linear array of stem-loop structures and a pseudoknot structure, respectively. Previously, switching between CPB and TL conformers has been proposed as a mechanism to regulate the competing processes of translation and replication of the viral RNA (R. C. L. Olsthoorn et al., EMBO J. 18:4856-4864, 1999). In the present study, the switch between CPB and TL conformers was further investigated. First, we showed that recognition of the AMV 3′ untranslated region (UTR) by a tRNA-specific enzyme (CCA-adding enzyme) in vitro is more efficient when the distribution is shifted toward the TL conformation. Second, the recognition of the 3′ UTR by the viral replicase was similarly dependent on the ratio of CBP and TL conformers. Furthermore, the addition of CP, which is expected to shift the distribution toward the CPB conformer, inhibited recognition by the CCA-adding enzyme and the replicase. Finally, we monitored how the binding affinity to CP is affected by this conformational switch in the yeast three-hybrid system. Here, disruption of the pseudoknot enhanced the binding affinity to CP by shifting the balance in favor of the CPB conformer, whereas stabilizing the pseudoknot did the reverse. Together, the in vitro and in vivo data clearly demonstrate the existence of the conformational switch in the 3′ UTR of AMV RNAs.
Alfalfa mosaic virus (AMV) is a plant virus that belongs to one of the five genera in the family Bromoviridae, whose genomes consist of three genomic RNAs (RNAs 1, 2, and 3) and one subgenomic RNA (RNA4) that are capped at the 5′ end and lack polyadenylation at the 3′ terminus (3). RNAs 1 and 2 encode the viral subunits P1 and P2 of the replicase, respectively. RNA3 encodes the movement protein and serves as a template for the synthesis of RNA4, which encodes the coat protein (CP).
The role of AMV CP has been the subject of extensive research in the past four decades. Initially, it was found that, in contrast to RNAs of the Bromo-, Cucumo-, and Oleavirus genera, the genomic RNAs of AMV and the closely related genus Ilarvirus were not infectious as such but required the presence of CP in the inoculum (15). This phenomenon was called genome activation and was long considered to compensate for the lack of a tRNA-like structure (TLS) at the 3′ end of their genomic RNAs, a prominent feature of bromo- and cucumovirus RNAs (3). However, in 1999 we demonstrated that the 3′ end of AMV RNAs can adopt an alternative conformation that shows many structural similarities to the TLS of other Bromoviridae, although it could not be charged with an amino acid (20). The tRNA-like (TL) conformation (Fig. 1) turned out to be the replicative form of the 3′ termini (19, 20), whereas the other, coat protein binding (CPB), conformer was subsequently shown to be required for translation (16-18). Although other models have been forwarded (9), we have proposed that switching between these two conformations, mediated by CP binding, plays a fundamental role in the life cycle of AMV and ilarviruses by regulating the competing processes of translation and replication of the viral RNAs.
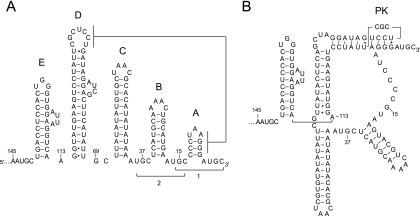
FIG. 1.
The CPB and the TL conformations of the AMV RNA3 3′ terminus. The two conformers of AMV RNA3 3′ 145 nt are shown. (A) CPB conformer. The two major CP binding sites are indicated by brackets. Base pairing between loop D and stem A promotes TL conformation. (B) Secondary structure of the TL conformer.
In the present study, the distribution between CPB and TL conformers was further investigated. We addressed how changes in this distribution would affect recognition of the AMV 3′ untranslated region (UTR) by a tRNA-specific enzyme (CCA-adding ezyme) and the viral polymerase in vitro. We also monitored how the binding affinity to CP is affected by this conformational switch in vivo using the yeast three-hybrid (Y3H) system (2, 11, 24). Together, the in vitro and in vivo data clearly demonstrate the existence and function of the conformational switch in the 3′ UTR of AMV RNAs.
MATERIALS AND METHODS
Construction of mutant templates for in vitro assays.
Mutations in loopD of RNA3 were engineered by PCR mutagenesis on the plasmid 3kWT, which harbors a cDNA copy of AMV RNA3 (27). This resulted in plasmids 3kDiC, which contains an additional C-residue in loopD, and 3k4U, which has a C-to-U mutation in loopD. Plasmids 3kWT, 3kDiC, and 3k4U were used as a template in PCR with the primers T7C (5′-AATTTAATACGACTCACTATAGGTTATATATGTGCTAACGC-3′) and AMTS1 (5′-CTACCTCCAGCATCTCCTATAAGGAGGCATTCAGTAGTATATTATATACTACTGGCACTTTATATATGTGCGTTAGC-3′). The resulting three PCR fragments were subsequently used as templates for a second PCR using the oligonucleotides T7C and TS3 (5′-GCATCTCCTATAAAGGAGGCAT-3′), TS1GA (5′-GCATCCCTTATAAAGGAGGCATTCATGCAG-3′), TS1UC (5′-GCATCTCCTATAAAAGGGGCATTCATGCAGTTTTGC-3′), or TS1GAUC (5′-GCATCCCTTATAAAAGGGGCATTCATGCAGTTTTGC-3′). The final 12 PCR fragments served as templates for transcription by T7 RNA polymerase. A wild-type template was obtained by PCR using oligonucleotides T7C and WT2 (5′-GCATCCCTTAGGGGCATTCAT-3′) on plasmid 3kWT.
Adenylation and replication assays.
Transcription reactions and purification of transcripts were performed as described previously (20). CCA-adding reactions were carried out at 4°C for 22 h, essentially as described previously (20). In the case of CP addition, RNA (4 pmol) was preincubated with 1 or 5 pmol of CP dimers for 15 min at room temperature. Replication assays were carried out as described previously (20) using 7 pmol RNA as a template. In the case of CP addition, 4 pmol RNA was preincubated with 2 or 20 pmol of CP dimers for 15 min at room temperature prior to the replication assay.
Yeast strain and plasmid constructs.
A DNA fragment containing the AMV CP gene was obtained by PCR on plasmid 3kWT using the primers 5′-ACGGAGCCCGGGTATGAGTTCTTCACAAAAGAAAGCTGGTGGG-3′ and 5′-CTGCAGCTCGAGTCAATGACGATCAAGATCGTCAGCTTCGTC-3′. After digestion by SmaI and XhoI, the CP gene was inserted into shuttle vector pGADT7 (Clontech) for protein expression. The cDNA fragments of wt and the variant 113-nt 3′ terminus of AMV RNA3 were synthesized by PCR on the template plasmid 3kWT by using combinations of variant forward and reverse primers. To construct wt and A4 series variants, the forward primers WtF (5′-CAACGCGCTAGCAGGTATGAAGTCCTATTCGCTCCTGATAGGATCGACTTCATATTGC-3′) and A4F (5′-CAACGCGCTAGCAGGTATGAAGTCCTATTAAAAGATAGGATCGAC TTCATATTGC-3′) and the reverse primers 1-7R (5′-GGTCGAAGATCTGCATCCCTTACCCGCATTCATGCAGTTTTGCATGAGC-3′), Ts-1R (5′-GGTCGAAGATCTGCATCTCCTATAAAAGGGGCATTCATGCAGTTTTGCATGAG C-3′), Ts-2R (5′-GGTCGAAGATCTGCATCTCCTATAAAGGGGCATTCATGCAGTTTTGCATGAGC-3′), Ts-3R (5′-GGTCGAAGATCTGCCTCCTATAAAAGGGGCATTCATGCAGTTTTGCATGAGC-3′), Ts-4R (5′-GGTCGAAGATCTCATCTCCTATAAAAGGGACATTCATGCAGTTTTGCATGAGCC-3′), and WtR (5′-GGTCGAAGATCTGCATCCCTTAGGGGCATTCATGCAGTTTTGCATGAGC-3′) were used to amplify cDNA fragments from plasmid 3kWT for cloning constructs Wt/Wt, A4/Wt, A4/Ts-1, A4/Ts-2, A4/Ts-3, A4/Ts4, A4/1-7, Wt/Ts-1, and Wt/1-7. These wt or variant sequences were inserted between NheI and BglII sites in a modified pIIIA/MS2.1 (13, 14), of which the unique SmaI and the flanking redundant sequences were removed by replacing the EcoRI fragment with the sequence 5′-AATTTATACTCACATGAGGATCACCCATGTAATTAACACTGAGGATCACCCAGTGGCTAGCTTCTAGAAAGATCTG-3′, creating unique NheI, XbaI, and BglII sites downstream of the MS2 CP binding sequences. Constructed RNA expressing vectors were cotransformed with pGADT7-AMV/CP into yeast strain L40-coat (24). Transformed cells inheriting both vectors were selected and cultured for subsequent assays measuring HIS3 activity.
Measurements of HIS3 activity.
To assay the reporter of RNA-protein interaction, expression of HIS3, we monitored the phenotypic growth of transformed L40 coat cells on 3-AT containing YNB medium in a way that slightly differed from what had been developed (13). Briefly, three independent colonies of each RNA-protein pair transformed cells were cultured 2 days in Leu−, Ade−, and Ura− YNB medium at 30°C. The cells were then subcultured, in 1-to-1,000 ratio, to an optical density at 600 nm of ∼0.2, followed by applying 2 μl onto Leu−, Ade−, Ura−, and His− YNB agar containing 0 to 100 mM 3-AT. After culture for 4 days at 30°C, the phenotypic growth of each clone was documented.
RESULTS
Recognition of the AMV 3′ UTR by CCA-adding enzyme and viral polymerase is sensitive to the distribution between hairpin and pseudoknot conformers.
We have previously shown that ATP(CTP):tRNA nucleotidyl transferase, the enzyme that adds CCA to the 3′ end of tRNAs, can add a radioactive AMP to the 3′ end of AMV RNA in vitro (20). To investigate the effect of changes in the distribution between hairpin and pseudoknot conformers on the adenylation efficiency we introduced mutations in either the pseudoknot, the hairpin A, or both in the 3′ 142 nucleotides (nt) of AMV RNA3 (Fig. 2A). Replacement of the wild-type (wt) hairpin (hp1) by a more stable variant (hp2) caused a reduction in adenylation (Fig. 2B, lanes 10 and 8). The predicted stacking energies of hp1 and hp2 are −6.6 and −10.9 kcal/mol at 37°C, respectively (or −13.4 if the terminal 3 nt are folding back as shown for hp2 in Fig. 2A). We did not take into account the energy contribution of the loop in order to make a better comparison with the pseudoknot stabilities for which no reliable loop energies exist (8). Reducing the stability of pk1 from −7.9 to −5.2 kcal/mol by a C-to-U change (Fig. 2A, pk2) in the context of hp2 led to undetectable levels of adenylation (Fig. 2B, lane 2). On the other hand, insertion of a C residue in the loop of hpD allows the formation of an extra base pair (bp) in the pseudoknot (Fig. 2A, pk3/hp2) and led to an increase in adenylation (Fig. 2B, lane 5). Thus, in the context of a certain stability of hairpin A (hp2) the recognition by the CCA enzyme can be improved by increasing the pseudoknot stability and hence the fraction of TL conformers.
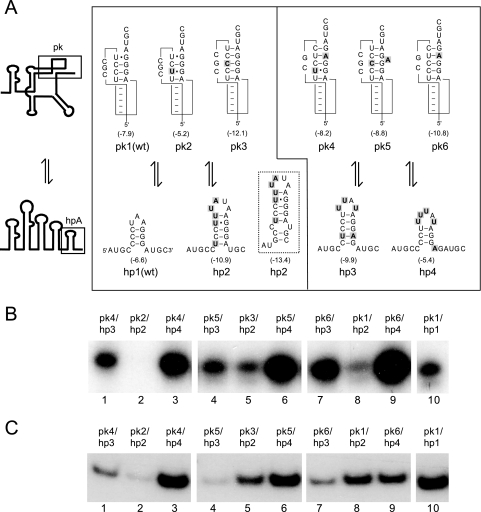
FIG. 2.
Adenylation and replication of pseudoknot and hairpin mutants (A) Structure of five different pseudoknot variants and four different hairpin variants are depicted. Note that pk1, pk2, and pk3 can only be tested in combination with hairpins hp1 and hp2 (left panel), whereas pk4, pk5, and pk6 can only be tested in combination with hp3 and hp4 (right panel). Values between brackets are the calculated stacking energies (31) for the “top” four or five base pairs in the pseudoknots or all base pairs in the hairpins. An alternative conformation of hp2 is indicated by the dashed box. (B) Denaturing gel showing products of the adenylation reactions with the indicated constructs (C) Products of minus-strand assays using the indicated transcripts as templates with AMV polymerase.
Conversely, if adenylation of RNA transcripts is dependent on the amount of RNA in the TL conformation, then, for a given pseudoknot, reducing hpA stability would increase labeling. This assumption was tested by pseudoknots pk4, pk5, and pk6 (Fig. 2A). In combination with a strong hairpin (hp3, −9.9 kcal/mol), labeling of these constructs was comparable to that of the wild type (Fig. 2B, compare lanes 1, 4, and 7 to lane 10), while weakening of the hairpin (hp4) led in all cases to an increase in adenylation (Fig. 2B, lanes 3, 6, and 9).
Since the replication of AMV RNA also depends on the formation of the TL structure (20), we expected to see a similar pattern regarding minus-strand synthesis by the viral replicase when the same set of variants was studied. Comparing Fig. 2B with 2C, the resemblance between the two assays is quite remarkable. Although quantitatively not the same, qualitatively we see the same trend. Again, if we compare the wt with pk1/hp2 we see a decrease in template activity (Fig. 2C, lanes 10 and 8), which is further decreased by weakening the pseudoknot pk2/hp2 (Fig. 2C, lane 2). Construct pk3/hp2 with a strong pseudoknot was a better template than pk2/hp2 but similar to pk1/hp2 (Fig. 2C, lane 5). The results for pseudoknots pk4, pk5, and pk6 parallel those of the adenylation assay. In combination with the stable hairpin hp3 (Fig. 2C, lanes 1, 4, and 7), the template activity of pk4 to pk6 was quite low; however, minus-strand synthesis was greatly improved by introduction of the weaker hp4 (Fig. 2C, lanes 3, 6, and 9).
In contrast to the adenylation assays, where four constructs were significantly better labeled than the wt, in the minus-strand assay none of the templates was better than the wt. A possible explanation for this is given in the discussion.
Coat protein antagonizes CCA addition and replication by shifting the distribution toward the hairpin conformation.
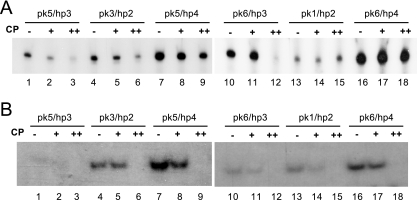
To understand how the recognition of TL conformation was affected by the presence of AMV CP, some of the constructs were incubated with low and high concentrations of CP (1:0.25 and 1:1.25 ratio of RNA to CP dimers) prior to adenylation. Low ratios of CP to RNA (0.25 to 1) had some inhibitory effect on the adenylation of pk5/hp3 but no significant effect on the other constructs (Fig. 3, lanes “+”). At a higher ratio of CP to RNA (1.25 to 1), adenylation of pk5/hp3, pk6/hp3 was severely reduced (Fig. 3, lanes 3 and 12). Adenylation of pk3/hp2 and pk5/hp4 was relatively moderately effected, while pk1/hp2 and pk6/hp4 seemed to be unaffected by the presence of CP (Fig. 3, lanes 3 to 9 and lanes 13 to 18). In constructs with a weak hairpin (hp4) CP was apparently unable to bind the RNA and induce a conformational shift (Fig. 3, pk5/hp4 and pk6/hp4). The relative resistance of constructs with hp2 to CP (Fig. 3, lanes 4 to 6 and lanes 13 to 15) can be explained by the extended base pairing of hp2 that interferes with CP binding to AUGC motifs (Fig. 2A). Constructs with hp3 are expected to bind CP very well, and these RNAs will easily be sequestered by CP and become unavailable for the CCA enzyme.
FIG. 3.
CP blocks adenylation and minus-strand synthesis In vitro. (A) Adenylation of the indicated transcripts by CCA-adding enzyme was done in the absence (−) or presence (+ and ++) of AMV CP. RNA/CP ratios were 1 to 0.25 (+) and 1 to 1.25 (++). (B) Minus-strand assay in the presence of CP. RNA/CP ratios were 1 to 0.5 (+) and 1 to 5 (++).
The effect of preincubating CP and templates before minus-strand synthesis is shown in Fig. 3B. At a CP/RNA ratio of 0.5 to 1 (lanes “+”), templates that are predicted to have strong pseudoknots, pk3 and pk6, suffered less from the addition of CP than those with the weaker pseudoknots, pk1 and pk5. Templates with the weaker hp4 were also relatively more resistant to CP than those with the stronger hp3, as also observed with the adenylation assay in Fig. 3A, although the intensities of the bands in lanes 1 and 2 of Fig. 3B do not allow us to draw firm conclusions about the pk5/hp3 template. Template pk1/hp2, which was resistant to CP addition in the adenylation assay, was sensitive to CP addition in the minus-strand assay (Fig. 3B, lanes 13 and 14). We note, however, that the CP/RNA ratios are different between the two assays and that the viral polymerase preparation is naturally contaminated with CP (22, 26), which would result in even higher CP/RNA ratios. Nevertheless, comparison of pk1/hp2 with pk3/hp2 shows that a stronger pseudoknot is capable of preventing CP of inhibiting minus-strand synthesis at a CP/RNA ratio of 0.5 to 1. (Fig. 3B, compare lanes 5 and 14). At the high CP/RNA ratio of 5 to 1, minus-strand synthesis was inhibited with all templates (Fig. 3B, lanes 3, 6, 9, 12, 15, and 18).
Interaction between AMV CP and RNA3 in the yeast three-hybrid system.
As independent support for the existence of the switch under in vivo conditions, in the absence of translation or replication, we applied the yeast three-hybrid system (2). For that purpose, AMV CP was fused to the Lex activation domain and expressed in parallel to chimeric transcripts composed of phage MS2 CP binding elements and sequences of the 3′ terminal 112 nt of the AMV RNA3 in yeast strain L40. L40 continuously expresses fusion proteins of MS2 phage CP and Lex DNA-binding domain. Figure 4A outlines the structure of the chimeric RNAs that were used in this assay. Wild-type AMV RNA3 3′ termini showed strong binding affinity to AMV CP, as indicated by growth of yeast cells on histidine-free (HIS−) medium supplemented with a 95 mM concentration of inhibitor 3-aminotriazole (3-AT) (Fig. 4B, rows 3 and 4). In the absence of AMV RNA, cells could not grow on HIS− medium in the presence of 2.5 mM or higher concentrations of 3-AT, but growth was observed when histidine was present, even in 100 mM 3-AT (Fig. 4B, rows 1 and 2). This indicated that 3-AT, up to 100 mM, did not interfere with growth when histidine was available. On the other hand, 2.5 mM can be taken as the threshold for the existence of an RNA protein in these cells.
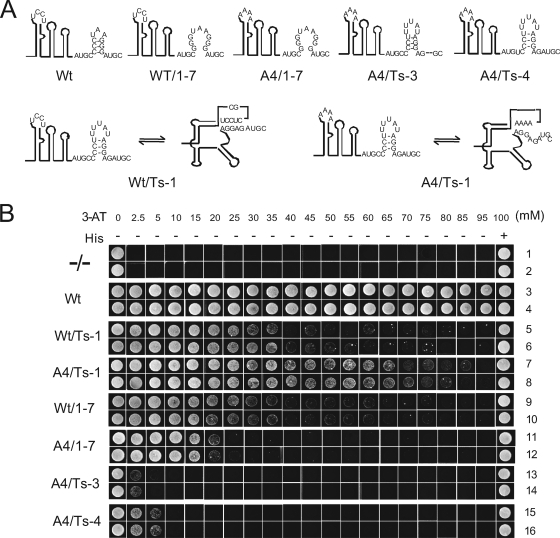
FIG. 4.
Conformational switch investigated in the yeast three-hybrid system. (A) Outline of the various constructs used in Y3H. Wt, 3′ 112 nt of wild-type RNA3; Wt/1-7, wt pseudoknot (pk) but disruption of hpA; A4/1-7, disruption of pk and hpA; A4/Ts-3 and A4/Ts-4, disruption of pk and mutation of the CP binding site; Wt/Ts-1, strong pk, weak hpA (see also pk5/hp4 Fig. 2A); A4/Ts-1, pk disrupted, weak hpA. (B) Growth of yeast cells transformed with the various constructs. Two independent colonies are grown for each construct. The negative control (−/−) lacked the 3′ terminus of the AMV RNA3. The concentration of 3-AT in the growth medium is indicated, as well as the presence (+) or absence (−) of histidine.
In this system we tested the effect of changes in the ratio between the two conformers on CP binding. Favoring the TL conformation by simultaneously strengthening the pseudoknot and weakening hpA (Fig. 4A, Wt/Ts1) severely reduced CP binding, as indicated by the at least threefold lower 3-AT tolerance (Fig. 4B, rows 5 and 6). We note that Wt/Ts1 actually has the same pk and hp as construct pk5/hp4 that was used for the in vitro assays (Fig. 2A). The reduced CP binding affinity of construct Wt/Ts-1 in the Y3H system thus provides independent support for the role of hpA stability for CP binding. Disruption of the pseudoknot of WT/Ts-1 by substitution of loopD residues by 4 A's (A4/Ts-1) raised 3-AT tolerance to 70 mM (Fig. 4B, rows 7 and 8). These data strongly suggest that the TL conformation is counteracting CP binding.
A more serious reduction in 3-AT tolerance was found when hpA was completely disrupted (Fig. 4A, construct Wt/1-7, Fig. 4B, rows 9 and 10). These results are in good agreement with previous in vitro data on CP binding to construct 1-7 (23). The A4 substitution in the context of the 1-7 construct did not result in a higher 3-AT tolerance (Fig. 4B, rows 11 and 12). In this case, blocking formation of TL conformers, as shown in A4/1-7, did not recover CP binding because hpA in 1-7 is a weak binder of CP anyway.
Two additional mutants demonstrated the role of the conserved AUGC motifs in CP binding. Deletion of AU from the terminal AUGC motif (Fig. 4A, A4/Ts-3), completely abolished CP binding (Fig. 4B, rows 13 and 14). 3-AT tolerance of these cells was less than 2.5 mM, i.e., almost as low as the negative control without any AMV RNA. A single C-to-U substitution in the second motif (Fig. 4A, A4/Ts4) also resulted in a dramatic reduction in 3-AT tolerance (Fig. 4B, rows 15 and 16). Both mutants demonstrate the sensitivity and specificity of the AMV RNA-CP interaction in yeast.
It has been reported that stretches of four U's or more in RNA transcripts may result in premature termination of RNA synthesis in the Y3H system (13). A stretch of four U's is present in the hpA of the Ts-1 constructs (Fig. 4A); however, removing one of the U's did not significantly alter CP affinity (data not shown). Therefore, the concern of premature termination was eliminated.
DISCUSSION
In this study, we have investigated the distribution between TL and CPB conformers at the 3′ terminus of AMV RNA3 by proteins that recognize either the TL or the CPB conformation. The data indicated that a tRNA-specific enzyme is very sensitive to changes in this distribution: mutations that favored the tRNA-like conformation resulted in better recognition by this enzyme, and vice versa, mutations that favored the CPB conformation were detrimental for recognition by this enzyme. The same trend was observed when these constructs were used as templates for the AMV polymerase. Moreover, both the tRNA-specific enzyme and the viral polymerase were inhibited by addition of AMV CP, which apparently diminished the fraction of TL conformers by sequestering RNA molecules in the CPB conformation. Based on these data we conclude that the TL conformation is the actual conformation that is required for minus-strand synthesis.
Although qualitatively similar, the adenylation and replication assays showed some quantitative differences. One difference is that most templates were as good or better than the wild type in adenylation but usually worse than the wild type in replication. This may reflect the different mechanism of the two enzymes. The CCA-adding enzyme leaves the RNA after addition of the AMP, whereas the replicase remains bound and has to move through the pseudoknot. A too-stable pseudoknot might interfere with transcription elongation by the replicase. Inhibition of transcription by secondary structure near the 3′ end of the template has been reported by Deiman et al. (6) using the Turnip yellow mosaic virus polymerase.
The switch between the two conformers was also studied in vivo by expressing AMV CP and RNA3 in the yeast three-hybrid (Y3H) system. The data obtained with this system were also in agreement with the conformational switch model. Stabilization of the pseudoknot or destabilization of the hairpin resulted in lower affinity for CP, while disruption of the pseudoknot caused the opposite.
It had been reported that the 3-AT tolerance obtained in Y3H is linearly correlated with the affinity between binding proteins and the coexpressed RNA ligands (11). Applied to AMV, the dissociation constant for the RNA-CP interaction is estimated to be 10 nM or lower since we have not tested concentrations of 3-AT higher than 95 mM. This value compares well to the Kd obtained from in vitro binding assays using purified virion CP (24). The requirements for CP binding in the Y3H were the same as found by in vitro studies. Reusken and Bol (23) and Houser-Scott et al. (12) have shown that a single mutation in the second AUGC motif (counting from the 3′ end) abolished CP binding, whereas two mutations in the terminal motif were required to abolish binding. The integrity of hpA (mutant 1-7) was also crucial for CP binding in vitro (23), as in our Y3H.
The Y3H system allowed us for the first time to study CP binding under in vivo conditions in the absence of translation and replication. This is a clear advantage over plant or protoplast systems where mutations usually interfere with replication or translation of the viral RNA. The presence of host proteins such as the CCA-enzyme or amino-acyl synthetases (aaRS), which are strongly conserved between yeast and plants, may also play a role in vivo (although we doubt that the CCA-enzyme is active on AMV RNAs in the absence of Mn2+ in yeast). We have indications, however, that TyrRS but not HisRS or AlaRS do interfere with replication of AMV RNA in vitro (R. C. L. Olsthoorn, unpublished data). Another advantage of Y3H is the relative ease of testing CP mutants without having to purify them or of randomizing the RNA and selecting for sequences that have high affinity for CP.
There are also some disadvantages of the Y3H. One is that the transcripts that are produced contain additional sequences derived from the cloning vector and hence do not have an authentic 3′ end. For some proteins, this may be important, although AMV CP can also bind to internal sites on the RNA (29, 30). These 3′ extensions could also affect pseudoknot stability or lead to alternative structures. This may be one of the reasons why the wild-type construct is such a good CP binder in this system. In addition, the absence of hairpin E (HpE, Fig. 1) in the Y3H transcripts may destabilize the TL conformation and thus favor CP binding. We have previously suggested the existence of an interaction between HpE and the TLS (19).
Recently, the existence of the conformational switch has been disputed by Gehrke and coworkers (21). By performing band-shift assays using a synthetic 26-amino-acid N-terminal peptide (CP26) representing the CP RNA binding domain and mutants of the 3′ terminal 170-nt AMV RNA, it was shown that the presence of the pseudoknot does not interfere with the binding of CP26. These data are in contrast to our earlier findings with wild-type AMV CP (20) and to those of Aparicio et al. (1) with a related ilarvirus for which we also proposed the existence of a TLS (4). This discrepancy may have been caused by the lack of specificity of CP26; the bandshifts observed by Petrillo et al. (21) are independent of the integrity of hpA since mutant C3 in which all base-pairs of hpA are disrupted binds this peptide as well as the wild type or any other RNA with an intact hpA. This means that CP26 is apparently binding elsewhere in the 3′ UTR and therefore may not be a good representative of the wild-type CP. The only experiments carried out by Petrillo et al. (21) with wild-type CP do in fact show that in the presence of Mg2+ twofold more CP is needed to shift the wild-type RNA than is needed for pseudoknot mutants A4 and G3. This is in agreement with our previous data which showed that the addition of Mg2+ can block CP binding to the wild-type 3′ UTR but not to pseudoknot mutant A4 (20). The effect of Mg2+ on CP binding was already observed by Zuidema (29), who showed that binding of CP to the 3′ UTR of RNA1 but not to internal sites was inhibited at Mg2+ concentrations greater than 5 mM.
Finally, Petrillo et al. (21) observed that RNA3 containing the pseudoknot compensatory mutant GC3 does not replicate in nontransgenic cells, in contrast to our previous data using transgenic cells expressing the viral replicase proteins (20). Although in transgenic cells the accumulation of GC3 was only ∼20% of wild type, it was still much higher than that of any of the single mutants, G3 and C3, whose accumulation was almost undetectable. It is possible that the assay of Petrillo et al. (21) was not sufficiently sensitive to detect a fivefold reduction in RNA accumulation. We think that the lower accumulation of GC3 is a reflection of the strong conservation of 5′ pyrimidine (Py)-3′ purine (Pu) base pairs in the 3′ stem of the pseudoknot of AMV and related ilarvirus RNAs (4). The strong preference for 5′Py-3′Pu base pairs in the pseudoknot stems of all members of the Bromoviridae may be evolutionarily conserved for other reasons.
In some of our experiments, we have noticed that addition of a small amount of CP seemed to increase adenylation (Fig. 3A, e.g., lane 11 or lane 17). We do not think this effect is significant enough to support the model of the Gehrke lab, which states that low amounts of CP actually favor minus-strand synthesis (9). Previous experiments have shown that minus-strand RNA3 synthesis and plus-strand RNA4 synthesis can be perfectly carried out using a CP-free polymerase (5, 10, 28). Addition of CP to reactions with CP-free polymerase was found to block minus-strand synthesis. Tiny amounts of CP were able to stimulate plus-strand synthesis, but this stimulation was also achieved by the addition of bovine serum albumin (10). Also, our present data do not support a stimulatory role for CP in minus-strand synthesis.
In summary, we can state that the in vitro and in vivo data that we have presented here clearly demonstrate the existence and action of the conformational switch in the 3′ UTR of AMV RNAs. Recently, a careful analysis of ilarvirus RNA sequences has confirmed the possible existence of a TLS in all known ilarvirus RNAs (4). Thus, the conformational switch may be a general phenomenon of alfamo- and ilarviruses.
Footnotes
Published ahead of print on 18 November 2009.
REFERENCES
- 1.Aparicio, F., M. Vilar, E. Perez-Payá, and V. Pallás. 2003. The coat protein of prunus necrotic ringspot virus specifically binds to and regulates the conformation of its genomic RNA. Virology 313:213-223. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, D. S., N. Buter, C. Stumpf, and M. Wickens. 2002. Analyzing RNA-protein complexes using a yeast three-hybrid system. Methods 26:123-141. [DOI] [PubMed] [Google Scholar]
- 3.Bol, J. F. 2005. Replication of alfamo- and ilarviruses: role of the coat protein. Annu. Rev. Phytopathol. 43:39-62. [DOI] [PubMed] [Google Scholar]
- 4.Chen, S. C., A. P. Gultyaev, C. W. A. Pleij, and R. C. L. Olsthoorn. 2009. A secondary structure model for the 3′-untranslated region of ilarvirus RNAs. In Z. Feng and M. Long (ed.), Viral genomes: diversity, properties and parameters. Nova Science Publishers, Hauppage, NY.
- 5.de Graaff, M., M. R. Man in't Veld, and E. M. J. Jaspars. 1995. In vitro evidence that the coat protein of alfalfa mosaic virus plays a direct role in the regulation of plus and minus RNA synthesis: implications for the life cycle of alfalfa mosaic virus. Virology 208:583-589. [DOI] [PubMed] [Google Scholar]
- 6.Deiman, B. A. L. M., P. W. G. Verlaan, and C. W. A. Pleij. 2000. In vitro transcription by the turnip yellow mosaic virus RNA polymerase: a comparison with the alfalfa mosaic virus and brome mosaic virus replicases. J. Virol. 74:264-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reference deleted.
- 8.Gultyaev, A. P., F. H. D. van Batenburg, and C. W. A. Pleij. 1999. An approximation of loop free energy values of RNA H-pseudoknots. RNA 5:609-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guogas, L. M., S. M. Laforest, and L. Gehrke. 2005. Coat protein activation of alfalfa mosaic virus replication is concentration dependent. J. Virol. 79:5752-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haasnoot, J. 2002. Requirements for promoter activity in alfalfa mosaic and brome mosaic viruses. Ph.D. thesis. Leiden University, Leiden, The Netherlands.
- 11.Hook, B., D. Bernstein, B. Zhang, and M. Wickens. 2005. RNA-protein interactions in the yeast three-hybrid system: affinity, sensitivity, and enhanced library screening. RNA 11:227-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houser-Scott, F., M. L. Baer, K. F. Liem, Jr., J. M. Cai, and L. Gehrke. 1994. Nucleotide sequence and structural determinants of specific binding of coat protein or coat protein peptides to the 3′ untranslated region of alfalfa mosaic virus RNA 4. J. Virol. 68:2194-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaeger, S., G. Eriani, and F. Martin. 2004. The results and prospects of the yeast three-hybrid system. FEBS Lett. 556:7-12. [DOI] [PubMed] [Google Scholar]
- 14.Jaeger, S., G. Eriani, and F. Martin. 2004. Critical residues for RNA discrimination of the histone hairpin binding protein (HBP) investigated by the yeast three-hybrid system. FEBS Lett. 556:265-270. [DOI] [PubMed] [Google Scholar]
- 15.Jaspars, E. M. J. 1999. Genome activation in alfamo- and ilarviruses. Arch. Virol. 144:843-863. [DOI] [PubMed] [Google Scholar]
- 16.Krab, I. M., C. Caldwell, D. R. Gallie, and J. F. Bol. 2005. Coat protein enhances translational efficiency of alfalfa mosaic virus RNAs and interacts with the eIF4G component of initiation factor eIF4F. J. Gen. Virol. 86:1841-1849. [DOI] [PubMed] [Google Scholar]
- 17.Neeleman, L., R. C. L. Olsthoorn, H. J. M. Linthorst, and J. F. Bol. 2001. Translation of a nonpolyadenylated viral RNA is enhanced by binding of viral coat protein or polyadenylation of the RNA. Proc. Natl. Acad. Sci. U. S. A. 98:14286-14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neeleman, L., H. J. M. Linthorst, and J. F. Bol. 2004. Efficient translation of alfamovirus RNAs requires the binding of coat protein dimers to the 3′ termini of the viral RNAs. J. Gen. Virol. 85:231-240. [DOI] [PubMed] [Google Scholar]
- 19.Olsthoorn, R. C. L., and J. F. Bol. 2002. Role of an essential triloop hairpin and flanking structures in the 3′ untranslated region of Alfalfa mosaic virus RNA in in vitro transcription. J. Virol. 76:8747-8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsthoorn, R. C. L., S. Mertens, F. T. Brederode, and J. F. Bol. 1999. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 18:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrillo, J. E., G. Rocheleau, B. Kelley-Clarke, and L. Gehrke. 2005. Evaluation of the conformational switch model for alfalfa mosaic virus RNA replication. J. Virol. 79:5743-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quadt, R., H. J. Rosdorff, T. W. Hunt, and E. M. J. Jaspars. 1991. Analysis of the protein composition of alfalfa mosaic virus RNA-dependent RNA polymerase. Virology 182:309-315. [DOI] [PubMed] [Google Scholar]
- 23.Reusken, C. B. E. M., and J. F. Bol. 1996. Structural elements of the 3′-terminal coat protein binding site in alfalfa mosaic virus RNAs. Nucleic Acids Res. 24:2660-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reusken, C. B. E. M., L. Neeleman, and J. F. Bol. 1994. The 3′-untranslated region of alfalfa mosaic virus RNA 3 contains at least two independent binding sites for viral coat protein. Nucleic Acids Res. 22:1346-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sengupta, D. J., B. Zhang, B. Kraemer, P. Pochart, S. Fields, and M. Wickens. 1996. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl. Acad. Sci. U. S. A. 93:8496-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Heijden, M. W., J. E. Carette, P. J. Reinhoud, A. Haegi, and J. F. Bol. 2001. Alfalfa mosaic virus replicase proteins P1 and P2 interact and colocalize at the vacuolar membrane. J. Virol. 75:1879-1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Vossen, E. A. G., L. Neeleman, and J. F. Bol. 1994. Early and late functions of alfalfa mosaic virus coat protein can be mutated separately. Virology 202:891-903. [DOI] [PubMed] [Google Scholar]
- 28.Vlot, A. C., L. Neeleman, H. J. M. Linthorst, and J. F. Bol. 2001. Role of the 3′-untranslated regions of alfalfa mosaic virus RNAs in the formation of a transiently expressed replicase in plants and in the assembly of virions. J. Virol. 75:6440-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuidema, D. 1983. Specific binding sites on RNAs and coat protein of alfalfa mosaic virus involved in genome activation. Ph.D. thesis. Leiden University, Leiden, The Netherlands.
- 30.Zuidema, D., R. H. Cool, and E. M. J. Jaspars. 1984. Minimum requirements for specific binding of RNA and coat protein of alfalfa mosaic virus. Virology 136:282-292. [DOI] [PubMed] [Google Scholar]
- 31.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]