Abstract
Chimpanzees and gorillas are the only nonhuman primates known to harbor viruses closely related to HIV-1. Phylogenetic analyses showed that gorillas acquired the simian immunodeficiency virus SIVgor from chimpanzees, and viruses from the SIVcpz/SIVgor lineage have been transmitted to humans on at least four occasions, leading to HIV-1 groups M, N, O, and P. To determine the geographic distribution, prevalence, and species association of SIVgor, we conducted a comprehensive molecular epidemiological survey of wild gorillas in Central Africa. Gorilla fecal samples were collected in the range of western lowland gorillas (n = 2,367) and eastern Grauer gorillas (n = 183) and tested for SIVgor antibodies and nucleic acids. SIVgor antibody-positive samples were identified at 2 sites in Cameroon, with no evidence of infection at 19 other sites, including 3 in the range of the Eastern gorillas. In Cameroon, based on DNA and microsatellite analyses of a subset of samples, we estimated the prevalence of SIVgor to be 1.6% (range, 0% to 4.6%), which is significantly lower than the prevalence of SIVcpzPtt in chimpanzees (5.9%; range, 0% to 32%). All newly identified SIVgor strains formed a monophyletic lineage within the SIVcpz radiation, closely related to HIV-1 groups O and P, and clustered according to their field site of origin. At one site, there was evidence for intergroup transmission and a high intragroup prevalence. These isolated hot spots of SIVgor-infected gorilla communities could serve as a source for human infection. The overall low prevalence and sporadic distribution of SIVgor could suggest a decline of SIVgor in wild populations, but it cannot be excluded that SIVgor is still more prevalent in other parts of the geographical range of gorillas.
Simian immunodeficiency viruses (SIVs) have been identified in approximately 40 African primate species, but chimpanzees and gorillas are the only nonhuman primates known to harbor viruses closely related to human immunodeficiency virus type 1 (HIV-1) (38). These viruses have been transmitted to humans on at least four occasions, leading to four different HIV-1 groups, M to P (14, 26). West central African chimpanzees (Pan troglodytes troglodytes) in southern Cameroon are recognized as the reservoir of the ancestors of HIV-1 group M, which resulted in the AIDS pandemic, and of HIV-1 group N, which has been identified in only a few individuals in Cameroon (15). Western lowland gorillas (Gorilla gorilla gorilla) are infected with SIVgor, which is closely related to the two other HIV-1 lineages, termed group O, which represents 1% of HIV-1 infections in west central Africa, and group P, recently described from a single Cameroonian patient residing in France (26, 36).
The phylogenetic relationships between SIVcpz, SIVgor, and HIV-1 show that chimpanzees are the original reservoir of SIVs found in gorillas and humans (31, 36). Pan troglodytes troglodytes apes were most likely the original source of SIVgor, because SIVgor is significantly more closely related to SIVcpzPtt, from Pan troglodytes troglodytes in west central Africa, than to SIVcpzPts, from Pan troglodytes schweinfurthii in east Africa. In addition, an ancestral SIVcpzPtt lineage from which SIVgor and HIV-1 group O viruses are derived has been identified in the form of mosaic pol fragments in present-day SIVcpzPtt recombinants (2, 31). However, the ways of transmission and the exact origin of SIVgor infection in gorillas are not yet resolved. Because of the extensive overlap in habitat and diet (6, 23, 29, 33, 40), direct encounters between gorillas and chimpanzees seem inevitable, but they have rarely been observed and have been described as primarily nonaggressive (17, 28). The primate source of HIV-1 groups O and P also remains unclear, since current data do not allow one to differentiate between a chimpanzee and a gorilla reservoir, especially for HIV-1 group O (26, 31, 36).
To determine the geographic distribution, prevalence, and species association of SIVgor, we performed a comprehensive survey of wild gorilla populations in west central (Gorilla gorilla gorilla) and east (Gorilla beringei graueri) Africa. We found an overall prevalence of SIVgor of 1.6%, with infection confirmed at only three field sites. At two of these sites, however, the prevalence of SIVgor was 4.6%, indicating efficient virus spread within and between different communities. The geographic distribution of SIVgor is thus far limited to only a few sites in Cameroon. However, isolated hot spots of infection do exist, which could serve as a source of human infection.
MATERIALS AND METHODS
Sample collection and study sites.
Fecal samples (2,992) were collected between June 2006 and June 2009 from wild-living apes in central Africa. In west central Africa, samples were obtained at 18 forest sites, located in the southern part of Cameroon (n = 13), the extreme southwest of the Central African Republic (CAR) (n = 3), the northeast of the Republic of Congo (n = 1), and western Gabon (n = 1) (Fig. 1). Eight of the 18 sites (CP, MM, LB, DD, ND, DS GT, and LO) were located in national parks or forest reserves, while the remainder were in nonprotected areas with considerable hunting pressure. In east central Africa, fecal samples were obtained at three sites, located in the northeastern part of the Democratic Republic of Congo (DRC). Overall, fecal samples were collected primarily around night nests or feeding sites. For almost all samples, the GPS position and estimated time of deposition were recorded, and the species origin was defined in the field according to nesting sites, prints, vocalizations, and morphological and physical aspects of the samples. Both gorilla and chimpanzee samples were collected at some sites. About 20 mg of dung was collected in a 50-ml tube containing 20 ml of RNAlater (Applied Biosystems/Ambion, Austin, TX). These tubes were kept at base camps at ambient temperature for a maximum of 3 weeks and subsequently transported to a central laboratory for storage at −20°C or −80°C.
FIG. 1.
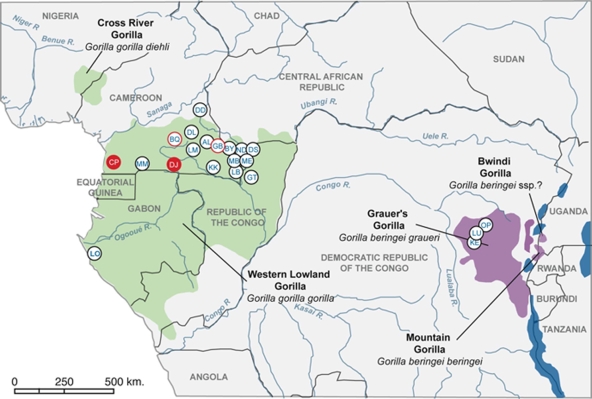
Locations of study sites of wild gorillas and/or chimpanzees in central Africa. Red filled circles indicate sampling sites where positive gorillas were identified in the current study. Red open circles indicate sites where SIVgor infection was previously identified (BQ) (36) or suspected (GB) (see Materials and Methods) and where no additional positive animals were identified in this survey. KE, LU, and OP are located in the range of Gorilla beringei graueri (purple), while all other sites are located in the range of Gorilla gorilla gorilla (dark green).
Detection of SIVgor and SIVcpz antibodies in ape fecal samples.
All gorilla and chimpanzee fecal samples were tested for the presence of HIV-1 cross-reactive antibodies, using the Inno-LIA HIV I/II score confirmation test (Innogenetics, Ghent, Belgium) and/or Western blot analysis (Maxim Biotech, Inc., Rockville, MD) as reported previously (15, 36). RNAlater-precipitated immunoglobulins were resolubilized by diluting the feces-RNAlater mixture (2 ml) with phosphate-buffered saline (PBS)-Tween 20 (7 ml), followed by an incubation for 1 h at 60°C, centrifugation (3,900 × g for 10 min) to clarify the solution, and then dialysis against PBS overnight at 4°C. The reconstituted extracts were then subjected to immunoblot analysis.
Nucleic acid extraction from fecal samples.
Total RNA was extracted from all antibody-positive fecal samples by use of an RNAqueous Midi kit (Applied Biosystems/Ambion, Austin, TX) as described previously (15). When the reverse transcription-PCR (RT-PCR) amplifications were negative using these RNA extracts, nucleic acid extraction was repeated using a NucliSens magnetic extraction kit (bioMérieux, Craponne, France), which utilizes magnetic silica particles to purify RNA (4). A 1.5-ml fecal sample was mixed for 1 min with 5 ml of PBS solution and centrifuged at 3,900 × g for 30 min. The supernatant was collected in a fresh tube and centrifuged again at 3,900 × g for 5 min. The new supernatant was passed through a gauze filter and incubated with 7 ml of NucliSens lysis buffer at room temperature for at least 10 min, followed by the magnetic extraction procedure according to the manufacturer's instructions, with a final elution volume of 50 μl. Fecal DNA was extracted using a QIAamp Stool DNA Mini kit (Qiagen, Valencia, CA) as previously described (15). Each extraction was performed using 2 ml of feces-RNAlater mixture and eluted in a final volume of 100 μl DNA solution.
Amplification of SIVgor and SIVcpz sequences from fecal RNA.
Fecal RNA was subjected to RT-PCR amplification using SIVgor/SIVcpz/HIV-1 consensus primers for env (gp41 ectodomain) (∼440 or 315 bp) and pol (∼330 bp or ∼245 bp) (Table 1). cDNA was synthesized using the R1 primer, followed by nested PCR using primers F1-R1 and F2-R2. All RT-PCRs were performed with Expand reverse transcriptase and an Expand long-template PCR system (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions. Briefly, 10 μl of fecal viral RNA was first incubated with 40 pmol of the outer reverse R1 primer (1 μl) for 10 min at 65°C, rapidly cooled on ice, and then added to the RT-PCR components, including 20 U of RNase inhibitor (Applied Biosystems/Ambion, Austin, TX), in a 20-μl reaction volume. The mixture was finally incubated for 60 min at 42°C, followed by 5 min at 95°C to inactivate the enzyme. For samples that could not be amplified with this protocol, a second type of RT-PCR was performed, using Superscript Retro-Transcriptase III (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Briefly, 10 μl of fecal RNA was incubated with 2 pmol of R1 primer and 2 μl of deoxynucleoside triphosphates (dNTPs) (10 mM) at 65°C for 5 min before being cooled on ice for 1 min. Buffer reagent (1×), 200 U of Superscript Retro-Transcriptase III, 20 U of RNase inhibitor, and 5 mM dithiothreitol were added to a final volume of 20 μl and incubated at 50°C for 90 min, followed by 15 min at 70°C to inactivate the enzyme. Ten microliters of cDNA was used for first-round PCR amplifications, and 5 μl of the first-round reaction product was used for nested PCR with the second-round primers, F2 and R2, by using the appropriate thermocycling conditions. Mostly, PCR amplifications included 45 cycles of denaturation (94°C, 20 s), annealing (50°C, 30 s), and elongation (68°C, 1 min) in a Peltier thermal cycler (PTC-200). For some amplifications, PCR conditions were slightly modified (with annealing temperatures varying from 45°C to 55°C and/or a touch-down PCR strategy). The resulting amplification products were gel purified using a GeneClean Turbo kit (Qbiogene, Inc., Carlsbad, CA) and directly sequenced using an automated sequencer (model 3130xl genetic analyzer; Applied Biosystems, Foster City, CA).
TABLE 1.
Primer sets used to amplify partial SIVcpz and SIVgor pol and env sequences
| Fragment name | Gene | Primera | Primer sequenceb | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| spol | pol | CPZ-pol-F1 | CCAGCNCACAAAGGNATAGGAGG | 823 | 37 |
| pol | CPZ-pol-R1 | ACBACYGCNCCTTCHCCTTTC | 37 | ||
| pol | CPZ-pol-F2 | GGAAGTGGATACTTAGAAGCAGAAGT | 340 | 37 | |
| pol | CPZ-pol-R2 | CCAATYCCYCCYYTTYKYTTAAAATT | 37 | ||
| polmini | pol | CON-POLmini-F1 | CATGTRGCHAGTGGNTWCMTAGARGCAGARGT | 518 | 31 |
| pol | CON-POLmini-R1 | ACBACYGCNCCTTCHCCTTTC | 31 | ||
| pol | CON-POLmini-F2 | AYAAYCCHCAAAGTCAAGGAGTRGT | 285 | 31 | |
| pol | CON-POLmini-R2 | GTCCTTTCCAAATDGGRTCTCTGCTGTC | 31 | ||
| gp41 | env | CPZ-gp41-F1 | TCTTAGGAGCAGCAGGAAGCACTATGGG | 594 | 37 |
| env | CPZ-gp41-R1 | AACGACAAAGGTGAGTATCCCTGCCTAA | 37 | ||
| env | CPZ-gp41-F2 | ACAATTATTGTCTGGTATAGTGCAACAGCA | 453 | 37 | |
| env | CPZ-gp41-R2 | TCCTACTATCATTATGAATATTTTTATATA | 37 | ||
| env | GOR-gp41-F1 | AGCARGAATTGCTGAGACTYTCTG | 359 | ||
| env | GOR-gp41-R1 | CCANTNTGTTATGTCAAGCCAAC | |||
| env | GOR-gp41-F2 | GGCATAAGACARCTCMGAGCTCGC | 315 | ||
| env | GOR-gp41-R2 | AAGCCAACTCCAAAGRTCTGC |
CPZ primers were designed according to SIVcpz/HIV-1 consensus sequences, GOR primers were designed according to SIVgor/HIV-1 group O consensus sequences, and CON primers were designed according to SIVcpz/SIVgor/HIV-1 consensus sequences. F1, first-round forward primer; F2, second-round forward primer; R1, first-round reverse primer; R2, second-round reverse primer.
K = G/T, M = A/C, R = A/G, W = A/T, Y = C/T, B = C/G/T, D = A/G/T, H = A/C/T, and N = A/C/T/G.
Phylogenetic analyses of SIVgor and SIVcpz sequences.
Newly derived SIVgor and SIVcpzPtt nucleotide sequences were compared to previously published SIVgor, SIVcpz, and HIV-1 reference sequences. Sequences were aligned using MEGA4 (32), and where necessary, minor manual adjustments were performed. Sites that could not be aligned unambiguously or that contained a gap in any sequence were excluded from the analyses. For the analyses of partial pol and env sequences, the number of nucleotides examined was 280 for pol and 355 or 266 for gp41. Maximum likelihood (ML) trees were constructed using PhyML (http://www.atgc-montpellier.fr/), with 1,000 bootstrap replicates (11). Phylogenies were also inferred by the Bayesian method (41), implemented in MrBayes, version 3.1 (27), run for 2,500,000 and 2,000,000 generations for partial pol and env fragments, respectively. Trees were sampled every 10 generations, with the first 25% being discarded as burn-in. Parameters were examined with the Tracer program (http://tree.bio.ed.ac.uk/software/tracer). The GTR model with a gamma distribution across sites was the most appropriate model according to TOPALI (22) and was used for both ML and Bayesian analyses.
GenBank accession numbers for additional partial pol and gp41 sequences used in comparative analyses are as follows: for SIVcpzPts, ANT (U42720), TAN1 (AF447763), TAN2 (DQ374657), and TAN3 (DQ374658); for SIVcpzPtt, MB897 (EF535994), LB7 (DQ373064), MB66 (DQ373063), EK505 (DQ373065), CAM5 (AJ271369), DP943 (EF535993), CAM3 (AF115393), US (AF103818), GAB2 (AF382828), GAB1 (X52154), CAM13 (AY169968), MT145 (DQ373066), CP1973 pol (FJ424869), and CP2680 pol (FJ424870); for SIVgor, BQ664 gp41 (AM296484), BQ664 pol (AM296488), CP684 (FJ424871), CP2135 (FJ424863), CP2139.2 (FJ424865), and CP1434 env (AM296487); for HIV-1 group M, subtype A, U455 (M62320); for HIV-1 group M, subtype B, HXB2 (K03455); for HIV-1 group N, YBF106 (AJ271370) and YBF30 (AJ006022); for HIV-1 group O, MVP5180 (L20571) and ANT70 (L20587); and for HIV-1 group P, RBF168 (GQ328744).
Species and subspecies determinations.
For all fecal samples positive by one of the serological assays, the species origin was also determined by mitochondrial DNA (mtDNA) analysis as described previously (15, 36). mtDNA analysis was also performed for a subset of negative samples from Cameroon and CAR, as well as for all samples from the DRC, Republic of Congo, and Gabon. Briefly, 5 μl of extracted fecal DNA was used for mitochondrial DNA amplification. An ∼450- to 500-bp mtDNA fragment spanning the hypervariable D loop was amplified from fecal DNA, using primers L15997 (5′-CACCATTAGCACCCAAAGCT-3′) and H16498 (5′-CCTGAAGTAGGAACCAGATG-3′). Phylogenetic analysis of these D loop sequences allowed identification of all chimpanzee samples and their subspecies classification (P. troglodytes troglodytes or P. troglodytes vellerosus). Whereas the majority of gorilla samples could also be identified with this approach, some samples yielded amplification products of poor quality and were reanalyzed by amplifying a 386-bp fragment spanning the 12S gene (using primers 12S-L1091 [5′-AAAAAGCTTCAAACTGGGATTAGATACCCCACTAT-3′] and 12S-H1478 [5′-TGACTGCAGAGGGTGACGGGCGGTGTGT-3′]) (35).
Microsatellite analyses.
Fecal DNA was extracted from all SIVgor and SIVcpz antibody-positive samples for microsatellite analysis to determine the number of infected individuals. Samples were genotyped at 8 loci (D18s536, D4s243, D10s676, D9s922, D2S1326, D2S1333, D4S1627, and D9S905) as previously described (15). For gender determination, a region of the amelogenin gene that contains a deletion in the X but not the Y chromosome (30) was amplified with a set of primers (AmelA-label [5′-CCCTGGGCTCTGTAAAGAATAGTG-3′] and AmelB [5′-ATCAGAGCTTAAACTGGGAAGCTG-3′]). All PCRs were initially performed in duplicate. Samples from individuals whose genotype appeared to be homozygous were amplified a minimum of four times to exclude allelic dropout. The resulting amplification products were analyzed using an automated sequencer (model 3130xl genetic analyzer; Applied Biosystems, Foster City, CA). Amplification products were visualized and sized using Genemapper 3.7 software (Applied Biosystems).
Estimation of individuals present in nonhabituated gorilla groups.
At the CP site in Cameroon, gorilla night nests were counted and the approximate time of construction was evaluated following the classification system of Tutin et al. (34). All nests belonging to the same category and distant from one another by less than 20 m were considered to be from the same group (8, 34). The number of individuals per group was estimated by nest counting and/or microsatellite analysis of samples collected around the nesting site. Since weaned gorillas generally make one nest every night, the number of nests corresponds to the number of weaned individuals present in the group; however, more than one nest per individual can occasionally be observed (5, 13). By performing microsatellite analysis, we were able to infer the number of individuals at a given nest site and to identify samples that were collected more than once from the same individual. However, the number of individuals detected is likely to be an underestimation of the total number of gorillas in a social group. The number of individuals estimated by fecal microsatellite analysis represents a minimal number of individuals in a group because it is likely that fecal samples are not collected for all gorillas.
Nucleotide sequence accession numbers.
All of the new SIVgor and SIVcpzPtt sequences are available at GenBank under accession numbers FN554923 to FN554939 for env (gp41), FN5554940 to FN554958 for the pol mini fragment, and FN554959 to FN554963 for the spol fragment.
RESULTS
Noninvasive sampling of wild-living gorillas and chimpanzees in central Africa.
Among the 2,992 fecal specimens analyzed in this study, 2,367 samples were collected from western lowland gorillas (G. gorilla gorilla) in west central Africa and 183 samples were from Grauer's gorillas (G. beringei graueri) in east central Africa. The remaining 442 samples were from chimpanzees in Cameroon and CAR. Collection sites are shown in Fig. 1, and numbers of collected samples are summarized in Table 2. In Cameroon, our survey included three sites where SIVgor infection had previously been documented (CP and BQ) (36) or suspected to be present (GB). At the latter site, a dried blood spot from a dead male gorilla, confiscated after being poached, was collected by an agent of the ministry of forestry and fauna in Cameroon in 2004. This sample was antibody positive, as indicated by a strongly positive Inno-LIA HIV test, but several attempts to amplify viral RNA remained unsuccessful.
TABLE 2.
SIV infection in wild gorilla and chimpanzee populations from different field sites in Central Africa
| Country | Collection sitea | No. of gorilla samplesb | No. of SIVgor antibody-positive samples | No. of SIVgor-infected gorillasc | No. of chimpanzee samplesb | No. of SIVcpz antibody-positive samples | No. of SIVcpz-infected chimpanzeesc |
|---|---|---|---|---|---|---|---|
| Cameroon | LM | 259 | 0 | 0 | 62 | 0 | 0 |
| BQ | 27 | 0 | 0 | 50 | 0 | 0 | |
| MM | 152 | 0 | 0 | 0 | |||
| CP | 473 | 29 | 11 | 180 | 9 | 2 | |
| GB | 88 | 0 | 0 | 4 | 0 | 0 | |
| MB | 117 | 0 | 0 | 14 | 2 | 1 | |
| KK | 18 | 0 | 0 | 1 | 0 | 0 | |
| LB | 135 | 0 | 0 | 4 | 0 | 0 | |
| DJ | 206 | 13 | 5 | 17 | 4 | 1 | |
| DI | 0 | 55 | 0 | 0 | |||
| AL | 46 | 0 | 0 | 0 | |||
| BY | 147 | 0 | 0 | 1 | 1 | 1 | |
| DD | 260 | 0 | 0 | 5 | 0 | 0 | |
| CAR | ME | 21 | 0 | 0 | 22 | 0 | 0 |
| ND | 171 | 0 | 0 | 27 | 0 | 0 | |
| DS | 93 | 0 | 0 | 0 | |||
| Congo | GT | 122 | 0 | 0 | 0 | ||
| Gabon | LO | 32 | 0 | 0 | 0 | ||
| DRC | KE | 128 | 0 | 0 | 0 | ||
| OP | 51 | 0 | 0 | 0 | |||
| LU | 4 | 0 | 0 | 0 | |||
| Total | 2,550 | 42 | 16 | 442 | 16 | 4 |
For all samples collected in CAR, Gabon, Congo, and DRC (n = 424), the species origin was confirmed by mtDNA analysis. A subset of the 2,321 samples from Cameroon (n = 662 [28.5%]) was also tested by mtDNA and/or 12S DNA analysis. These samples were selected to confirm the species derivation for at least one sample per nesting group or collection site with different GPS coordinates. The results showed 97.3% concordance between field observations and sequence analysis. Thus, for the remainder of the samples, the species origin determined by the field assistants was assumed to be correct.
SIVgor infection in wild gorillas.
The immunoblot results for the 2,550 gorilla samples are summarized in Table 2. New SIVgor antibody-positive samples were identified only in Cameroon. These included 29 samples from the CP site, in the extreme southwest, where two SIVgor-positive gorillas were initially discovered (36), and 13 samples from the DJ site, located just south of the Dja reserve in south-central Cameroon. All positive samples exhibited strong cross-reactivity with HIV-1 gp41 and/or p24 antigen in the Inno-LIA assay and with HIV-1 p24, p31, and/or one or more Env proteins on HIV-1 Western blots. Microsatellite analyses revealed that the 29 antibody-positive CP samples represented 11 different gorillas (Table 3), none of whom corresponded to the two initially identified SIVgor-infected individuals (36). The 13 immunoblot-positive samples from the DJ site corresponded to 5 different individuals.
TABLE 3.
Characteristics of SIVgor-infected gorillas in Cameroona
| Individual IDb | Fecal sample | Date of collection (day/mo/yr) | Groupc | Location | Size of amplified SIV virion RNA (bp)d |
Alleles in locus by microsatellite analysise |
Sexf | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spol | Minipol | gp41 | FL | D18S536 | D4S243 | D10S676 | D9S922 | D2S1326 | D2S1333 | D4S1627 | D9S905 | ||||||
| CPg-ID1 | CP456 | 18/04/04 | CP-GR | − | − | − | 146/150 | − | − | − | − | − | 236/236 | 274/278 | F | ||
| CP684 | 18/04/04 | CP-GR | 330 | 220 | 394 | 9,143 | 146/150 | 185/193 | − | − | 251/251 | 298/318 | 236/236 | 274/278 | F | ||
| CP685 | 19/04/04 | 280 | − | 394 | 146/150 | 185/193 | − | − | 251/251 | 298/318 | 236/236 | 274/278 | F | ||||
| CPg-ID2 | CP1434 | 31/03/06 | CP-GR | − | − | 388 | 150/158 | 181/193 | − | − | 255/267 | 294/294 | 236/244 | 278/278 | F | ||
| CP1436 | 31/03/06 | − | − | − | 150/158 | 181/193 | − | − | 255/267 | 294/294 | 236/244 | 278/278 | F | ||||
| CPg-ID3 | CP2071 | 10/02/07 | CP-GR | − | − | − | 146/150 | 181/193 | 196/200 | 268/280 | − | 314/334 | 236/240 | 278/278 | F | ||
| CPg-ID4 | CP2072 | 10/02/07 | A | CP-GR | − | 202 | 394 | 146/146 | 185/193 | 196/200 | 268/268 | 267/275 | 322/334 | 232/232 | − | F | |
| CP2074 | 11/02/07 | − | 202 | 394 | 146/146 | 185/193 | 196/200 | 268/268 | 267/275 | 322/334 | − | − | F | ||||
| CP2109 | 11/02/07 | − | 202 | − | 146/146 | 185/193 | 196/200 | 268/268 | 267/275 | 322/334 | 232/232 | 274/278 | F | ||||
| CP2117 | 11/02/07 | 306 | 202 | 442 | 146/146 | 185/193 | 196/200 | 268/268 | 267/275 | 322/334 | 232/232 | − | F | ||||
| CP2118 | 11/02/07 | − | 202 | 412 | 146/146 | 185/193 | 196/200 | 268/268 | 267/275 | 322/334 | 232/232 | 274/278 | F | ||||
| CPg-ID5 | CP2110 | 11/02/07 | A | CP-GR | − | − | 290 | 146/150 | 177/185 | 192/200 | 260/280 | 251/267 | 322/326 | 232/244 | 278/278 | F | |
| CP2111 | 11/02/07 | − | − | − | 146/150 | 177/185 | 192/200 | 260/280 | 251/267 | 322/326 | 232/244 | 278/278 | F | ||||
| CP2119 | 11/02/07 | − | − | − | 146/150 | 177/185 | 192/200 | 260/280 | 251/267 | 322/326 | 232/244 | 278/278 | F | ||||
| CP2123 | 11/02/07 | − | − | − | 146/150 | 177/185 | 192/200 | 260/280 | 251/267 | 322/326 | 232/244 | 278/278 | F | ||||
| CPg-ID6 | CP2094 | 11/02/07 | B | CP-GR | − | 202 | − | 146/150 | − | 196/200 | 266/280 | 267/275 | 322/334 | 244/ | 278/278 | F | |
| CP2095 | 11/02/07 | − | − | − | 146/150 | − | 196/200 | 266/280 | 267/275 | 322/334 | − | − | |||||
| CPg-ID7 | CP2098 | 11/02/07 | B | CP-GR | − | − | − | 146/150 | 185/185 | 196/200 | − | 251/− | − | 240/244 | 278/278 | F | |
| CPg-ID8 | CP2087 | 11/02/07 | B | CP-GR | − | − | − | 146/150 | − | 196/200 | − | 267/271 | 354/374 | 232/236 | 278/278 | ||
| CPg-ID9 | CP2139 | 11/02/07 | C | CP-GR | 330 | 220 | 394 | 9,252 | 146/146 | 177/189 | 192/200 | 276/280 | 251/271 | 318/334 | 228/240 | 278/278 | F |
| CPg-ID10 | CP2126 | 11/02/07 | C | CP-GR | − | − | − | 146/154 | 173/193 | 180/180 | 276/280 | 255/271 | 298/322 | 228/232 | 278/278 | F | |
| CP2132 | 11/02/07 | 306 | 202 | 412 | 146/154 | 173/193 | 180/180 | 276/280 | 255/271 | 298/322 | 228/232 | 278/278 | F | ||||
| CP2133 | 11/02/07 | 286 | 202 | 394 | 146/154 | 173/193 | 180/180 | 276/280 | 255/271 | 298/322 | 228/232 | 278/278 | F | ||||
| CP2134 | 11/02/07 | − | 202 | − | 146/154 | 173/193 | 180/180 | 276/280 | 255/271 | 298/322 | 228/232 | − | F | ||||
| CP2135 | 11/02/07 | 330 | 220 | 394 | 9,246 | 146/154 | 173/193 | 180/180 | 276/280 | 255/271 | 298/322 | 228/232 | 278/278 | F | |||
| CP2141 | 11/02/07 | ND | ND | ND | 146/154 | 173/193 | 180/180 | 276/280 | 255/271 | 298/322 | 228/232 | 278/278 | F | ||||
| CPg-ID11 | CP2740 | 08/11/07 | CP-OV | − | − | − | 146/150 | 181/189 | 196/200 | 264/280 | − | 314/318 | 236/236 | 278/278 | |||
| CP2744 | 08/11/07 | − | − | − | 146/150 | 181/189 | 196/200 | 264/280 | − | 314/318 | 236/236 | 278/278 | M | ||||
| CP2746 | 08/11/07 | − | − | − | 146/150 | 181/189 | 196/200 | 264/280 | − | 314/318 | 236/236 | 278/278 | M | ||||
| CP2747 | 08/11/07 | − | − | − | 146/150 | 181/189 | 196/200 | 264/280 | − | 314/318 | 236/236 | 278/278 | M | ||||
| CP2749 | 08/11/07 | − | − | − | 146/150 | 181/189 | 196/200 | 264/280 | − | 314/318 | 236/236 | 278/278 | M | ||||
| CPg-ID12 | CP2994 | 21/04/08 | CP-MV | − | − | − | 150/154 | 185/185 | 184/196 | 268/280 | 267/267 | 298/306 | − | 278/278 | F | ||
| CPg-ID13 | CP3018 | 23/04/08 | CP-MV | − | − | − | 150/154 | 185/185 | 180/180 | 268/280 | − | − | − | − | M | ||
| CPg-ID13? | CP3019 | 23/04/08 | − | − | − | 150/154 | 185/185 | − | 268/280 | − | − | − | − | M | |||
| DJg-ID1 | DJ3795 | 28/05/08 | DJ | − | − | 409 | 146/146 | − | 200/200 | − | 271/279 | 318/318 | 236/236 | 294/294 | F | ||
| DJg-ID2 | DJ4099 | 09/04/09 | DJ | 328 | 240 | 287 | 146/146 | 185/189 | 196/204 | 272/280 | 271/275 | 318/322 | 232/244 | 278/278 | F | ||
| DJ4229 | 30/05/09 | − | − | 323 | 146/146 | 185/189 | 196/204 | 272/280 | 271/275 | 318/322 | 232/244 | 278/278 | F | ||||
| DJ4230 | 30/05/09 | − | − | − | 146/146 | 185/189 | 196/204 | 272/280 | 271/275 | 318/322 | 232/244 | 278/278 | F | ||||
| DJ4237 | 30/05/09 | − | 285 | 435 | 146/146 | 185/189 | 196/204 | 272/280 | 271/275 | 318/322 | 232/244 | 278/278 | F | ||||
| DJ4240 | 30/05/09 | − | − | − | 146/146 | 185/189 | 196/204 | 272/280 | 271/275 | 318/322 | 232/244 | 278/278 | F | ||||
| DJ4258 | 30/05/09 | − | 257 | 377 | 146/146 | 185/189 | 196/204 | 272/280 | 271/275 | 318/322 | 232/244 | 278/278 | F | ||||
| DJ4259 | 31/05/09 | − | 252 | 299 | 146/146 | 185/189 | 196/204 | 272/280 | 271/275 | 318/322 | 232/244 | 278/278 | F | ||||
| DJ4273 | 31/05/09 | − | − | − | 146/146 | 185/189 | 196/204 | 272/280 | 271/275 | 318/322 | 232/244 | 278/278 | F | ||||
| DJg-ID3 | DJ4102 | 09/04/09 | DJ | − | − | − | 146/146 | 189/193 | 196/204 | 264/280 | 271/275 | 293/318 | 232/244 | 278/278 | M | ||
| DJ4112 | 09/04/09 | − | 245 | − | 146/146 | 189/193 | 196/204 | 264/280 | 271/275 | 293/318 | − | 278/278 | |||||
| DJg-ID4 | DJ4114 | 09/04/09 | DJ | − | − | − | 146/146 | − | 192/200 | 280/280 | 275/275 | 298/314 | − | 282/282 | M | ||
| DJg-ID5 | DJ4257 | 30/05/09 | DJ | − | − | − | 146/146 | 189/193 | 196/204 | 264/280 | 271/275 | 318/318 | 232/240 | 278/278 | F | ||
| BQg-ID1 | BQ664 | 09/08/04 | BQ | 892 | − | 394 | 150/154 | 177/193 | ND | ND | 263/263 | 314/322 | 240/248 | 278/278 | F | ||
Data in bold represent previously reported samples (CPg-ID1, CPg-ID2, and BQg-ID1) (36). All samples tested positive in two serological assays, namely, the fecal Inno-LIA HIV test and fecal HIV Western blot analysis.
Individuals are numbered in accordance with a previous survey (36) in which three SIVgor-positive gorillas were identified. Overall, the first two letters refer to the collection site, followed by “g” for gorilla and the ID number.
Gorilla groups were identified as described in Materials and Methods.
Spol, partial pol fragment; minipol, second partial pol fragment; gp41, partial env fragment; FL, full-length genome; −, RT-PCR negative; ND, not done.
Eight STR loci were amplified from fecal DNA. Two alleles per locus are shown. −, repetitively negative; ND, not done.
F, female; M, male.
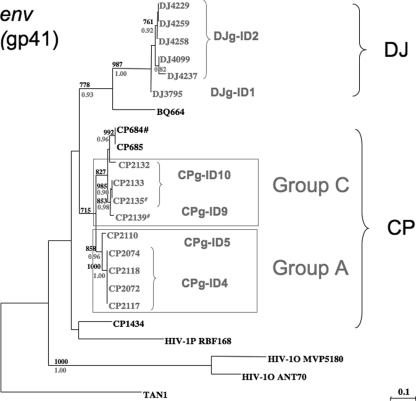
For all antibody-positive samples, RNA was extracted and subjected to RT-PCR amplification, using consensus env (gp41 region) and pol primers. SIVgor sequences were amplified from 12 CP samples, corresponding to 5 new gorillas, and from 7 DJ samples, corresponding to 3 infected gorillas (Table 3). For 2 of the 5 CP gorillas, a full-length SIVgor sequence (SIVgorCP2135 and SIVgorCP2139) has previously been reported (31). Despite strong antibody reactivity, samples from the remaining six CP (n = 11) and two DJ (n = 2) gorillas were repeatedly virion RNA negative. To compare the evolutionary relationships of the newly derived SIVgor viruses to each other and to previously characterized SIVgor, SIVcpz, and HIV-1 strains, phylogenetic trees were constructed using partial pol (286 bp) and gp41 (355 bp) sequences (Fig. 2). In both trees, the new SIVgor strains from the CP and DJ sites fell into a well-supported clade which included the previously characterized SIVgor strain, HIV-1 group O, and the recently described HIV-1 group P sequence. All new SIVgor strains from the CP site clustered with the previously reported CP strains, SIVgorCP684 and SIVgorCP1434. The two new DJ viruses also formed an independent lineage. Thus, as previously reported for SIVcpzPtt, SIVgor viruses cluster according to their field site of origin. Interestingly, HIV-1 group P and SIVgor formed a sister clade to HIV-1 group O.
FIG. 2.
Phylogenetic analysis of partial pol (polymerase gene; 286 bp) (a) and env (gp41 envelope transmembrane region; 355 bp) (b) sequences of newly identified SIVgor and SIVcpzPtt strains. Representative HIV-1 group M (U455 and HXB2), N (YBF30 and YBF106), O (Ant70 and MVP5180), and P (RBF168) sequences are included, as well as SIVcpzPts sequences (ANT and TAN1-3), which form the outgroup. Trees were inferred by maximum likelihood phylogeny (PhyML) with previously characterized SIVcpz/SIVgor/HIV-1 strains (31, 36, 37). The support values in black above the branches are from 1,000 maximum likelihood bootstraps (shown as ‰; only values above 700‰ are shown), and posterior probabilities (only values above 0.80) from nucleotide Bayesian analysis are represented by gray asterisks below the branches. The scale bar represents the number of substitutions per site. HIV-1 strains are shown in gray italic letters. New SIVgor strains identified in this epidemiologic survey are highlighted in black boxes. For SIVgor strains highlighted by a pound sign, the full-length genome sequence has been reported (31). New SIVcpzPtt strains identified in this study are highlighted in gray boxes.
During this survey, we also identified antibody-positive samples from chimpanzees at the CP (n = 2), DJ (n = 1), MB (n = 1), and BY (n = 1) sites (Tables 1 and 4). Although SIVcpzPtt-infected chimpanzees were identified within 10 km of SIVgor-infected gorillas at both the CP and DJ sites, their molecular characterization showed that the new viruses formed distinct lineages within the SIVcpzPtt radiation that were not closely related to SIVgor or HIV-1 group O and P viruses (Fig. 2).
TABLE 4.
Characteristics of SIVcpz-infected chimpanzees identified in Cameroona
| Individual IDb | Fecal sample | Date of collection (day/mo/yr) | Location | Size of amplified SIV virion RNA (bp)c |
Alleles in locus by microsatellite analysisd |
Sexe | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spol | Minipol | gp41 | D18S536 | D4S243 | D10S676 | D9S922 | D2S1326 | D2S1333 | D4S1627 | D9S905 | |||||
| CPc-ID1 | CP1973 | 17/12/06 | CP-MV | 232 | 202 | − | 154/166 | 225/225 | 168/180 | 273/295 | − | − | − | − | |
| CP1974 | 17/12/06 | 232 | 202 | − | 154/166 | 225/225 | 168/180 | − | − | − | − | − | |||
| CPc-ID2 | CP2678 | 28/08/07 | CP-MV | − | 202 | − | 171/171 | 221/245 | 168/180 | 292/304 | 223/251 | 318/326 | 224/232 | 286/286 | F |
| CP2679 | 28/08/07 | − | − | − | 171/171 | 221/245 | 168/180 | 292/304 | 223/251 | 318/326 | 224/232 | 286/286 | F | ||
| CP2680 | 28/08/07 | − | 202 | − | 171/171 | 221/245 | 168/180 | 292/304 | 223/251 | 318/326 | 224/232 | 286/286 | F | ||
| CP2685 | 28/08/07 | − | 202 | − | 171/171 | 221/245 | 168/180 | 292/304 | 223/251 | 318/326 | 224/232 | 286/286 | F | ||
| CP2686 | 28/08/07 | − | 202 | − | 171/171 | 221/245 | 168/180 | 292/304 | 223/251 | 318/326 | 224/232 | 286/286 | F | ||
| CP2687 | 28/08/07 | − | 202 | 1,260 | 171/171 | 221/245 | 168/180 | 292/304 | 223/251 | 318/326 | 224/232 | 286/286 | F | ||
| CP2688 | 28/08/07 | − | − | − | 171/171 | 221/245 | 168/180 | 292/304 | 223/251 | 318/326 | 224/232 | 286/286 | |||
| CP2689 | 28/08/07 | − | − | − | 171/171 | 221/245 | 168/180 | 292/304 | 223/251 | 318/326 | 224/232 | 286/286 | F | ||
| DJc-ID1 | DJ3259 | 28/05/08 | DJ | − | − | − | 158/170 | 197/229 | 184/184 | 288/292 | 235/243 | 314/318 | 216/224 | 286/290 | M |
| DJ3260 | 28/05/08 | − | − | − | 158/170 | 197/229 | 184/184 | 288/292 | 235/243 | 314/318 | 216/224 | 286/290 | M | ||
| DJ3261 | 28/05/08 | − | 245 | 429 | 158/170 | 197/229 | 184/184 | 288/292 | 235/243 | 314/318 | 216/224 | 286/290 | M | ||
| DJ3262 | 28/05/08 | − | − | − | 158/170 | 197/229 | 184/184 | 288/292 | 235/243 | 314/318 | 216/224 | − | M | ||
| MBc-ID1 | MB2334 | 11/04/07 | MB | − | ND | 404 | 146/154 | 225/225 | 176/188 | 296/304 | ND | ND | ND | ND | ND |
| MBc-ID2 | MB2340 | 11/04/07 | MB | 328 | ND | 381 | 158/158 | 245/245 | 168/176 | 304/316 | ND | ND | ND | ND | ND |
Data in bold represent previously reported samples (CPc-ID1) (31). All samples tested positive in two serological assays, namely, the fecal Inno-LIA HIV test and fecal HIV Western blot analysis.
The first two letters refer to the collection site, followed by “c” for chimpanzee and the ID number.
Spol, partial pol fragment; minipol, second partial pol fragment; gp41, partial env fragment; −, RT-PCR negative; ND, not done.
Eight STR loci were amplified from fecal DNA. Two alleles per locus are shown. −, repetitively negative; ND, not done.
F, female; M, male.
Prevalence and genetic diversity of SIVgor strains in nonhabituated gorilla groups with overlapping home ranges.
In Campo Ma'an National Park (CP), where SIVgor was initially discovered (36), a total of 473 gorilla fecal samples were collected at six different locations (Fig. 3b). The 11 infected CP gorillas were found at just three sites (Table 5; Fig. 3b), in the center of the park. The other collection sites were separated from these three focal sites by either the Ntem river (NG and ML) or a distance of at least 50 km (ON).
FIG. 3.
Localization of the Campo Ma'an Reserve (a) and the six different collection sites (b). Black circles represent the three sites where new SIVgor-infected gorillas were discovered (MV, GR, and OV). (c) The positions of the 13 identified SIVgor-positive gorillas at the MV, GR, and OV areas within the CP site are shown in detail. Each individual was identified by microsatellite analysis and is represented by an ID, corresponding to CPg-IDx in Table 3. The numbers in parentheses correspond to the samples collected from each individual. Previously collected samples are represented with striped gray symbols, and new samples are shown with black symbols. Circles represent samples collected at nesting sites, while squares represent those collected on trails. The three different identified groups (A, B, and C) are highlighted in gray boxes. Triangles represent SIVcpz-positive chimpanzees (Table 4).
TABLE 5.
SIV infection in wild gorilla and chimpanzee populations from different prospected areas in the CP site, southwest Cameroon
| Collection sitea | No. of gorilla samplesb | No. of SIVgor antibody-positive samples | No. of SIVgor-infected gorillasc | No. of chimpanzee samplesb | No. of SIVcpz antibody-positive samples | No. of SIVcpz-infected chimpanzeesc |
|---|---|---|---|---|---|---|
| CP-GR | 111 | 21 | 8 | 5 | 0 | 0 |
| CP-OV | 83 | 5 | 1 | 15 | 0 | 0 |
| CP-MV | 119 | 3 | 2 | 95 | 9 | 2 |
| CP-ML | 7 | 0 | 0 | 49 | 0 | 0 |
| CP-ON | 116 | 0 | 0 | 12 | 0 | 0 |
| CP-NG | 37 | 0 | 0 | 4 | 0 | 0 |
| Total | 473 | 29 | 11 | 180 | 9 | 2 |
To examine the transmission pattern of SIVgor, we compared the prevalence rates and genetic diversity of SIVgor strains among gorilla groups with overlapping home ranges at the CP-GR site (Fig. 3c). Field data indicated that two nesting groups (groups A and B in Fig. 3c and Table 3) corresponded to two different gorilla groups, since nests were built on the same day (nests of category 1) and were 2 km apart. In addition, microsatellite analyses of positive fecal samples from both groups confirmed that they were from different individuals. Microsatellite analyses of fecal samples from the third nesting site showed that they also belonged to a different group, group C (Fig. 3c). However, for the fourth group of nests, the positive sample (CP2074) corresponded to a member (ID4) of group A. Thus, nest sites A and D most likely represented the same gorilla group. We estimated the prevalence of SIVgor infection in groups A, B, and C. In group A, 2 gorillas were positive among 8 sampled, indicating an infection rate of 25%; assuming that none of the other individuals of that group of 12 (based on nest counts) were infected, the prevalence in group A was at least 17% (2/12 individuals). In group B, 3 gorillas were positive in a group of 12 (based on nest counts), indicating a prevalence of at least 25%. Finally, in group C, 2 gorillas were positive among 9 sampled, suggesting that 22% of this group could be infected. At the CP site, SIVgor infection was identified in at least two other groups, but at the CP-OV site, prevalence could not be estimated because this constituted a feeding site where different groups passed through during the day. At the CP-MV site, samples were collected only around a single nest.
SIVgor sequences from members of different gorilla groups formed separate clusters in phylogenetic tree analyses (Fig. 4), whereas those from members of the same group were closely related, suggesting epidemiologically linked infections. The tree in Fig. 4 also identified two relatively more divergent viruses in a single individual (CPg-ID10), suggesting a case of coinfection with epidemiologically linked viruses. A similar case was also identified at the DJ collection site (DJg-ID2). Interestingly, most new SIVgor infections were identified in females: at both the CP and DJ sites, only 2 of 10 and 2 of 5 positive gorillas for whom gender could be determined were males, respectively.
FIG. 4.
Phylogenetic analysis of partial env (gp41; 266 bp) nucleotide sequences of new SIVgor strains (in gray) identified in this survey. The strains for which the full-length genome has been reported previously (31) are highlighted with a pound sign. The tree was inferred by maximum likelihood phylogeny (PhyML) with previously characterized SIVcpzPts/SIVgor/HIV-1 strains (26, 31, 36). The support values in black above the branches are from 1,000 maximum likelihood bootstraps (shown as ‰; only values above 700‰ are shown), and posterior probabilities (shown as proportions; only values above 0.80 are shown) from nucleotide Bayesian analysis are shown in gray below the branches. The scale bar represents the number of substitutions per site. Groups A and C correspond to identified groups (Fig. 3). Samples identified in this study are highlighted in gray.
Comparison of SIVgor and SIVcpzPtt prevalence rates in Cameroon.
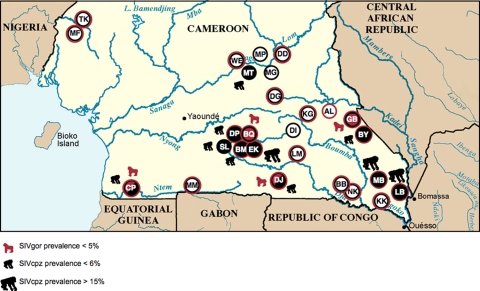
To compare the prevalence rates of SIV infection in gorillas and chimpanzees, we compiled all data from this study with results from previous studies on wild ape populations in Cameroon (15, 36, 37). Since 2003, ape fecal samples have been collected at 27 different sites in Cameroon, which are depicted in Fig. 5, with sample numbers shown in Table 6. A total of 1,217 chimpanzee samples were collected at 25 sites, and 2,239 gorilla samples were collected at 21 sites. Prevalence rates were estimated based on the proportions of antibody-positive fecal samples, while also adjusting for repeat sampling, sample degradation, and potential misidentification of species. Microsatellite analyses of a subset of samples revealed that each chimpanzee was sampled approximately 1.7 times (15) and each gorilla was sampled 1.8 (118 samples/64 gorillas) times. This suggests that, on average, 649 chimpanzees and 1,194 gorillas were sampled. Table 6 lists the SIVcpzPtt and SIVgor prevalence rates for each field site as well as for the entire sampled population. The mean SIVcpzPtt prevalence in chimpanzees was 5.9% (ranging from 0% to 32%), while the mean SIVgor prevalence in gorillas was 1.6% (ranging from 0% to 4.6%). Thus, there are three times more SIVcpz-infected chimpanzees than SIVgor-infected gorillas. Moreover, positive gorillas were identified at only 3 of the 21 sites surveyed, whereas positive chimpanzees were found at 10 of 25 collection sites. Finally, we did not find sites where SIVgor infection rates exceeded 5%, although in some gorilla groups at least one-fourth of group members were infected.
FIG. 5.
Locations of study sites of wild chimpanzees and gorillas from this and previous surveys in Cameroon. Red and black circles indicate sampling sites where gorillas and chimpanzees were sampled, respectively. Red filled circles indicate sites where SIVgor-positive gorillas were identified, and black filled circles indicate sites where SIVcpzPtt-positive chimpanzees were identified. Circles with half red and half black indicate sites where SIVgor-infected gorillas and SIVcpzPtt-infected chimpanzees were identified. MF, TK, MP, and WE are located in the range of Pan troglodytes vellerosus, while all other sites are located in the range of Pan troglodytes troglodytes. TK was located in the area of the cross-river gorillas (Gorilla gorilla dielhi), and all other locations were in the range of western lowland gorillas (Gorilla gorilla gorilla).
TABLE 6.
SIV infection in wild chimpanzee and gorilla populations from different field sites in this and previous surveys in Cameroon
| Collection sitea | No. of chimpanzee samplesb | No. of SIVcpz antibody-positive samples | No. of SIVcpz antibody-positive chimpanzeesc | No. of SIVcpz PCR-positive chimpanzeesc | SIVcpz prevalence in chimpanzees (95% CI)d | No. of gorilla samplesb | No. of SIVgor antibody-positive samples | No. of SIVgor antibody-positive gorillasc | No. of SIVgor PCR-positive gorillasc | SIVgor prevalence (%) in gorillas (95% CI)d |
|---|---|---|---|---|---|---|---|---|---|---|
| WE | 25 | 0 | 0 | 0 | 0.0 (0.0-23.1)* | 1 | 0 | 0 | 0 | — |
| MT | 81 | 10 | 2 | 2 | 5.4 (1.0-16.0)* | 0 | — | |||
| DG | 29 | 0 | 0 | 0 | 0.0 (0.0-20.3)* | 25 | 0 | 0 | 0 | 0.0 (0.0-22.8) |
| DP | 160 | 17 | 4 | 4 | 4.7 (1.8-11.5) | 16 | 0 | 0 | 0 | 0.0 (0.0-29.9) |
| BQ | 142 | 0 | 0 | 0 | 0.0 (0.0-4.8) | 58 | 1 | 1 | 1 | 3.2 (0.6-16.2) |
| EK | 46 | 6 | 4 | 4 | 16.3 (6.4-34.7) | 12 | 0 | 0 | 0 | 0.0 (0.0-39.0) |
| BB | 34 | 0 | 0 | 0 | 0.0 (0.0-16.4)* | 7 | 0 | 0 | 0 | — |
| MB | 101 | 31 | 17 | 16 | 31.6 (20.7-44.8) | 119 | 0 | 0 | 0 | 0.0 (0.0-5.8) |
| LB | 38 | 5 | 5 | 4 | 24.7 (11.2-46.9) | 207 | 0 | 0 | 0 | 0.0 (0.0-3.4) |
| CP | 232 | 9 | 2 | 2 | 1.6 (0.4-5.7) | 525 | 34 | 13 | 7 | 4.6 (2.7-7.8) |
| TK | 1 | 0 | 0 | 0 | — | 21 | 0 | 0 | 0 | 0.0 (0.0-25.9) |
| MF | 39 | 0 | 0 | 0 | 0.0 (0.0-15.5) | 0 | ||||
| MP | 15 | 0 | 0 | 0 | 0.0 (0.0-32.5) | 0 | ||||
| MG | 24 | 0 | 0 | 0 | 0.0 (0.0-22.8) | 0 | ||||
| KG | 15 | 0 | 0 | 0 | 0.0 (0.0-32.5) | 15 | 0 | 0 | 0 | 0.0 (0.0-32.5) |
| SL | 44 | 5 | 1 | 1 | 4.3 (0.8-21.0) | 0 | ||||
| BM | 38 | 2 | 1 | 1 | 4.9 (0.9-23.6) | 1 | 0 | 0 | 0 | — |
| NK | 8 | 0 | 0 | 0 | — | 25 | 0 | 0 | 0 | 0.0 (0.0-22.8) |
| LM | 62 | 0 | 0 | 0 | 0.0 (0.0-10.4) | 293 | 0 | 0 | 0 | 0.0 (0.0-2.4) |
| MM | 0 | 152 | 0 | 0 | 0 | 0.0 (0.0-4.5) | ||||
| GB | 4 | 0 | 0 | 0 | — | 85 | 0 | 0 | 0 | 0.0 (0.0-7.9) |
| KK | 1 | 0 | 0 | 0 | — | 18 | 0 | 0 | 0 | 0.0 (0.0-27.8) |
| DJ | 17 | 4 | 1 | 1 | 11.0 (2.0-43.5) | 206 | 13 | 5 | 3 | 4.6 (0.9-7.7) |
| DI | 55 | 0 | 0 | 0 | 0.0 (0.0-11.7) | 0 | ||||
| AL | 0 | 46 | 0 | 0 | 0 | 0.0 (0.0-13.3) | ||||
| BY | 1 | 1 | 1 | 0 | — | 147 | 0 | 0 | 0 | 0.0 (0.0-4.7) |
| DD | 5 | 0 | 0 | 0 | — | 260 | 0 | 0 | 0 | 0.0 (0.0-2.6) |
| Total | 1,217 | 90 | 38 | 35 | 5.9 (4.3-7.9) | 2,239 | 48 | 19 | 12 | 1.6 (1.0-2.4) |
Locations of sites are shown in Fig. 5.
Determined by mitochondrial and 12S DNA analysis of fecal DNA and/or species identification in the field (see Materials and Methods).
CI, confidence interval. —, prevalence rates were not calculated because the number of samples collected was too small. *, SIVcpzPtt prevalence rates for these sites have been reported previously.
DISCUSSION
The objective of this study was to determine to what extent gorillas are infected with SIVgor. We thus performed a comprehensive survey of wild gorilla populations in west central Africa and, to a lesser extent, eastern Africa. Our results show a low and uneven distribution of SIVgor infection in western lowland gorillas (G. gorilla gorilla) and an absence of infection in eastern lowland gorillas (G. beringei graueri). We confirmed the presence of SIVgor at the CP site, where SIVgor was initially discovered, and identified a new site, DJ, with at least five infected gorillas. No additional positive gorillas were identified at the previously reported BQ site and in an area, GB in Fig. 1, where we suspected the presence of SIVgor infection based on serological cross-reactivity of a dried blood spot from a dead male gorilla in 2004. It should be noted that since the discovery of the first positive gorilla at the BQ site, the gorilla population decreased in that area due to an anthrax outbreak in 2004 (12, 18). In addition, intensive poaching and logging made ape tracking at the GB site extremely difficult. Thus, it is possible that infected individuals were missed. However, by combining data from this and previous studies (15, 36, 37), we have now screened more than 1,200 western lowland gorillas in Cameroon and adjacent areas and found an overall prevalence of only 1.6%, which is significantly lower than the overall prevalence of SIVcpz in chimpanzees. Moreover, SIVgor is less evenly distributed, since we found evidence of infection at only three sites. Thus, SIVgor infection of gorillas is much less widespread than SIVcpz infection of chimpanzees, at least in Cameroon. However, the fact that SIVgor infection was observed at three (or possibly four) sites, located 60 to 400 km apart, and the observation that at some sites SIVgor prevalence was almost 5% indicate that this infection is not new in gorillas. It is also possible that SIVgor infection may be declining in gorilla populations in Cameroon. However, because of the sporadic distribution of SIVgor and the unknown influences of gorilla population decline on the spread of this virus, it remains possible that SIVgor is still more prevalent in other parts of the geographical range of western lowland gorillas.
A total of 19 SIV-positive gorillas have now been identified, and SIVgor strains have been characterized for 11 of them, from three different locations in southern Cameroon. All SIVgor strains form a monophyletic lineage closely related to HIV-1 group P, and the SIVgor/HIV-1O/HIV-1P clade falls within the SIVcpz radiation. SIVgor strains also cluster according to their collection site of origin. This is not surprising, since the movement of gorilla groups, like that of chimpanzees, is influenced by major geographical barriers (15). The importance of these geographical barriers for the spread of SIVgor is particularly obvious in the Campo Ma'an Reserve, where we found a concentration of positive gorillas in the center of the park but not at other sites separated by the Ntem river. Although our sampling size of positive gorillas is limited, we did not observe cocirculation of divergent SIVgor strains at single field sites. This differs from our observations in chimpanzees, where we previously reported cocirculation of genetically highly diverse SIVcpz strains within a single site (16, 37).
The newly identified SIVgor strains confirm that gorillas acquired their infection after a single cross-species transmission from chimpanzees, followed by intraspecies transmissions within the gorilla population (31). Our data are also in line with previous calculations of the most recent common ancestor (MRCA) of the current SIVgor clade, which estimated that SIVgor strains started to diverge at least 100 to 200 years ago (31). Given our new data, it is clear that cross-species transmissions from chimpanzees to gorillas are rare events. Gorillas do not harbor local SIVcpzPtt strains, and thus far, eastern gorillas seem to be free of SIVgor infection. This also corresponds with observations that encounters between gorillas and chimpanzees are rare and seem to be primarily nonaggressive (28). Western lowland gorillas and central chimpanzees are sympatric species. Some fruits are eaten by both species, but they differ in their foraging strategies (17, 23, 33, 40) as well as in their choice of nesting sites (6, 29, 34). This has led to niche differentiation, probably to avoid competition. However, it cannot be excluded that physical encounters between chimpanzees and gorillas, possibly involving biting or other forms of aggression, occurred in the past. Moreover, sharing the same habitat could also lead to indirect contact with the virus of the other species, e.g., during disease outbreaks such as anthrax, Ebola fever, etc. (39).
Although we have now screened a substantial number of chimpanzees in Cameroon (∼649 individuals), we have not identified SIVcpzPtt strains that are so closely related to SIVgor or HIV-1 group O or P as to constitute their source. It is thus possible that the ancestral SIVcpz lineage that gave rise to SIVgor has become extinct or that the gorillas or chimpanzees that harbor the ancestors of group O are located in areas not yet sampled. HIV-1 group P is the closest lineage to SIVgor and is most likely the result of a cross-species transmission from gorillas to humans; however, the actually identified SIVgor strains are not the precursors of group P, and it cannot be excluded that the ancestor of group P still circulates in chimpanzees, which transmitted it directly to humans. The actual SIVgor strains are also still too divergent to be the direct ancestors of HIV-1 group O.
Our data from the CP site indicate that SIVgor is transmitted both within and between different groups of gorillas. The exact modes of virus spread are not known, but the presence of phylogeographic clusters of SIVgor similar to those of SIVcpzPtt in chimpanzees suggests a very similar epidemiology. Interestingly, we observed a predominance of SIVgor infection in females, and whether this is related to a sample bias and/or the social organization of gorilla groups needs to be studied further. Western lowland gorillas live in social breeding groups usually formed by one silverback male, three adult females (on average), and their offspring (7, 9, 20, 24). Before reaching sexual maturity, male gorillas leave their natal group and go through a bachelor stage that can last several years (9, 19). In the field, it is easier to localize a group of gorillas than single individuals. However, the social organization of gorilla groups and the presence of different groups in a 15- to 20-km2 area clearly provide opportunities for SIV to spread both within and between neighboring communities, as shown here and as confirmed by a follow-up survey in 2009, where eight new SIVgor-infected individuals were identified in the CP-GR area (C. Neel and L. Etienne, unpublished results).
It was recently shown that SIVcpz infection has a negative impact on the health reproduction and survival of chimpanzees in the wild (16). Phylogenetic analysis revealed that gorillas acquired SIVs by cross-species transmission from chimpanzees. The lower prevalence and lower genetic diversity among circulating SIVgor strains are consistent with a more recent infection in gorillas but could also be an indication that SIVgor has a negative impact on the life span of infected gorillas and their ability to propagate virus spread.
Importantly, gorillas and chimpanzees are highly endangered species which continue to be hunted, especially in west central Africa, and remain a potential source of human infections (21). SIVs from apes are now known to have crossed the species barrier to humans on at least four occasions in west central Africa (14, 26). Given the recent discovery of the HIV-1 group P lineage in a Cameroonian woman in France, it would not be surprising if additional cases of SIVcpz or SIVgor cross-species transmissions had occurred, especially in geographic regions where these viruses are most prevalent and where hunting pressure is high. As shown for the group P virus-infected patient, SIVcpz/SIVgor infections are likely to be detected by commercial HIV antibody screening assays. However, HIV screening in Africa is still suboptimal, and serological tests with low sensitivities for group M, and especially for HIV-1 group O, are still used in this part of the world (1, 10, 25, 42). In addition to the use of assays with low sensitivities, poor serological results are also related to the high turnover of trained personnel, suboptimal storage conditions, failure of the cold chain during transport, use of tests after the expiration date, etc. (3). It is thus possible that in regions where the risk for cross-species transmissions with new SIVcpz or SIVgor strains is high, the currently used HIV serological assays will not detect such new infections, and that infection with divergent HIV strains can go unrecognized for a long time.
We showed here that gorillas are the natural hosts of SIVgor infection and that despite an overall low prevalence, gorilla communities with high infection rates exist. We did not identify sites with a high SIVgor prevalence close to 30%, as observed in chimpanzee communities harboring the group M and N ancestors (15). However, it is clear that wild-living gorillas can represent a reservoir for human infections (29). Noninvasive studies of chimpanzees and gorillas in the subregion are therefore still needed to identify the primate origin of HIV-1 groups O and P, the additional reservoirs for new HIV variants, and the impact of SIV infection on survival of gorillas and chimpanzees.
Acknowledgments
We thank the staff from PRESICA for logistical support in Cameroon; the Cameroonian Ministries of Health, Environment and Forestry, and Research for permission to collect samples in Cameroon; the Wildlife Conservation Society for collaboration in the Deng Deng Reserve in Cameroon; the Department of Ecology and Management of Plant and Animal Resources (University of Kisangani) for authorization to collect samples in the Democratic Republic of Congo; the Republic of Congo Ministry of Science and Technology and Ministry of Forest Economy for permission to collect samples in northern Republic of Congo; and the Water and Forest Ministry of the Central African Republic for permission to collect samples. The Wildlife Conservation Society's Congo Program and Global Health Program provided logistical support in the Republic of Congo. We thank Shelly Masi and the WWF Dzanga-Sangha Project for assistance in the field in the Central African Republic and the Institut Pasteur of Bangui for assistance with sample export. We thank the Agence Nationale des Parcs Nationaux (ANPN) and the Centre National de la Recherche Scientifique et Technique (CENAREST) of Gabon for permission to conduct our research in the Loango National Park, and we thank the SCD and the Wildlife Conservation Society for logistical support.
The gorilla sample collection process was supported by grants from the National Institutes of Health (R01 AI50529, R01 AI58715, and P30 AI 27767), the Bristol Myers Freedom to Discover Program, the Agence National de Recherche sur le SIDA, France (ANRS 12125 and ANRS 12182), the Institut de Recherche pour le Développement (IRD), the U.S. Fish and Wildlife Service, the Columbus Zoo and Aquarium, the National Geographic Society, the Brevard Zoo, The Wallace Global Fund, the Société pour la Conservation et le Développement (SCD), and the Max Planck Society. Philip Kranzusch was supported under grant NIH T32 AI007245-25. Lucie Etienne was supported by a Ph.D. grant from Sidaction.
Footnotes
Published ahead of print on 11 November 2009.
REFERENCES
- 1.Aghokeng, A. F., E. Mpoudi-Ngole, H. Dimodi, A. Atem-Tambe, M. Tongo, C. Butel, E. Delaporte, and M. Peeters. 2009. Inaccurate diagnosis of HIV-1 group M and O is a key challenge for ongoing universal access to antiretroviral treatment and HIV prevention in Cameroon. PLoS One 4:e7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibollet-Ruche, F., F. Gao, E. Bailes, S. Saragosti, E. Delaporte, M. Peeters, G. M. Shaw, B. H. Hahn, and P. M. Sharp. 2004. Complete genome analysis of one of the earliest SIVcpzPtt strains from Gabon (SIVcpzGAB2). AIDS Res. Hum. Retrovir. 20:1377-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birx, D., M. de Souza, and J. N. Nkengasong. 2009. Laboratory challenges in the scaling up of HIV, TB, and malaria programs: the interaction of health and laboratory systems, clinical research, and service delivery. Am. J. Clin. Pathol. 131:849-851. [DOI] [PubMed] [Google Scholar]
- 4.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. Van Der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley, B. J., D. M. Doran-Sheehy, and L. Vigilant. 2008. Genetic identification of elusive animals: re-evaluating tracking and nesting data for wild western gorillas. J. Zool. 275:333-340. [Google Scholar]
- 6.Devos, C., C. Sanz, D. Morgan, J. R. Onononga, N. Laporte, and M. C. Huynen. 2008. Comparing ape densities and habitats in northern Congo: surveys of sympatric gorillas and chimpanzees in the Odzala and Ndoki regions. Am. J. Primatol. 70:439-451. [DOI] [PubMed] [Google Scholar]
- 7.Doaudi, M., S. Gatti, F. Levrero, G. Duhamel, M. Bermejo, D. Vallet, N. Menard, and E. Petit. 2007. Sex-biased dispersal in western lowland gorillas (Gorilla gorilla gorilla). Mol. Ecol. 16:2247-2259. [DOI] [PubMed] [Google Scholar]
- 8.Furuichi, T., H. Ignaki, and S. Angoue-Ovongo. 1997. Population density of chimpanzees and gorillas in the Petit Loango Reserve, Gabon: employing a new method to distinguish between nests of the two species. Int. J. Primatol. 18:1029-1046. [Google Scholar]
- 9.Gatti, S., F. Levrero, N. Menard, and A. Gautier-Hion. 2004. Population and group structure of western lowland gorillas (Gorilla gorilla gorilla) at Lokoue, Republic of Congo. Am. J. Primatol. 63:111-123. [DOI] [PubMed] [Google Scholar]
- 10.Gautheret-Dejean, A., S. Mesmin-Poho, J. Birguel, V. Lemée, J. M. Huraux, and J. C. Plantier. 2008. Unequal detection of HIV type 1 group O infection by simple rapid tests. Clin. Infect. Dis. 46:1936-1937. [DOI] [PubMed] [Google Scholar]
- 11.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 12.Guislain, P., and J. Dupain. 2005. Sudden great ape die-off in the periphery of the Dja Biosphere Reserve. Gorilla J. 30:28-30. [Google Scholar]
- 13.Guschanski, K., L. Vigilant, A. McNeilage, M. Grayd, E. Kagodae, and M. M. Robbins. 2009. Counting elusive animals: comparing field and genetic census of the entire mountain gorilla population of Bwindi Impenetrable National Park, Uganda. Biol. Conserv. 142:290-300. [Google Scholar]
- 14.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 15.Keele, B. F., F. Van Heuverswyn, Y. Li, E. Bailes, J. Takehisa, M. L. Santiago, F. Bibollet-Ruche, Y. Chen, L. V. Wain, F. Liegeois, S. Loul, E. Mpoudi-Ngole, Y. Bienvenue, E. Delaporte, J. F. Brookfield, P. M. Sharp, G. M. Shaw, M. Peeters, and B. H. Hahn. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313:523-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keele, B. F., J. H. Jones, K. A. Terio, J. D. Estes, R. S. Rudicell, M. L. Wilson, Y. Li, G. H. Learn, T. M. Beasley, J. Schumacher-Stankey, E. Wroblewski, A. Mosser, J. Raphael, S. Kamenya, E. V. Lonsdorf, D. A. Travis, T. Mlengeya, M. J. Kinsel, J. G. Else, G. Silvestri, J. Goodall, P. M. Sharp, G. M. Shaw, A. E. Pusey, and B. H. Hahn. 2009. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature 460:515-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuroda, S., T. Nishihara, S. Suzuki, and R. A. Oko. 1996. Sympatric chimpanzees and gorillas in the Ndoki Forest, Congo, p. 71-81. In W. C. McGrew, L. F. Marchant, and T. Nishida (ed.), Great ape societies. Cambridge University Press, Cambridge, United Kingdom.
- 18.Leendertz, F. H., F. Lankester, P. Guislain, C. Néel, O. Drori, J. Dupain, S. Speede, P. Reed, N. Wolfe, S. Loul, E. Mpoudi-Ngole, M. Peeters, C. Boesch, G. Pauli, H. Ellerbrok, and E. M. Leroy. 2006. Anthrax in western and central African great apes. Am. J. Primatol. 68:928-933. [DOI] [PubMed] [Google Scholar]
- 19.Levréro, F., S. Gatti, N. Ménard, E. Petit, D. Caillaud, and A. Gautier-Hion. 2006. Living in nonbreeding groups: an alternative strategy for maturing gorillas. Am. J. Primatol. 68:275-291. [DOI] [PubMed] [Google Scholar]
- 20.Magliocca, F., S. Querouil, and A. Gautier-Hion. 1999. Population structure and group composition of western lowland gorillas in north-western Republic of Congo. Am. J. Primatol. 48:1-14. [DOI] [PubMed] [Google Scholar]
- 21.Matthews, A., and A. Matthews. 2004. Survey of gorillas (Gorilla gorilla gorilla) and chimpanzees (Pan troglodytes troglodytes) in southwestern Cameroon. Primates 45:15-24. [DOI] [PubMed] [Google Scholar]
- 22.Milne, I., D. Lindner, M. Bayer, D. Husmeier, G. McGuire, D. F. Marshall, and F. Wright. 2009. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 25:126-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan, D., and C. Sanz. 2006. Chimpanzee feeding ecology and comparisons with sympatric gorillas in the Goualougo Triangle, Republic of Congo, p. 97-122. In G. Hohmann, M. M. Robbins, and C. Boesch (ed.), Feeding ecology in apes and other primates: ecological, physical and behavioral aspects. Cambridge University Press, New York, NY.
- 24.Parnell, R. J. 2002. Group size and structure in western lowland gorillas (Gorilla gorilla gorilla) at Mbeli Bai, Republic of Congo. Am. J. Primatol. 57:193-206. [DOI] [PubMed] [Google Scholar]
- 25.Plantier, J. C., M. Djemai, V. Lemée, A. Reggiani, M. Leoz, L. Burc, A. Vessière, D. Rousset, J. D. Poveda, C. Henquell, A. Gautheret-Dejean, and F. Barin. 2009. Census and analysis of persistent false-negative results in serological diagnosis of human immunodeficiency virus type 1 group O infections. J. Clin. Microbiol. 47:2906-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plantier, J. C., M. Leoz, J. E. Dickerson, F. De Oliveira, F. Cordonnier, V. Lemée, F. Damond, D. L. Robertson, and F. Simon. 2009. A new human immunodeficiency virus derived from gorillas. Nat. Med. 15:871-872. [DOI] [PubMed] [Google Scholar]
- 27.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1571-1574. [DOI] [PubMed] [Google Scholar]
- 28.Stanford, C. B., and J. B. Nkurunungi. 2003. Sympatric ecology of chimpanzees and gorillas in Bwindi Impenetrable National Park, Uganda. Int. J. Primatol. 24:901-918. [Google Scholar]
- 29.Stanford, C. B. 2006. The behavioral ecology of sympatric African apes: implications for understanding fossil hominoid ecology. Primates 47:91-101. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan, K. M., A. Mannucci, C. P. Kimpton, and P. Gill. 1993. A rapid and quantitative DNA sex test: fluorescence-based PCR analysis of X-Y homologous gene amelogenin. Biotechniques 15:636-638. [PubMed] [Google Scholar]
- 31.Takehisa, J., M. H. Kraus, A. Ayouba, E. Bailes, F. Van Heuverswyn, J. M. Decker, Y. Li, R. S. Rudicell, G. H. Learn, C. Neel, E. Mpoudi-Ngole, G. M. Shaw, M. Peeters, P. M. Sharp, and B. H. Hahn. 2009. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J. Virol. 83:1635-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 33.Tutin, C. E., M. Fernandez, M. E. Rogers, E. A. Williamson, and W. C. McGrew. 1991. Foraging profiles of sympatric lowland gorillas and chimpanzees in the Lopé Reserve, Gabon. Philos. Trans. R. Soc. Lond. B 334:179-185. [DOI] [PubMed] [Google Scholar]
- 34.Tutin, C. E., R. J. Parnell, L. J. White, and M. Fernandez. 1995. Nest building by lowland gorillas in the Lope Reserve, Gabon: environmental influences and implications for censusing. Int. J. Primatol. 16:53-76. [Google Scholar]
- 35.van der Kuyl, A. C., C. L. Kuiken, J. T. Dekker, and J. Goudsmit. 1995. Phylogeny of African monkeys based upon mitochondrial 12S rRNA sequences. J. Mol. Evol. 40:173-180. [DOI] [PubMed] [Google Scholar]
- 36.Van Heuverswyn, F., Y. Li, C. Neel, E. Bailes, B. F. Keele, W. Liu, S. Loul, C. Butel, F. Liegeois, Y. Bienvenue, E. Mpoudi-Ngole, P. M. Sharp, G. M. Shaw, E. Delaporte, B. H. Hahn, and M. Peeters. 2006. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature 444:164. [DOI] [PubMed] [Google Scholar]
- 37.Van Heuverswyn, F., Y. Li, E. Bailes, C. Neel, B. Lafay, B. F. Keele, K. S. Shaw, J. Takehisa, M. H. Kraus, S. Loul, C. Butel, F. Liegeois, B. Yangda, P. M. Sharp, E. Mpoudi-Ngole, E. Delaporte, B. H. Hahn, and M. Peeters. 2007. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology 368:155-171. [DOI] [PubMed] [Google Scholar]
- 38.Van Heuverswyn, F., and M. Peeters. 2007. The origins of HIV and implications for the global epidemic. Curr. Infect. Dis. Rep. 9:338-346. [DOI] [PubMed] [Google Scholar]
- 39.Walsh, P. D., T. Breuer, C. Sanz, D. Morgan, and D. Doran-Sheehy. 2007. Potential for Ebola transmission between gorilla and chimpanzee social groups. Am. Nat. 169:684-689. [DOI] [PubMed] [Google Scholar]
- 40.Yamagiwa, J., and A. K. Basabose. 2006. Diet and seasonal changes in sympatric gorillas and chimpanzees at Kahuzi-Biega National Park. Primates 47:74-90. [DOI] [PubMed] [Google Scholar]
- 41.Yang, Z., and B. Rannala. 1997. Bayesian phylogenetic inference using DNA sequences: a Markov chain Monte Carlo method. Mol. Biol. Evol. 14:717-724. [DOI] [PubMed] [Google Scholar]
- 42.Zouhair, S., S. Roussin-Bretagne, A. Moreau, S. Brunet, S. Laperche, M. Maniez, F. Barin, and M. Harzic. 2006. Group O human immunodeficiency virus type 1 infection that escaped detection in two immunoassays. J. Clin. Microbiol. 44:662-665. [DOI] [PMC free article] [PubMed] [Google Scholar]