Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) infection of swine leads to a serious disease characterized by a delayed and defective adaptive immune response. It is hypothesized that a suboptimal innate immune response is responsible for the disease pathogenesis. In the study presented here we tested this hypothesis and identified several nonstructural proteins (NSPs) with innate immune evasion properties encoded by the PRRS viral genome. Four of the total ten PRRSV NSPs tested were found to have strong to moderate inhibitory effects on beta interferon (IFN-β) promoter activation. The strongest inhibitory effect was exhibited by NSP1 followed by, NSP2, NSP11, and NSP4. We focused on NSP1α and NSP1β (self-cleavage products of NSP1 during virus infection) and NSP11, three NSPs with strong inhibitory activity. All of three proteins, when expressed stably in cell lines, strongly inhibited double-stranded RNA (dsRNA) signaling pathways. NSP1β was found to inhibit both IFN regulatory factor 3 (IRF3)- and NF-κB-dependent gene induction by dsRNA and Sendai virus. Mechanistically, the dsRNA-induced phosphorylation and nuclear translocation of IRF3 were strongly inhibited by NSP1β. Moreover, when tested in a porcine myelomonocytic cell line, NSP1β inhibited Sendai virus-mediated activation of porcine IFN-β promoter activity. We propose that this NSP1β-mediated subversion of the host innate immune response plays an important role in PRRSV pathogenesis.
Porcine reproductive and respiratory syndrome virus (PRRSV) is a member of family Arteriviridae which, along with the Coronaviridae, are classified in the order Nidovirales (9). PRRSV is an enveloped, single-stranded RNA virus with positive-sense genome of ∼15 kb. The genome has nine open reading frames (ORFs), namely, ORF1a, ORF1b, ORF2a, ORF2b, and ORF3-7. ORF1a and ORF1b are synthesized as a single polyprotein and later processed to 14 different nonstructural proteins (NSPs), e.g., NSP1α, NSP1β, and NSP2 to NSP12 by the viral proteases. NSP1α, NSP1β, NSP2, and NSP4 are the viral proteases which carry out this function. The viral genome replication and transcription is carried out by NSP9, which encodes the viral RNA-dependent RNA polymerase, and NSP10, which encodes a helicase (5, 49).
The ailment caused by PRRSV is considered to be the most economically significant infectious disease of swine worldwide. It causes an annual loss of approximately $560 million in the United States (38). The clinical signs of PRRSV infection include late-term reproductive failure in sows and respiratory illness in growing pigs. Most cases of PRRSV infection in pigs are complicated by secondary opportunistic bacterial infections, which have been attributed to the immunosuppressive nature of the virus (13). Upon initial virus infection, viremia lasts for a few weeks, after which it resolves. However, the virus can be detected in secondary lymphoid organs for several months, indicating failure of host immune response to clear the virus (2).
The immune response to PRRSV infection is characterized by a delayed appearance of neutralizing antibodies (31), a short cell-mediated immune response (34, 59), and slow development of a virus-specific gamma interferon (IFN-γ) response (35). We and others have identified various factors that are likely to play multiple roles in delayed clearance of PRRSV from the host. These include weak innate immune response (1, 35), presence of decoy epitopes (39), and glycan shielding of envelope proteins (3). Previous studies have shown that very low or negligible levels of IFN-α are produced upon PRRSV infection in pulmonary alveolar macrophages (PAMs) and PRRSV permissive monkey kidney cells (MARC-145) in vitro (1, 36). IFN-α production in the lungs of pigs acutely infected with PRRSV was either almost undetectable or 100- to 200-fold lower than that induced by porcine respiratory coronavirus (PRCV) (8, 57). PRRSV has also been found to suppress IFN-α production by transmissible gastroenteritis corona virus (TGEV), a known inducer of IFNs in infected alveolar macrophages (1). At the same time, externally provided IFN-α or IFN-β have been able to reduce viral replication in cultured alveolar macrophages (1, 40). The virus was also found to inhibit the dsRNA-mediated upregulation of IFN-β gene transcription (36). A microarray analysis of PAMs infected with Lelystad virus (European type PRRSV) showed no significant change in the IFN-α from the control at 12 h postinfection (17). Considering the broad role of IFNs in establishing an effective adaptive immune response, we hypothesize that the suboptimal induction of type I IFN may be one of the determining factors in deficient development of acquired immunity.
In the constant struggle to outsmart the host, viruses have developed strategies to evade and/or inhibit key elements of host immune response. Sometimes, a substantial part of the viral genome is dedicated toward suppressing IFN signaling pathways, IFN-stimulated gene (ISG) functions, or pathways for RNA processing and translation (7, 16). In the present study we sought to identify the PRRSV proteins that are responsible for mediating the inhibition of IFN production. We observed that several PRRSV NSPs inhibit IFN regulatory factor 3 (IRF3)-mediated activation of IFN-β promoter. Detailed investigation of the mechanism of inhibition of one of those viral proteins, NSP1β, revealed that it inhibits double-stranded RNA (dsRNA)-mediated IRF3 phosphorylation and nuclear translocation.
MATERIALS AND METHODS
Cells and viruses.
HEK293-TLR3 (Invivogen), HeLa (American Type Culture Collection [ATCC]), HT1080 (ATCC), MARC-145 (obtained from Will Laegreid, USMARC, USDA/ARS) (25), and ST (ATCC) cells were maintained in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum and 50 μg of gentamicin (Sigma)/ml. HEK293-TLR3 and HT1080 cells stably expressing PRRSV NSP1α, NSP1β, and NSP11 were established by cotransfecting them with corresponding PRRSV protein expression plasmid and pBabePuro. After selection with 1 μg of Puromycin (Mediatech)/ml, individual colonies were picked and screened for stable expression of PRRSV NSPs. For the swine macrophage experiments, mononuclear cells were isolated from blood of donor pigs by Ficoll density centrifugation using lymphocyte separation medium (Mediatech) (32). These cells were then plated in a glass petri dish for 1 day, followed by washing with phosphate-buffered saline (PBS) to enrich the monocytes. The attached monocytes were cultured in RPMI 1640 medium supplemented with 10% BVDV-free fetal bovine serum in the presence of the growth factor macrophage colony-stimulating factor (5 ng/ml) (Sigma) for 7 days for maturation into macrophages. These monocyte-derived macrophages were then harvested by cell dissociation buffer (Gibco) and plated in appropriate tissue culture plate with RPMI 1640 and 10% fetal bovine serum (FBS) for virus infection. Porcine monocytic cell line 3D4/31 (ATCC) was maintained in RPMI 1640 medium supplemented with 10% BVDV-free fetal bovine serum and 50 μg of gentamicin (Sigma)/ml. Highly virulent North American PRRSV strain FL12 (56) was propagated in MARC-145 cells to prepare the virus stocks for the present study. TGEV (Purdue strain) (1) was obtained from the Veterinary Diagnostic Center (University of Nebraska) and propagated in ST cells to prepare the virus stocks for infection in macrophages. Sendai virus (SeV; Cantell strain) was obtained from Charles River Laboratories and was used as described below.
Reporter assays.
IFN-β-CAT assays were performed in HeLa cells transfected with 1 μg of IFN-β-CAT reporter plasmid, 0.5 μg of IRF3 plasmid (Luwen Zhang, University of Nebraska), and 1 μg of indicated viral protein coding plasmid using Lipofectamine-2000 (Invitrogen) according to the manufacturer's instructions. At 40 h posttransfection, the cells were lysed by freeze-thawing, and a chloramphenicol acetyltransferase (CAT) assay was performed as described earlier (44). CAT activity is expressed as the fold induction relative to the empty vector control. A minimum of three independent experiments was performed to confirm the results. For ISG56-luciferase reporter assays, HEK293-TLR3 cells were transfected with 0.4 μg of ISG56-luciferase reporter plasmid and 10 ng of pRL-TK (Promega), along with the indicated protein expression vectors using Fugene 6 (Roche Applied Science) according to the manufacturer's protocol. For the NF-κB luciferase assays, HEK293-TLR3 cells were transfected with 0.5 μg of 5X-NF-κB-luciferase reporter plasmid (48) and 25 ng of pRL-TK (Promega), along with the indicated protein expression vector using Fugene 6 as described above. At 24 h posttransfection, the medium was replaced with fresh medium. At 40 h posttransfection, the cells were either treated with 10 μg of poly(I-C) (GE Healthcare)/ml or PBS for 6 h, and luciferase assays were performed by using a dual luciferase assay kit (Promega). For the porcine IFN-β-luciferase assay, 3D4/31 cells were transfected with 0.4 μg of porcine IFN-β-luc plasmid and 10 ng of pRL-TK (Promega), along with the indicated protein expression vectors using Ingenio electroporation solution (Miru Bio) in an Amaxa Nucleofector (Lonza) according to the manufacturer's instructions. Then, at 40 h posttransfection, the cells were either mock infected or infected with SeV (Cantell strain) for 8 h, after which the cells were lysed, and luciferase assay was performed. Firefly luciferase activities were expressed in terms of the fold change with respect to controls after normalization with the Renilla luciferase activities.
Real-time PCR and enzyme-linked immunosorbent assay (ELISA).
Total RNA was isolated from cells by using TRIzol (Invitrogen) reagent and cDNA was prepared by using Moloney murine leukemia virus reverse transcriptase (Invitrogen). This cDNA was used for quantification of the indicated mRNA copy number by using SmartCycler (Cepheid). Porcine IFN-α, IFN-β, and β-actin (internal control) real-time PCR was performed using a TaqMan probe based method. Human interleukin-8 (IL-8) and human ribosomal protein L32 (RPL32, internal control) copy numbers were calculated by Sybr-green based quantitative PCR. The following primer sets and, where indicated, probes were used in the real-time PCRs. For porcine IFN-α, we used forward primer 5′-AGAGCCTCCTGCACCAGTTCT-3′, reverse primer 5′-CTGCATGACACAGGCTTCCA-3′, and probe 5′-ACTGGACTGGATCAGCAGCTCAGGGA-3′; for porcine β-actin, we used forward primer 5′-CTCCTTCCTGGGCATGGA-3′, reverse primer 5′-CCACGTCGCACTTCATGATC-3′, and probe 5′-TGCGGCATCCACGAGACCACCT-3′; and for human IL-8, we used forward primer 5′-TAGCCAGGATCCACAAGTCC-3′ and reverse primer 5′-GCTTCCACATGTCCTCACAA-3′. The primer and probe sequences for swine IFN-β (17) and human RPL-32 (33) were as described earlier. IFN-α, IFN-β, and IL-8 mRNA levels in virus-infected or poly(I-C)-stimulated cells were expressed in copy numbers relative to mock-infected or unstimulated cells according to the method described earlier (51). The porcine IFN and human IL-8 mRNA levels were normalized across the experiments using porcine β-actin and human RPL-32 levels, respectively. Porcine IFN-α protein levels were measured by ELISA using F17 monoclonal antibody (MAb; PBL Biomedical Labs) and peroxidase-conjugated K9 MAb (PBL Biomedical Labs) as coating and detection antibodies, respectively, as described previously (29). Briefly, flat-bottom 96-well plates were coated overnight at 4°C with F17 at a concentration of 2 μg/μl in bicarbonate coating buffer (pH 9.6). After blocking with 1% bovine serum albumin, for 1 h at 37°C, the plates were washed three times with 0.05% Tween 20 in PBS. Samples (100 μl) were added into each well, followed by incubation for 4 h at 37°C. After three washes, 100 μl of peroxidase-conjugated K9 at a concentration of 0.5 μg/ml was added to each well. After 2 h of incubation and three washes, 100 μl of tetramethylbenzidine (TMB) substrate solution was added for 30 min, the reaction was stopped with 1 N HCl, and the optical density was measured at a 450-nm wavelength by an ELISA plate reader (Bio-Tek). Quantified recombinant porcine IFN-α (R&D Systems) was used as a standard, and IFN-α concentrations were calculated based upon a standard curve.
Immunoblotting.
For immunoblotting, cells were washed twice with PBS and then pelleted by centrifugation. In the case of isolation of a nuclear fraction, these pellets were processed as described previously (45). For whole-cell protein extraction, PBS-washed cell pellets were resuspended in lysis buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM sodium orthovanadate, complete protease inhibitor [Roche]). The cell suspension was incubated on ice for 15 min, followed by clarification by centrifugation at 17,000 × g at 4°C for 10 min. The total protein concentrations in the supernatants were quantified by using a Bradford assay kit (Bio-Rad). Equal amounts of total protein were separated by SDS-PAGE, transferred to polyvinylidene difluoride membrane, and probed with appropriate antibodies. Protein bands were detected by using an ECL detection system (Pierce).
Plasmids and antibodies.
All PRRSV (FL-12 strain) NSPs were amplified by using pFL12 (56) as a template and the primers as listed in Table 1. Since the exact site of cleavage between NSP1α and NSP1β is not known, the putative cleavage site (nucleotides [nt] 678 to 692) was included in both expression constructs. The NSP1α and NSP1β expression constructs contain the FL12 (GenBank accession no. AY545985) coding sequence from nt 192 to 710 and nt 651 to 1340, respectively. They were cloned into a mammalian expression vector pIHA-FLAG, which was derived from pHyg-EGFP (Clontech) by removing the Hygro-EGFP part and inserting an oligonucleotide containing 5′ FLAG epitope tag and a few restriction enzyme sites to enable the expression of an N-terminal FLAG-tagged gene. NSP1β was cloned in the same vector but with an N-terminal hemagglutinin (HA) tag. Porcine IFN-β-luciferase plasmid was constructed by placing porcine IFN-β promoter (from nt −281 to + 19; the +1 position refers to the transcription start site of swine IFN-β gene GenBank accession no. M86762.1) upstream of the firefly luciferase gene in a pGL3-Basic vector (Promega). pIFN-β-CAT (44), pISG56-luc (45), p5X-NF-κB-luciferase, pEFBOS-N-RIG-I (52), pmyc-IKKɛ (53), pHA-TRIF (23), and pCMV2C-bICP0 (44) were described previously. Immunoblotting and indirect immunofluorescence were performed with the following antibodies: anti-FLAG M2 MAb (Sigma), anti-HA MAb (Covance), anti-β-actin MAb (Santa Cruz Biotech), rabbit anti-IRF3 polyclonal antibody (pAb) (obtained from Michael David, University of California at San Diego), rabbit anti-ISG56 pAb (19), anti-IRF3-p396 MAb (Cell Signaling), rabbit anti-DRBP76 (45), anti-tubulin MAb (Santa-Cruz Biotech), and peroxidase-conjugated goat anti-mouse and goat anti-rabbit pAbs (KPL). Protein bands were visualized by using ECL Plus Western blotting detection reagent (Pierce), followed by exposure to X-ray film.
TABLE 1.
Primers used to generate individual NSP-expressing plasmids
| Primer | Sequence (5′-3′)a | Genomic positionb |
|---|---|---|
| NSP1F | ATATATCTCGAGATGTCTGGGATACTTGATCGG | 192-212 |
| NSP1αR | ATATATGCGGCCGCTTAAAAGTCTTCAGGCTTA | 694-710 |
| NSP1βF | ATATATGCTAGCATGTACCCATACGATGTTCCAGATTACGCTGGAGCAACTCATGTGC | 651-666 |
| NSP1R | ATATATGCGGCCGCTTAACCGTACCATTTGTGACTGC | 1321-1340 |
| NSP2F | ATATATACGCGTGCTGGAAAGAGAGCAAGGAA | 1341-1360 |
| NSP2R | ATATATGCGGCCGCTTAGCCCAGTAACCTGCCAAGA | 4261-4280 |
| NSP3F | ATATATCTCGAGGGGGCACGCTACATCTGGC | 4281-4299 |
| NSP3R | ATATATGCGGCCGCTTACTCAAGAAGGGACCCAAGC | 5599-5618 |
| NSP4F | ATATCTCGAGATGGGCGCTTTCAGAACTC | 5619-5634 |
| NSP4R | TATATCTAGATCATTCCAGTTCGGGTTTG | 6214-6230 |
| NSP5F | ATATATCTCGAGGGAGGCCTCTCCACCGTC | 6231-6248 |
| NSP5R | ATATATGCGGCCGCTTACTCGGCAAAGTATCGC | 6724-6740 |
| SP7F | ATATATCTCGAGTCGCTGACTGGTGCCCTC | 6788-6806 |
| NSP7R | ATATATGCGGCCGCTTATTCCCATTGGTTTTTTTTGCTC | 7543-7565 |
| NSP8F | ATATATCTCGAGGCTGCGAAGCTTTCCGTG | 7566-7583 |
| NSP8R | ATATATGCGGCCGCTTACTTAGTCAGGCCCTGGA | 7665-7683 |
| NSP9F | ATATATCTCGAGGGAGCAGTGTTTAAACTGCTAGC | 7684-7706 |
| NSP9R | ATATATGCGGCCGCTTACTCATGATTGGATCTGAGTTTTTC | 9595-9619 |
| NSP10F | ATATATCTCGAGGGGAAGAAGTCCAGAATGTG | 9620-9639 |
| NSP10R | ATATATGCGGCCGCTTATTCCAGATCTGCACAAATG | 10923-10942 |
| NSP11F | ATATCTCGAGATGTCGAGCTCCCCGCTC | 10946-10960 |
| NSP11R | TATATCTAGATCATTCAAGTTGAAAATAGGC | 11593-11611 |
| NSP12F | ATATATCTCGAGGGCCGCCATTTCACCTGGTA | 11612-11631 |
| NSP12R | ATATATGCGGCCGCTTATCAATTCAGGCCTAAAGTTGGTTC | 12049-12073 |
The restriction enzyme sites used for cloning are underlined. The N-terminal HA tag in the NSP1βF primer is indicated in boldface.
The genomic positions for the primers are based on GenBank accession number AY545985.
Indirect immunofluorescence.
Cells grown in coverslips were treated as indicated, washed with PBS twice, and then fixed with 4% paraformaldehyde for 20 min at room temperature. After two washes in PBS, the cells were then permeabilized with 0.2% Triton X-100 for 15 min and blocked in PBS containing 3% bovine serum albumin, 5% glycine, 10% nonimmune horse serum, and 0.05% Tween 20 for 2 h at room temperature. The coverslips were then incubated with a rabbit polyclonal antiserum against human IRF3 (1:500) or with anti-HA tag MAb (1:1,000) in the same blocking buffer at 4°C overnight. After three PBS washes, coverslips were incubated with Alexa Fluor 568-labeled donkey anti-mouse (Invitrogen) and Alexa Fluor 488-labeled goat anti-rabbit (Invitrogen) antibodies at room temperature for 1 h. The coverslips were washed and treated with DAPI (4′,6′-diamidino-2-phenylindole) to view the nuclei (Sigma), followed by washing. The coverslips were then mounted in aqueous mounting medium (Sigma) and observed under Olympus FV500/IX81 inverted confocal microscope.
RESULTS
PRRSV inhibits type I IFN induction.
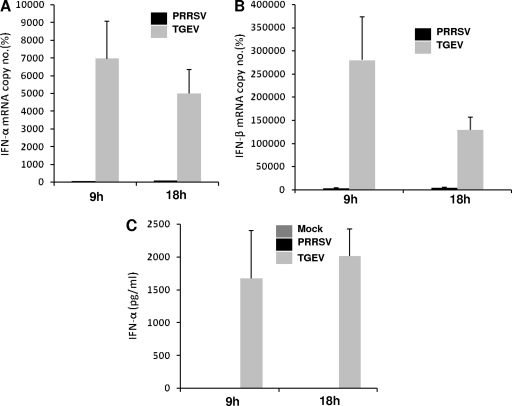
PRRSV has been found to inhibit IFN induction (1, 29, 36). In order to confirm and characterize this feature of PRRSV in a well-known model, we used our well-characterized, infectious clone-derived PRRS virus strain, FL12 (27, 56), corresponding to a highly virulent North American PRRSV strain (NVSL no. 97-7895). The FL12 virus strain has been extensively used in our laboratory for studies on virulence and immune response in infected animals (3, 11, 27, 56). The FL12 strain was used at a multiplicity of infection of 1.0 to infect macrophages derived from monocytes isolated from the peripheral blood mononuclear cells of healthy pigs. Transmissible gastroenteritis virus (TGEV), a well-known inducer of type I IFN, was used as a positive control for type I IFN induction. As shown in Fig. 1, very little or no IFN-α (Fig. 1A) or IFN-β (Fig. 1B) mRNA was detected in the cells infected with PRRSV, whereas TGEV infection showed strong induction of IFN-α and IFN-β mRNA in the same types of cells at both 9 and 18 h postinfection (p.i.). This observation was confirmed at the protein levels by measuring porcine IFN-α in infected cell culture supernatants by ELISA (Fig. 1C). Productive PRRSV infection in these cells was confirmed by an immunofluorescence assay conducted at 24 h p.i. using the monoclonal antibody SDOW-17, which is specific for PRRSV nucleocapsid protein (data not shown). Similar differences in the type I IFN induction were observed between PRRSV and TGEV infections in pulmonary alveolar macrophages obtained from lung lavage of adult animals (data not shown).
FIG. 1.
Inhibition of type I IFN production after PRRSV infection. Porcine monocyte-derived macrophages were mock infected or infected with PRRSV (FL-12) or TGEV at a multiplicity of infection of 1. Supernatants and cells were collected after 9 and 18 h p.i., respectively. Total RNA isolated from cells was reverse transcribed, and real-time PCR was performed for the detection of porcine IFN-α (A) and IFN-β (B) mRNA. The mRNA copy numbers were calculated after normalization with the β-actin copy number and are expressed as percentages relative to the mock control. Bars show the average of mRNA copy numbers ± the standard error of the mean (SEM) from three independent experiments using cells isolated from three different pigs. (C) Supernatants from the same experiments were used to detect secreted IFN-α by ELISA. Quantified recombinant porcine IFN-α was used as a standard, and IFN-α levels were calculated based upon a standard curve.
Several of the PRRSV NSPs exhibit strong to moderate inhibition of IFN-β promoter activation.
Viruses have been known to code for proteins that antagonize the type I IFN response (7). It has been hypothesized that the poor induction of type I IFN by PRRSV may be an active pathogenic process triggered by the virus when infecting the host (1). To investigate the role of PRRS viral proteins in active IFN antagonism, we focused on NSPs of PRRSV. NSPs comprise almost 80% of the PRRSV genome coding capacity. All of the PRRSV NSPs, except NSP6 (a very short peptide of 16 amino acids) derived from the viral genome (FL12 strain), were cloned as N-terminal tagged fusion protein in a mammalian expression vector and verified for expression by transient transfection and immunofluorescence in HeLa cells. We were able to obtain satisfactory expression levels for 10 NSPs (data not shown). We failed to achieve high-level expression for NSP3, which may be due to its instability when expressed alone, since it has been most often reported to be part of polyprotein complex consisting of other NSPs in case of equine arteritis virus (EAV), the prototypic family member of Arteriviridae (41). Each of these NSP constructs was cotransfected into HeLa cells, along with IRF3 and human IFN-β-CAT constructs, and CAT assays were performed. As shown in Fig. 2A, IFN-β promoter activation by IRF3 was suppressed to various degrees by NSP1, NSP2, NSP4, and NSP11. NSP1 exhibited the strongest inhibitory effect, followed by others (Fig. 2A). The bovine herpesvirus protein bICP0 was used as a positive control, which is known to inhibit IFN-β promoter induction by IRF3 (44). This result suggested that several of the PRRSV NSPs are capable of suppressing IFN-β promoter activation. To confirm this inhibition of IFN-β promoter activation, we cotransfected increasing concentrations of each of the corresponding NSP plasmids into HeLa cells by using the same assay. Figure 2B shows the dose-dependent inhibition of IFN-β promoter activity with increasing concentrations of NSPs (Fig. 2B, bottom panel).
FIG. 2.
Specific PRRSV proteins are involved in downregulation of IFN-β promoter. (A) HeLa cells were cotransfected with pIFN-β-CAT plasmid (1 μg), IRF3 expression plasmid (0.5 μg), and the viral nonstructural protein expression plasmids (1 μg) or bovine herpesvirus type 1 protein bICP0. Empty vector was added to the transfection mixture to keep the total DNA amount constant. Cell extracts collected at 40 h posttransfection were analyzed for CAT activity as described in Materials and Methods. The value of the CAT activity in the presence of empty vector was set at 100%, and those of the viral proteins were normalized accordingly. (B) Increasing amounts of indicated NSP coding plasmids were transfected, and CAT activities were measured as described earlier. The bars represent the means ± the SEM from three independent experiments. The bottom panels show increasing levels of individual NSP expression upon transfection (actin was used as a loading control).
PRRSV NSP1 and NSP11 inhibit IRF3-mediated gene induction.
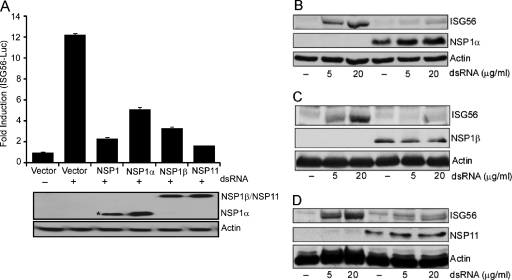
Viral nucleic acids are detected by the host innate immune system using Toll-like receptors (TLR) and RIG-like receptors (RLR). One such unique nucleic acid is dsRNA, a common by-product or intermediate in viral genome replication. In mammals, TLR3, retinoic acid inducible gene I (RIG-I), and melanoma differentiation-associated gene 5 (MDA5) are the three known sensors of dsRNA. Engagement of TLR3 or RLR with dsRNA induces signaling cascades, leading to the activation of multiple transcription factors such as NF-κB and IRF3. These activated transcription factors induce the expression of a set of cytokine genes, among which are type I IFNs, the hallmark of the antiviral response. IFNs signal via cell surface receptors and regulate many genes (IFN-stimulated genes [ISGs]) which sensitize cells for detection of invading pathogens, inhibit protein synthesis, and limit viral replication (46). We used dsRNA-mediated TLR3 activation and IFN induction as a model system to understand the mechanism of IFN antagonism by PRRSV NSPs. We decided to focus on NSP1 and NSP11 since they consistently exhibited the strong inhibition property (Fig. 2B). NSP1 is proteolytically processed during the infection by its own papainlike protease domain into NSP1α and NSP1β. Therefore, the three proteins (NSP1α, NSP1β, and NSP11) were tested for their IFN antagonistic activity using an ISG56-luciferase reporter assay. Unlike a complex promoter such as IFN-β, the ISG56 promoter has a simpler structure and can be activated by IRF3/IRF7. HEK293 cells stably expressing human TLR3 (HEK293-TLR3) were used in order to focus on the TLR3-IRF3 arm of the dsRNA signaling pathway. These cells were cotransfected with the NSP expression constructs, ISG56-luciferase, and Renilla-luciferase vectors. At 40 h after transfection, the cells were stimulated with dsRNA for 6 h, followed by a luciferase assay. As expected, all of the three proteins showed significant downregulation of dsRNA-mediated ISG56 promoter activation compared to an empty vector control (Fig. 3A).
FIG. 3.
Inhibition of IRF3-mediated gene induction by PRRSV proteins. (A) HEK-293TLR3 cells were cotransfected with the indicated viral NSP expression plasmids (1 μg) or empty vector, ISG56-luciferase plasmid (0.4 μg), and pRLTK (0.01 μg). At 40 h posttransfection the cells were treated with 10 μg of dsRNA/ml or PBS for 6 h and assayed for luciferase activity. The bars represent the average fold inductions ± the SEM for luciferase activities compared to PBS-treated vector control. The panel below the bar graph shows the expression of the respective NSPs. The asterisk (*) at NSP1 lane indicates the detection of proteolytically processed NSP1 to NSP1α, which carries the N-terminal FLAG tag. (B to D) HEK293-TLR3 cells stably expressing NSP1α (B), NSP1β (C), and NSP11 (D) (lanes 4 to 6) were stimulated with the indicated amount of dsRNA for 6 h, and the ISG56 level was measured by immunoblotting and compared to empty vector-transfected, puromycin-resistant control cells (lanes 1 to 3). Expressions of NSPs were detected by probing with anti-FLAG/HA antibody.
For a more direct confirmation of the inhibition of dsRNA signaling by these proteins, we established HEK293-TLR3 cell lines stably expressing these three PRRSV NSPs. Upon treatment with dsRNA, these cell lines induced no or very low levels of endogenous ISG56 compared to the empty vector-expressing cells (Fig. 3B, C, and D). A different dsRNA-responsive cell line, HT1080 (a human fibrosarcoma cell line), stably expressing these viral proteins also showed similar results upon dsRNA treatment (data not shown). This indicates that the inhibitory effect observed is independent of the cell line being used. Taken together, our results from two different assays suggest that PRRSV NSP1α, NSP1β, and NSP11 are involved in the inhibition of IRF3-mediated IFN signaling.
PRRSV NSP1β inhibits NF-κB-mediated gene induction.
In addition to IRF3, the TLR3 signaling pathway also activates NF-κB. The next series of experiments were directed to investigate the effect of these NSPs on NF-κB-mediated gene induction by dsRNA. Cells were cotransfected with NSPs, along with 5X-NF-κB-luciferase and Renilla luciferase reporters. Upon dsRNA treatment, the empty vector-transfected cells showed strong induction of luciferase activity compared to the untreated cells. Expression of NSP1β and NSP11 downregulated the induction of luciferase activity to the level of the untreated control (Fig. 4A). Interestingly, NSP1α expression did not have a significant negative effect on NF-κB promoter activity. This suggests differential inhibition of NSP1α compared to NSP1β and NSP11. To confirm the negative effect of NSP1β on NF-κB signaling, we assessed the induction of endogenous IL-8 mRNA, a cytokine whose expression is principally driven by NF-κB. In accordance with the reporter assay result, IL-8 mRNA induction upon dsRNA treatment was significantly reduced in HEK293-TLR3 cells stably expressing NSP1β compared to empty vector control cells (Fig. 4B).
FIG. 4.
Inhibition of NF-κB-mediated gene induction by PRRSV proteins. (A) HEK293-TLR3 cells were cotransfected with the indicated viral NSP expression plasmids or empty vector (1 μg), 5X NF-κB-luciferase plasmid (0.5 μg), and pRLTK (0.025 μg). At 40 h posttransfection the cells were treated with dsRNA for 6 h and assayed for luciferase activity. The fold induction of the average luciferase activity compared to the untreated vector control from a representative experiment is shown. The immunoblot panel indicates the expression of individual NSPs in the luciferase assay. (B) IL-8 mRNA levels were measured by real-time PCR in HEK293-TLR3 cells stably expressing NSP1β or vector control cells with or without dsRNA treatment. The mRNA copy numbers were calculated after normalizing them with the RPL32 (internal control) copy number and are expressed as percentages relative to the uninduced vector control.
TRIF or IKKɛ do not rescue NSP1β-mediated inhibition of IRF3 activation.
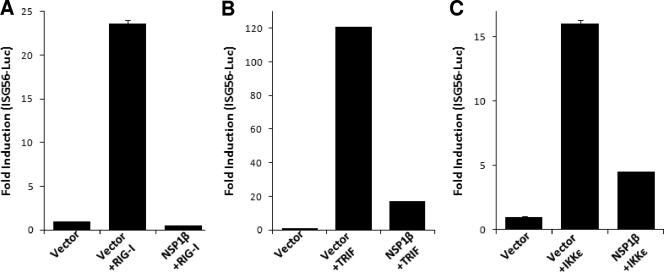
Besides TLR3, cytoplasmic dsRNA is sensed by DExD/H box RNA helicases: RIG-I and MDA5 (24). A number of RNA viruses induce signaling through dsRNA receptor RIG-I (30). Having established that PRRSV NSP1β inhibits TLR3-mediated IRF3 and NF-κB signaling, we investigated the effect of NSP1β on RIG-I signaling pathways. We took advantage of expressing the N-terminal CARD domain of RIG-I, which constitutively activates the RIG-I signaling pathway (14), and tested the effect of NSP1β. As shown in Fig. 5A, the constitutively active RIG-I, when coexpressed with ISG56-luciferase, induced strong luciferase activity compared to the vector control. However, in the presence of NSP1β the induction is completely inhibited showing that, in addition to the TLR3 pathway, NSP1β also inhibits the RIG-I-mediated signaling pathway.
FIG. 5.
NSP1β inhibits IRF3 signaling by acting downstream of IRF3 kinases. HEK293-TLR3 cells were cotransfected with ISG56-luciferase plasmid, NSP1β expression plasmids, or empty vector. IRF3 signaling pathway was induced by including in the transfection constitutively active RIG-I (A), TRIF (B), and IKKɛ (C). Cells were lysed 40 h posttransfection and assayed for luciferase activity. Bars indicate the mean fold inductions ± the SEM of luciferase activity compared to the vector control from three independent experiments.
Oftentimes, a very similar approach is used to identify the step(s) of the signaling pathway that is being inhibited (33). We used expression constructs for TRIF and IKKɛ, two major components of TLR3 signaling pathway, to examine whether the NSP1β-mediated inhibition could be rescued. Our results show that IRF3 activity was induced by transfection of TRIF (Fig. 5B) and IKKɛ (Fig. 5C) in an ISG56-luciferase reporter assay. The presence of NSP1β significantly downregulated the ISG-56 promoter activation induced by these molecules. These results indicate that part of the inhibitory activity of NSP1β affects the downstream signaling of IRF3 kinases, probably by directly affecting IRF3. Exogenous expression of TRIF could not recover activation of a NF-κB-dependent promoter in the presence of NSP1β either (data not shown).
NSP1β inhibits dsRNA-induced IRF3 phosphorylation and nuclear translocation.
In the resting state, IRF3 shuttles between the cytoplasm and the nucleus. Activation by dsRNA causes its phosphorylation and translocation to the nucleus (58). The binding and recruitment of IRF3 on ISRE promoters is stabilized by CBP/p300, which prevents its export from the nucleus. To investigate the mechanism of NSP1β-mediated inhibition of IRF3 activation, we assayed several hallmark steps of the IRF3 activation process, such as IRF3 phosphorylation (on Ser396) and nuclear translocation, upon dsRNA treatment. As expected, dsRNA treatment induced a marked increase in the level of phosphorylated IRF3 in the control cells. However, in NSP1β-expressing cells, IRF3 phosphorylation was reduced to the basal level (Fig. 6A). IRF3 nuclear translocation was similarly affected by NSP1β expression. As shown in Fig. 6B, the level of IRF3 present in the nuclear fraction increased upon dsRNA treatment. However, in the presence of NSP1β IRF3 failed to translocate to the nucleus. This was consistent with the previous result showing the reduction of IRF3 phosphorylation in the presence of NSP1β. This phenomenon was further verified by visualization of the subcellular location of IRF3 by indirect immunofluorescence. HEK293-TLR3 cells transfected with empty vector showed efficient IRF3 nuclear translocation 2 h after dsRNA treatment but, in the presence of NSP1β, most of the IRF3 was retained in the cytoplasm (Fig. 6C) in a manner similar to empty vector-transfected cells without dsRNA treatment (top left panel). These results suggest that NSP1β inhibits dsRNA-mediated phosphorylation of IRF3 and its nuclear translocation.
FIG. 6.
NSP1β inhibits dsRNA-induced IRF3 phosphorylation and nuclear translocation. (A) HEK293-TLR3 cells stably expressing NSP1β- or vector-transfected cells (vector) were either mock treated or treated with dsRNA for 2 h. Phosphorylated IRF3 (pIRF3) and total IRF3 levels were measured by immunoblotting. (B) Nuclear fractions (N.P) from HEK293-TLR3 cells stably expressing NSP1β either mock treated or treated with dsRNA were blotted for IRF3. DRBP76 and tubulin are markers for nuclear and cytoplasmic fractions, respectively. WCE, whole-cell extract. (C) IRF3 subcellular localization was determined by confocal microscopy in HEK-293TLR3 cells stably transfected with empty vector (vector) or NSP1β after dsRNA stimulation. For this purpose, cells were either treated or left untreated with 100 μg of dsRNA/ml for 2 h, followed by immunofluorescence for endogenous IRF3 (green) and NSP1β (red) with rabbit anti-IRF3 and mouse anti-HA antibody, as indicated at the top of the panel. Position of nucleus is indicated by DAPI (blue) staining in the merge image.
NSP1β inhibits SeV-induced IFN-β promoter activation.
To establish the innate immune evasion property of NSP1β in a physiological context, we tested SeV-mediated ISG56 induction in NSP1β-expressing cells. The innate immune response against SeV infection is primarily mediated by RIG-I (30). We tested SeV-mediated ISG56 induction in NSP1β-expressing cells. HEK293-TLR3 cells stably expressing NSP1β- or empty vector-transfected control cells were either mock infected or infected with SeV (40 HA unit/ml) for 8 h. As shown in Fig. 7A, the presence of NSP1β significantly inhibited SeV-mediated ISG56 induction compared to control cells. Finally, to replicate the effect of NSP1β on porcine IFN-β induction, we used the porcine monocytic cell line 3D4/31. This myelomonocytic cell line has been recently characterized to support PRRSV replication and infectious virus production upon transfection with PRRSV genomic RNA, but it lacks the necessary receptor in order to enable spread of virus from cell to cell (22). The 3D4/31 cells were transfected by electroporation with porcine IFN-β-luciferase reporter and NSP1 or NSP1β. SeV infection of vector-transfected cells showed induction of porcine IFN-β-luciferase activity. However, in the presence of NSP1 or NSP1β, porcine IFN-β-luciferase activity was inhibited in a dose-dependent manner (Fig. 7B). This result confirms our findings that NSP1β plays a crucial role in innate immune evasion function of PRRSV in a physiologically relevant environment.
FIG. 7.
NSP1 and NSP1β inhibit SeV-induced porcine IFN-β promoter activation and human ISG56 induction. (A) HEK293-TLR3 cells stably transfected with empty vector (vector) or NSP1β were either mock infected or infected with SeV (40 HA units/ml) for 8 h. Endogenous ISG56 levels were detected by immunoblotting. NSP1β protein was detected by anti-HA antibody. (B) Porcine monocytic cells (3D4/31 cell line) were cotransfected with increasing concentrations of NSP1/NSP1β expression plasmids or empty vector (vector), along with porcine IFN-β-luciferase plasmid (0.4 μg) and pRLTK (0.01 μg). At 40 h after the transfection cells were either mock infected or infected with 80 HA units of SeV/ml for 8 h and assayed for luciferase activity. Bars indicate the average fold induction of luciferase activities ± the SEM compared to uninfected vector control cells. The bottom panel shows immunoblots for the level of each NSP expression in respective transfection. The asterisk indicates detection of N-terminally FLAG-tagged NSP1α, the cleavage product of NSP1.
DISCUSSION
Viruses have developed a number of mechanisms to subvert or prevent IFN induction and response (7). In the case of PRRSV, both North American and European PRRSV strains have been shown to be poor inducers of IFN in vitro and in vivo. Infection of PAMs with the North American prototype PRRSV VR-2332 significantly reduced their ability to produce type I IFN (36, 37). Furthermore, the absence of IFN-α production in TGEV-infected swine alveolar macrophages, which had been previously infected with PRRSV, implicates an active inhibition of IFN production by PRRSV. In summary, all of the studies are consistent with the fact that PRRSV exerts an active suppression of type I IFN response. In agreement with these, we have shown here that PRRSV infection of monocyte-derived macrophages showed inhibition of IFN-α and IFN-β production both at the mRNA and the protein levels. Our data show that five PRRSV NSPs—the NSP1α, NSP1β, NSP2, NSP4, and NSP11—have roles in IFN antagonism and inhibit activation of the IFN-β promoter.
In the present study we used TLR3- and RIG-I-mediated dsRNA signaling pathways as model systems to understand the mechanism of innate immune evasion by PRRSV. NSP1α, NSP1β, and NSP11, when stably expressed in HEK293-TLR3 cells, were able to inhibit upregulation of endogenous ISG56 by dsRNA. Moreover, stable expression of NSP1β inhibited endogenous ISG56 induction by SeV. Investigation of IRF3 phosphorylation status after dsRNA stimulation showed that NSP1β inhibits IRF3 (Ser396) phosphorylation and subsequent nuclear translocation. Although the biochemical nature of this inhibition by PRRSV NSP1β remains to be characterized, there are several mechanisms by which different viruses have been shown to inhibit IRF3 activation. Certain paramyxoviral V proteins interact with IKKɛ/TBK1 and function as a competitive inhibitor for IRF3 phosphorylation (33). The W protein of Nipah virus specifically interacts with karyopherin α3 and α4 through its nuclear localization signal and prevents IRF3 translocation (47). It has been reported that NSP2 of EAV, the prototypic member of Arteriviridae family, antagonizes host innate immune response by deconjugating ubiquitin and ISG15 from their cellular substrates (15). Therefore, it is possible that NSP1β, a member of cysteine protease family that includes most deubiquitinases, might also use a similar strategy of deconjugating important ubiquitin modifications of IRF3 or RIG-I (6). All of these possible mechanisms of inhibition of dsRNA signaling by NSP1β are currently under investigation.
NSP1β and NSP11 expression also inhibited transcription from an NF-κB-responsive promoter, but NSP1α expression has no inhibitory effect. The suppressive role of NSP1β in NF-κB-mediated signaling was further evident from the low level of IL-8 induction in dsRNA-treated HEK293-TLR3 cells stably expressing NSP1β. This finding contradicts a previous report that found PRRSV infection of MARC-145 cells and alveolar macrophages resulting in the activation of the NF-κB pathway through the degradation of IκB (28). The disagreement might be due to the fact that the NF-κB activation in that study was assayed at a late phase of virus infection (24 to 48 h after virus infection). The activation of NF-κB is an immediate-early event that occurs within minutes after exposure to a stimulus and results in a strong transcriptional stimulation of several cytokines (21). Considering that the PRRSV replication cycle in macrophages is completed by 12 h, this effect may be a secondary effect of virus infection. Inhibition of NF-κB pathway by PRRSV NSPs, especially during the early infection phase, would help the virus not only to subvert the host innate immune response but also to produce enough progeny virus before being released by apoptosis as proposed by others (10).
PRRSV NSP1 contributes two accessory proteinases NSP1α and NSP1β for the processing of replicase polyprotein. NSP1α and NSP1β both possess two papainlike cysteine protease (PCP) domains, PCPα and PCPβ, which mediate the release of NSP1α and NSP1β from the polyproteins (12). Previous reverse genetics experiments showed that PCPα activity is essential for subgenomic mRNA synthesis but dispensable for genome replication. However, mutation of active-site residues of PCPβ resulted in no genome replication and a failure in virus recovery after electroporation of in vitro-transcribed RNA, underscoring its important role (26). Whether the NSP1β active site residues (Cys and His) are involved in its IFN antagonistic activity in the context of whole virus infection remains to be investigated. However, it may not be feasible to structurally separate proteolytic function of NSP1β from its IFN antagonistic function because of the general multifunctional nature of viral proteins. This multifunctional nature of NSP1 has been already established in EAV, and it has been found that the structural integrity of NSP1 is essential for transcription (54, 55). In such case, given the essential function of PRRSV NSP1β in virus viability, it may not be possible to recover by reverse genetics a replication-capable virus that lacks anti-IFN activity.
The inadequate immunological response to PRRSV infection is underscored by the fact that an infected animal exhibits prolonged viremia, persistent infection, and continuous virus shedding. The poor type I IFN production at the site of PRRSV infection (e.g., lungs) has long been considered the root cause behind defective or suboptimal initiation and elaboration of the antigen-specific adaptive immune response (1, 35). Influenza virus carrying truncated nonstructural 1 (NS1) protein, which counteracts the host type I IFN response, is highly attenuated in mice, swine, and chicken models (42, 50). Another important example of correlation between anti-IFN activity and virulence is Ebola virus VP35 protein, which inhibits IRF3 activation. Single amino acid mutation (R312A) of VP35 protein, which compromises its ability to inhibit IRF3 activation, renders the virus highly attenuated in mice (20). It is possible that the strong anti-IFN activity of the NSPs of PRRSV may have a direct effect in the paradoxical immune response observed in PRRSV infection and would explain the inability of the host to clear PRRSV. Such an important immunomodulatory effect of these NSPs may also be the basis for the pathogenesis and virulence of this virus. On the other hand, pestiviruses, which include classical swine fever virus (CSFV) and bovine viral diarrhea virus, have been shown to induce proteasomal degradation of IRF3 by the viral autoprotease Npro, which subsequently inhibits IFN-α/β production (4, 18). A recent investigation into the correlation between the ability of CSFV for IRF3 degradation and viral virulence found that impairment of the IFN antagonistic activity did not give rise to an attenuated virus (43). The authors of that study suggest that the inhibition of type I IFN may have a role in longer persistence of the virus in pigs. Previous results from our laboratory indicated that viral NSP3-8 might play a major role in PRRSV virulence (27). Taken together with our findings reported here, this suggests that the pathogenesis of PRRSV may be a complex summation of several major effectors contributing to the overall pathogenesis of PRRSV. The NSP1β acts as a suppressor of innate immune response, and NSP3-8 determines the virulence and invasive capacity, which helps the virus to translocate and cross placenta and infect fetuses. Highly pathogenic viruses such as SARS-CoV and Ebola viruses encode multiple proteins targeting different steps in IFN signaling. This not only ensures complete inhibition of the IFN response but also provides a fail-safe mechanism for the virus in case a single protein became dysfunctional. Our current findings that five NSPs of PRRSV are involved in IFN antagonism, though of variable ability, point toward PRRSV's pathogenic nature.
Acknowledgments
This research has been supported by a grant from the USDA NRICGP (project 2008-00903 USDA-NRICGP).
The primary cells obtained ex vivo for these experiments were collected according animal protocols reviewed and approved by the Institutional Animal Care Committee of the University of Nebraska-Lincoln under protocol IACUC no. 07-10-048C.
We thank Luwen Zhang (UNL) and Michael David (UCSD) for providing IRF3 expression plasmid and IRF3 antibody, respectively. We thank Terry Fangman for help in confocal microscopy. The invaluable help of Kazima Saira and Vu Hiep in the preparation and execution of some of these experiments is greatly appreciated.
Footnotes
Published ahead of print on 18 November 2009.
REFERENCES
- 1.Albina, E., C. Carrat, and B. Charley. 1998. Interferon-alpha response to swine arterivirus (PoAV), the porcine reproductive and respiratory syndrome virus. J. Interferon Cytokine Res. 18:485-490. [DOI] [PubMed] [Google Scholar]
- 2.Allende, R., W. W. Laegreid, G. F. Kutish, J. A. Galeota, R. W. Wills, and F. A. Osorio. 2000. Porcine reproductive and respiratory syndrome virus: description of persistence in individual pigs upon experimental infection. J. Virol. 74:10834-10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansari, I. H., B. Kwon, F. A. Osorio, and A. K. Pattnaik. 2006. Influence of N-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J. Virol. 80:3994-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauhofer, O., A. Summerfield, Y. Sakoda, J. D. Tratschin, M. A. Hofmann, and N. Ruggli. 2007. Classical swine fever virus Npro interacts with interferon regulatory factor 3 and induces its proteasomal degradation. J. Virol. 81:3087-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bautista, E. M., K. S. Faaberg, D. Mickelson, and E. D. McGruder. 2002. Functional properties of the predicted helicase of porcine reproductive and respiratory syndrome virus. Virology 298:258-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhoj, V. G., and Z. J. Chen. 2009. Ubiquitylation in innate and adaptive immunity. Nature 458:430-437. [DOI] [PubMed] [Google Scholar]
- 7.Bowie, A. G., and L. Unterholzner. 2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 8:911-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buddaert, W., K. Van Reeth, and M. Pensaert. 1998. In vivo and in vitro interferon (IFN) studies with the porcine reproductive and respiratory syndrome virus (PRRSV). Adv. Exp. Med. Biol. 440:461-467. [DOI] [PubMed] [Google Scholar]
- 9.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 10.Costers, S., D. J. Lefebvre, P. L. Delputte, and H. J. Nauwynck. 2008. Porcine reproductive and respiratory syndrome virus modulates apoptosis during replication in alveolar macrophages. Arch. Virol. 153:1453-1465. [DOI] [PubMed] [Google Scholar]
- 11.de Lima, M., B. Kwon, I. H. Ansari, A. K. Pattnaik, E. F. Flores, and F. A. Osorio. 2008. Development of a porcine reproductive and respiratory syndrome virus differentiable (DIVA) strain through deletion of specific immunodominant epitopes. Vaccine 26:3594-3600. [DOI] [PubMed] [Google Scholar]
- 12.den Boon, J. A., K. S. Faaberg, J. J. Meulenberg, A. L. Wassenaar, P. G. Plagemann, A. E. Gorbalenya, and E. J. Snijder. 1995. Processing and evolution of the N-terminal region of the arterivirus replicase ORF1a protein: identification of two papainlike cysteine proteases. J. Virol. 69:4500-4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng, W., S. M. Laster, M. Tompkins, T. Brown, J. S. Xu, C. Altier, W. Gomez, D. Benfield, and M. B. McCaw. 2001. In utero infection by porcine reproductive and respiratory syndrome virus is sufficient to increase susceptibility of piglets to challenge by Streptococcus suis type II. J. Virol. 75:4889-4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foy, E., K. Li, R. Sumpter, Jr., Y. M. Loo, C. L. Johnson, C. Wang, P. M. Fish, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc. Natl. Acad. Sci. U. S. A. 1 02:2986-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frias-Staheli, N., N. V. Giannakopoulos, M. Kikkert, S. L. Taylor, A. Bridgen, J. Paragas, J. A. Richt, R. R. Rowland, C. S. Schmaljohn, D. J. Lenschow, E. J. Snijder, A. Garcia-Sastre, and H. W. t. Virgin. 2007. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2:404-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Sastre, A., and C. A. Biron. 2006. Type 1 interferons and the virus-host relationship: a lesson in detente. Science 312:879-882. [DOI] [PubMed] [Google Scholar]
- 17.Genini, S., P. L. Delputte, R. Malinverni, M. Cecere, A. Stella, H. J. Nauwynck, and E. Giuffra. 2008. Genome-wide transcriptional response of primary alveolar macrophages following infection with porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 89:2550-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gil, L. H., I. H. Ansari, V. Vassilev, D. Liang, V. C. Lai, W. Zhong, Z. Hong, E. J. Dubovi, and R. O. Donis. 2006. The amino-terminal domain of bovine viral diarrhea virus Npro protein is necessary for alpha/beta interferon antagonism. J. Virol. 80:900-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo, J., K. L. Peters, and G. C. Sen. 2000. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology 267:209-219. [DOI] [PubMed] [Google Scholar]
- 20.Hartman, A. L., B. H. Bird, J. S. Towner, Z. A. Antoniadou, S. R. Zaki, and S. T. Nichol. 2008. Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of Ebola virus. J. Virol. 82:2699-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiscott, J., H. Kwon, and P. Genin. 2001. Hostile takeovers: viral appropriation of the NF-κB pathway. J. Clin. Invest. 107:143-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, Y. W., Y. Fang, and X. J. Meng. 2009. Identification and characterization of a porcine monocytic cell line supporting porcine reproductive and respiratory syndrome virus (PRRSV) replication and progeny virion production by using an improved DNA-launched PRRSV reverse genetics system. Virus Res. 145:1-8. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, Z., T. W. Mak, G. Sen, and X. Li. 2004. Toll-like receptor 3-mediated activation of NF-κB and IRF3 diverges at Toll-IL-1 receptor domain-containing adapter inducing IFN-beta. Proc. Natl. Acad. Sci. U. S. A. 101:3533-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 25.Kim, H. S., J. Kwang, I. J. Yoon, H. S. Joo, and M. L. Frey. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 133:477-483. [DOI] [PubMed] [Google Scholar]
- 26.Kroese, M. V., J. C. Zevenhoven-Dobbe, J. N. Bos-de Ruijter, B. P. Peeters, J. J. Meulenberg, L. A. Cornelissen, and E. J. Snijder. 2008. The nsp1alpha and nsp1 papain-like autoproteinases are essential for porcine reproductive and respiratory syndrome virus RNA synthesis. J. Gen. Virol. 89:494-499. [DOI] [PubMed] [Google Scholar]
- 27.Kwon, B., I. H. Ansari, A. K. Pattnaik, and F. A. Osorio. 2008. Identification of virulence determinants of porcine reproductive and respiratory syndrome virus through construction of chimeric clones. Virology 380:371-378. [DOI] [PubMed] [Google Scholar]
- 28.Lee, S. M., and S. B. Kleiboeker. 2005. Porcine arterivirus activates the NF-κB pathway through IκB degradation. Virology 342:47-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, S. M., S. K. Schommer, and S. B. Kleiboeker. 2004. Porcine reproductive and respiratory syndrome virus field isolates differ in in vitro interferon phenotypes. Vet. Immunol. Immunopathol. 102:217-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loo, Y. M., J. Fornek, N. Crochet, G. Bajwa, O. Perwitasari, L. Martinez-Sobrido, S. Akira, M. A. Gill, A. Garcia-Sastre, M. G. Katze, and M. Gale, Jr. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez, O. J., and F. A. Osorio. 2004. Role of neutralizing antibodies in PRRSV protective immunity. Vet. Immunol. Immunopathol. 102:155-163. [DOI] [PubMed] [Google Scholar]
- 32.Loving, C. L., S. L. Brockmeier, and R. E. Sacco. 2007. Differential type I interferon activation and susceptibility of dendritic cell populations to porcine arterivirus. Immunology 120:217-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, L. L., M. Puri, C. M. Horvath, and G. C. Sen. 2008. Select paramyxoviral V proteins inhibit IRF3 activation by acting as alternative substrates for inhibitor of κB kinase epsilon (IKKe)/TBK1. J. Biol. Chem. 283:14269-14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mateu, E., and I. Diaz. 2008. The challenge of PRRS immunology. Vet. J. 177:345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meier, W. A., J. Galeota, F. A. Osorio, R. J. Husmann, W. M. Schnitzlein, and F. A. Zuckermann. 2003. Gradual development of the interferon-gamma response of swine to porcine reproductive and respiratory syndrome virus infection or vaccination. Virology 309:18-31. [DOI] [PubMed] [Google Scholar]
- 36.Miller, L. C., W. W. Laegreid, J. L. Bono, C. G. Chitko-McKown, and J. M. Fox. 2004. Interferon type I response in porcine reproductive and respiratory syndrome virus-infected MARC-145 cells. Arch. Virol. 149:2453-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, L. C., K. M. Lager, and M. E. Kehrli, Jr. 2009. Role of Toll-like receptors in activation of porcine alveolar macrophages by porcine reproductive and respiratory syndrome virus. Clin. Vaccine Immunol. 16:360-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neumann, E. J., J. B. Kliebenstein, C. D. Johnson, J. W. Mabry, E. J. Bush, A. H. Seitzinger, A. L. Green, and J. J. Zimmerman. 2005. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J. Am. Vet. Med. Assoc. 227:385-392. [DOI] [PubMed] [Google Scholar]
- 39.Ostrowski, M., J. A. Galeota, A. M. Jar, K. B. Platt, F. A. Osorio, and O. J. Lopez. 2002. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. J. Virol. 76:4241-4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overend, C., R. Mitchell, D. He, G. Rompato, M. J. Grubman, and A. E. Garmendia. 2007. Recombinant swine beta interferon protects swine alveolar macrophages and MARC-145 cells from infection with porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 88:925-931. [DOI] [PubMed] [Google Scholar]
- 41.Posthuma, C. C., K. W. Pedersen, Z. Lu, R. G. Joosten, N. Roos, J. C. Zevenhoven-Dobbe, and E. J. Snijder. 2008. Formation of the arterivirus replication/transcription complex: a key role for nonstructural protein 3 in the remodeling of intracellular membranes. J. Virol. 82:4480-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richt, J. A., P. Lekcharoensuk, K. M. Lager, A. L. Vincent, C. M. Loiacono, B. H. Janke, W. H. Wu, K. J. Yoon, R. J. Webby, A. Solorzano, and A. Garcia-Sastre. 2006. Vaccination of pigs against swine influenza viruses by using an NS1-truncated modified live-virus vaccine. J. Virol. 80:11009-11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruggli, N., A. Summerfield, A. R. Fiebach, L. Guzylack-Piriou, O. Bauhofer, C. G. Lamm, S. Waltersperger, K. Matsuno, L. Liu, M. Gerber, K. H. Choi, M. A. Hofmann, Y. Sakoda, and J. D. Tratschin. 2009. Classical swine fever virus can remain virulent after specific elimination of the interferon regulatory factor 3-degrading function of Npro. J. Virol. 83:817-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saira, K., Y. Zhou, and C. Jones. 2007. The infected cell protein 0 encoded by bovine herpesvirus 1 (bICP0) induces degradation of interferon response factor 3 and, consequently, inhibits beta interferon promoter activity. J. Virol. 81:3077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarkar, S. N., K. L. Peters, C. P. Elco, S. Sakamoto, S. Pal, and G. C. Sen. 2004. Novel roles of TLR3 tyrosine phosphorylation and PI3 kinase in double-stranded RNA signaling. Nat. Struct. Mol. Biol. 11:1060-1067. [DOI] [PubMed] [Google Scholar]
- 46.Sarkar, S. N., and G. C. Sen. 2004. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol. Ther. 103:245-259. [DOI] [PubMed] [Google Scholar]
- 47.Shaw, M. L., W. B. Cardenas, D. Zamarin, P. Palese, and C. F. Basler. 2005. Nuclear localization of the Nipah virus W protein allows for inhibition of both virus- and Toll-like receptor 3-triggered signaling pathways. J. Virol. 79:6078-6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sizemore, N., N. Lerner, N. Dombrowski, H. Sakurai, and G. R. Stark. 2002. Distinct roles of the IκB kinase alpha and beta subunits in liberating nuclear factor κB (NF-κB) from IκB and in phosphorylating the p65 subunit of NF-κB. J. Biol. Chem. 277:3863-3869. [DOI] [PubMed] [Google Scholar]
- 49.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79(Pt. 5):961-979. [DOI] [PubMed] [Google Scholar]
- 50.Steel, J., A. C. Lowen, L. Pena, M. Angel, A. Solorzano, R. Albrecht, D. R. Perez, A. Garcia-Sastre, and P. Palese. 2009. Live attenuated influenza viruses containing NS1 truncations as vaccine candidates against H5N1 highly pathogenic avian influenza. J. Virol. 83:1742-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stordeur, P., L. F. Poulin, L. Craciun, L. Zhou, L. Schandene, A. de Lavareille, S. Goriely, and M. Goldman. 2002. Cytokine mRNA quantification by real-time PCR. J. Immunol. Methods 259:55-64. [DOI] [PubMed] [Google Scholar]
- 52.Sumpter, R., Jr., Y. M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tenoever, B. R., S. L. Ng, M. A. Chua, S. M. McWhirter, A. Garcia-Sastre, and T. Maniatis. 2007. Multiple functions of the IKK-related kinase IKKɛ in interferon-mediated antiviral immunity. Science 315:1274-1278. [DOI] [PubMed] [Google Scholar]
- 54.Tijms, M. A., D. D. Nedialkova, J. C. Zevenhoven-Dobbe, A. E. Gorbalenya, and E. J. Snijder. 2007. Arterivirus subgenomic mRNA synthesis and virion biogenesis depend on the multifunctional nsp1 autoprotease. J. Virol. 81:10496-10505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tijms, M. A., L. C. van Dinten, A. E. Gorbalenya, and E. J. Snijder. 2001. A zinc finger-containing papain-like protease couples subgenomic mRNA synthesis to genome translation in a positive-stranded RNA virus. Proc. Natl. Acad. Sci. U. S. A. 98:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Truong, H. M., Z. Lu, G. F. Kutish, J. Galeota, F. A. Osorio, and A. K. Pattnaik. 2004. A highly pathogenic porcine reproductive and respiratory syndrome virus generated from an infectious cDNA clone retains the in vivo virulence and transmissibility properties of the parental virus. Virology 325:308-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Reeth, K., G. Labarque, H. Nauwynck, and M. Pensaert. 1999. Differential production of proinflammatory cytokines in the pig lung during different respiratory virus infections: correlations with pathogenicity. Res. Vet. Sci. 67:47-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao, Z., L. Batista, S. Dee, P. Halbur, and M. P. Murtaugh. 2004. The level of virus-specific T-cell and macrophage recruitment in porcine reproductive and respiratory syndrome virus infection in pigs is independent of virus load. J. Virol. 78:5923-5933. [DOI] [PMC free article] [PubMed] [Google Scholar]