Abstract
Flavivirus NS1 is a nonstructural protein involved in virus replication and regulation of the innate immune response. Interestingly, a larger NS1-related protein, NS1′, is often detected during infection with the members of the Japanese encephalitis virus serogroup of flaviviruses. However, how NS1′ is made and what role it performs in the viral life cycle have not been determined. Here we provide experimental evidence that NS1′ is the product of a −1 ribosomal frameshift event that occurs at a conserved slippery heptanucleotide motif located near the beginning of the NS2A gene and is stimulated by a downstream RNA pseudoknot structure. Using site-directed mutagenesis of these sequence elements in an infectious clone of the Kunjin subtype of West Nile virus, we demonstrate that NS1′ plays a role in viral neuroinvasiveness.
Flaviviruses are positive-strand RNA viruses infecting more than 100 million people a year with the main representatives including West Nile virus (WNV), dengue virus (DENV), Japanese encephalitis virus (JEV), yellow fever virus (YFV), and tick-borne encephalitis virus (TBEV). The viruses from the Japanese encephalitis virus subgroup of this genus include JEV, Murray Valley encephalitis virus (MVEV), WNV, and St. Louis encephalitis virus (SLEV). The characteristic feature of these viruses is that they may cause neuroinvasive disease.
The flavivirus genome is ∼11 kb in length and contains a single long open reading frame that encodes three structural (C, prM, and E) and seven nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins (9, 14, 23). Flavivirus NS1 is a multifunctional protein shown to play a role in virus replication and assembly (13, 15, 18, 20), as well as in the modulation of the innate immune response (6, 7, 24). However, the mechanism(s) of its contribution to viral pathogenesis is not fully understood. Interestingly, NS1 appears in Western blots with anti-NS1 antibodies as at least two heterogeneous clusters of proteins of different molecular masses (NS1 and NS1′) (2, 3, 5, 19). It was suggested that the slower migration of NS1′ proteins may be due to either different glycosylation patterns or the generation of an alternative cleavage product with the cleavage site likely to reside within the downstream NS2A protein (3, 10, 15, 19). Recently, it was proposed—based on computational analysis of RNA sequence and structure of the members of the Japanese encephalitis virus subgroup—that the NS1′ protein is produced as the result of a −1 ribosomal frameshift (11). This analysis revealed the presence of conserved canonical frameshift-stimulating motifs—namely, a slippery heptanucleotide and a 3′-adjacent potential pseudoknot—near the beginning of the NS2A gene (1, 4, 11, 22) (Fig. 1A). However, experimental evidence demonstrating −1 ribosomal frameshifting for NS1′ production was lacking.
FIG. 1.
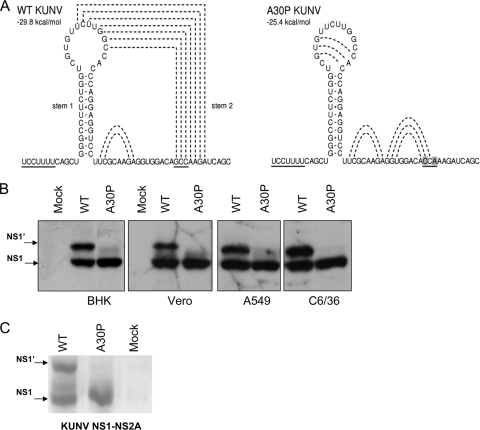
Disruption of the predicted pseudoknot structure by the alanine-to-proline mutation at position 30 of the NS2A gene abolishes NS1′ production. (A) The frameshift motif and pseudoknot structure predicted for WT and A30P KUNV using pknotsRG software (21). The frameshift heptanucleotide and codon 30 of the NS2A gene are underlined, and the proposed interactions between bases in the predicted pseudoknot are shown as dashed lines. Altered nucleotides in codon 30 of the A30P mutant are highlighted. (B) Detection of NS1 and NS1′ in lysates of BHK, Vero, A549, and C6/36 cells 18 h after infection with WT or A30P KUNV viruses at an MOI of 5. Infected cells were pulsed with 50 μCi of [35S]methionine in the methionine-free medium for 60 min, and then labeled medium was replaced with medium containing an excess of unlabeled methionine and cells were incubated for an additional 90 min. On completion of pulse-chase labeling, cells were lysed in 1% NP-40 lysis buffer and labeled NS1-containing proteins were immunoprecipitated with the anti-NS1 monoclonal antibody 4G4. Precipitated proteins were separated by SDS-PAGE and visualized on X-ray film. (C) Vero cells were transfected with DNA constructs coding for WT or A30P-mutated NS1-NS2A gene cassettes and incubated for 24 h prior to pulse-chase labeling and immunoprecipitation with 4G4 antibodies as in panel B.
We previously described an Ala-to-Pro mutation at amino acid position 30 in the NS2A gene of the Kunjin strain (KUNV) of West Nile virus that allows persistent noncytopathic replication of replicon RNA in several mammalian cell lines (16, 17). Introduction of this mutation into a KUNV infectious clone resulted in increased transcription of the beta interferon (IFN-β) promoter in response to virus infection, and the mutant virus exhibited significantly reduced neuroinvasiveness in mice (16, 17). However, the exact mechanism of how the A30P mutation in NS2A changed the properties of the mutant virus was not clear. Given the location of the mutation within the pseudoknot structure predicted to be involved in −1 ribosomal frameshifting (11), it was reasonable to assume that the mutation could abolish the formation of NS1′, which may be at least partially responsible for the attenuated phenotype of the mutant virus.
The A30P mutation in the NS2A gene abolishes production of NS1′.
RNA structure and sequence analysis showed that the A30P mutation in NS2A indeed disrupted the predicted frameshift-stimulating pseudoknot structure (Fig. 1A). To examine whether this pseudoknot-disrupting mutation affects generation of NS1′ and whether this effect is cell specific, we infected different cell lines (BHK21, Vero76, A549, and C6/36) with KUNV or mutant KUNV A30P and investigated the production of NS1 and NS1′ in cell lysates 18 h postinfection. Pulse-chase 35S-labeling experiments followed by immunoprecipitation with the NS1-specific monoclonal antibody 4G4 (8, 9) showed NS1 and NS1′ production in lysates of all cells infected with wild-type (WT) KUNV (Fig. 1B). However, only NS1 and no NS1′ was detected in lysates of cells infected with the A30P mutant of KUNV. Further, no NS1′ was detected in immunoprecipitates from lysates of Vero cells transfected with plasmid DNA encoding a KUNV NS1-NS2A gene cassette with the A30P mutation (Fig. 1C). The same results were obtained with an A30P expression cassette from MVEV (M. Lobigs, personal communication). Our results in virus-infected or plasmid DNA-transfected cells indicate that the putative pseudoknot structure in the NS2A gene is indeed required for the production of NS1′ and that the disruption of this RNA structure abolishes production of NS1′.
NS1′ contains peptides resulting from a −1 ribosomal frameshift.
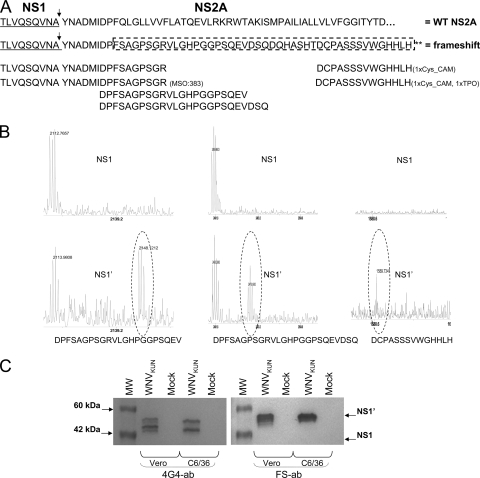
If the NS1′ protein produced during KUNV infection is the result of a ribosomal frameshift, then one should be able to detect, by mass spectrometry of digested NS1′ protein, the peptides corresponding to the putative −1 frameshift region and these peptides should not be present in the digested NS1 protein (Fig. 2A). To determine the presence/absence of such peptides, we infected BHK21 cells with KUNV (multiplicity of infection [MOI] = 5) and then NS1 and NS1′ proteins were immunoprecipitated with 4G4 antibodies at 24 h postinfection from cell lysates treated with NP-40 lysis buffer (1% NP-40, 50 mM Tris-HCl, 150 mM NaCl, 2 mM EDTA). The precipitated proteins were separated by electrophoresis in 10% polyacrylamide gels (polyacrylamide gel electrophoresis ); NS1 and NS1′ proteins were excised, destained with acetonitrile (50 mM), reduced with dithiothreitol (DTT; 10 mM) followed by alkylation with iodoacetamide (55 mM), and digested overnight with trypsin or AspN (80 ng). The resulting peptide mixes were analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry using a Voyager DE STR mass spectrometer. In the NS1′-derived digestion, several peptides matching the mass of peptides expected to arise as a result of the predicted −1 ribosomal frameshift were identified (11) (Fig. 2B). These peptides covered most of the frameshift region, and one of them (TLVQSQVNAYNADMIDPFSAGPSGR) showed the complete transition from the last 9 amino acids of NS1 (TLVQSQVNA) through the first 9 amino acids of NS2A (YNADMIDPF) and then the first 7 amino acids of the frameshift region (SAGPSGR) (Fig. 2A and B). Moreover, several peptides from the NS1′-derived digestion were mapped to the NS1 region (data not shown), demonstrating again that NS1′ consists of NS1 with an additional C-terminal extension. In the NS1-derived digestions none of the putative frameshift peptides were detected but several peptides mapping to the NS1 region were identified (Fig. 2B and data not shown).
FIG. 2.
Mass spectrometry analysis of NS1 and NS1′ proteins shows that NS1′ is the product of −1 ribosomal frameshifting. (A) Amino acid sequence of the C-terminal region of NS1 (underlined) and N-terminal region of NS2A with the potential frameshift sequence. Arrows indicate cleavage between the NS1 and NS2A proteins. Frameshift amino acids are enclosed in the dashed box, and the sequences of frameshift peptides detected by mass spectrometry are indicated after trypsin and AspN digestion. (B) Peaks of three of the frameshift peptides detected after AspN digestion of NS1′ (bottom panels) but not of NS1 (top panels). (C) Western blot detection with NS1′ frameshift peptide-specific (FS ab) and 4G4 antibodies. Lysates from Vero and C6/36 cells harvested at 3 and 5 days after WNV infection were separated by 10% PAGE, transferred onto nitrocellulose membranes, and incubated with 4G4 (NS1/NS1′ specific) or FS ab (NS1′ specific).
Additional compelling evidence of the presence of the frameshift region in NS1′ was obtained by using polyclonal antibodies (FS ab) raised against a peptide from the frameshift region (GHPGGPSQEVDSQD). Western blot analysis of WNV-infected cells with this antibody detected NS1′ but not NS1 (Fig. 2C). To our knowledge, this is the first clear experimental detection of efficient programmed −1 ribosomal frameshifting for any member of the Flaviviridae family.
NS1′ production requires a conserved heptanucleotide frameshift motif and a stable pseudoknot structure.
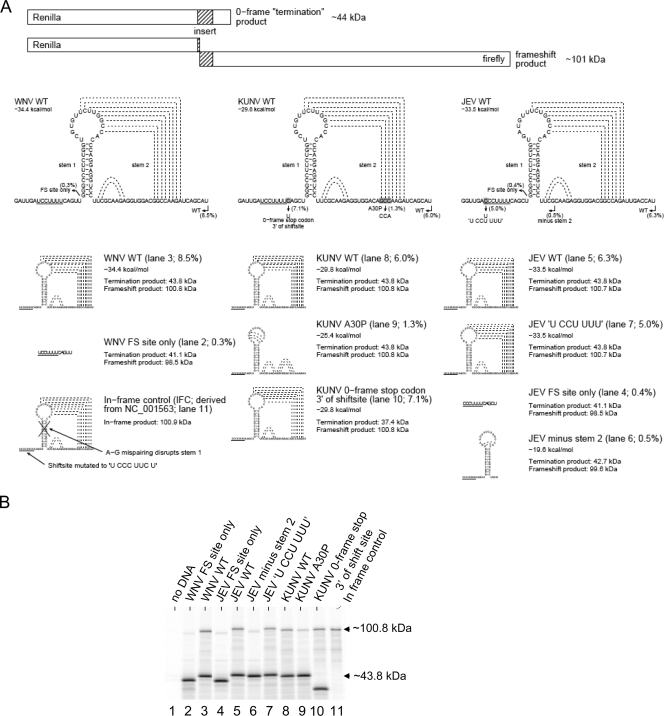
Next we elucidated the specific sequence requirements for NS1′ production using an in vitro dual-luciferase reporter system described previously (12). Briefly, the constructs contain sequences encoding two luciferase proteins (Renilla reniformis luciferase and firefly luciferase) separated by an insert containing the putative frameshift-inducing sequences or mutant versions. The second reporter-encoding sequence lacks an initiation codon and is in the −1 frame with respect to the first reporter sequence. Expression of the second reporter gene is possible only if the inserted sequence induces a −1 ribosomal frameshift (Fig. 3A). Therefore, frameshifting results in a larger (∼101-kDa) translation product while no frameshifting results in translation of just the first reporter gene, which results in the production of a smaller (∼44-kDa) product (Fig. 3A). We generated a series of constructs with different inserts containing wild-type or mutant frameshift cassettes from WNV, KUNV, and JEV. Inserts containing the slippery heptanucleotide and pseudoknot structure of WNV, KUNV, or JEV induced −1 ribosomal frameshifting in 6 to 8.5% of translation events (Fig. 3B). Wild-type levels of frameshifting in these in vitro experiments required the presence of both the slippery heptanucleotide and the pseudoknot sequences; insertion of only the slippery heptanucleotide motif from WNV or JEV induced frameshifting in just 0.3 to 0.4% of translation events (Fig. 3B), while partial deletion of the pseudoknot sequence (construct JEV minus stem 2; 0.5%) or introduction of the A30P mutation (construct KUNV A30P; 1.3%) also greatly diminished frameshifting. KUNV naturally has a −1 frame stop codon 5′ adjacent to the slippery heptanucleotide. When a zero-frame stop codon was introduced 3′ adjacent to the heptanucleotide by mutating CAG to UAG, the frameshifting efficiency was as high as WT, thus confirming (alongside the mass spectrometry results) that the slippery heptanucleotide is the site of frameshifting (Fig. 3B, construct KUNV 0-frame stop 3′ of shift site). The efficiency of frameshifting appeared to be lower in our dual luciferase in vitro system compared to results in vivo (compare Fig. 3B with Fig. 1B and C). In experiments performed using 35S-labeled virus-infected cells followed by immunoprecipitation with the 4G4 antibodies, the efficiency of frameshifting appeared to be higher (∼20 to 50%, depending on the cell line [Fig. 1B]). Such differences are a common feature of this type of analysis and may result from different salt concentrations, tRNA abundances, and other differences between the in vitro system and virus-infected cells. The lower frameshifting efficiency in vitro may also be due to the absence of additional flanking sequences outside the insert contained in the constructs.
FIG. 3.
Sequence requirements for −1 ribosomal frameshift in vitro. (A) Diagram of the dual-luciferase reporter system and predicted structures of the WT and mutant sequences used to evaluate the role of WNV, KUNV, and JEV viral sequences in −1 ribosomal frameshifting. Production of an ∼101-kDa product is observed if the inserted sequence induces frameshifting. Locations of different stimulatory elements and mutations are shown as underlined or highlighted sequences. Putative interactions between bases are shown as dashed lines. Frameshifting efficiencies calculated from the intensity of labeled bands in panel B, after normalization for the number of methionine residues in each product, are shown next to the lane numbers in parentheses. (B) SDS-PAGE of in vitro translation products of indicated constructs performed in reticulocyte lysates. The molecular masses of Renilla luciferase (no frameshift, ∼44 kDa) and the frameshift product (∼101 kDa; fusion of Renilla and firefly luciferases) are indicated.
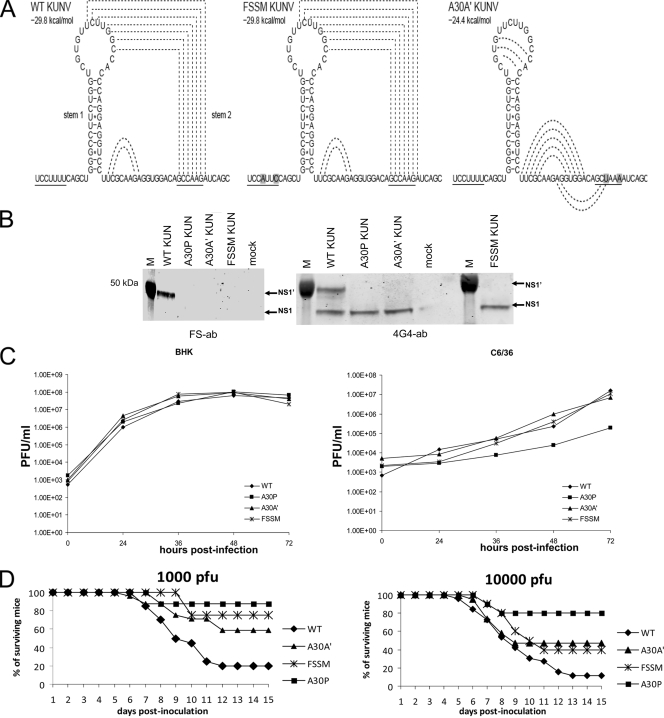
To confirm the role of the slippery heptanucleotide and pseudoknot-forming sequences in the generation of NS1′ during KUNV infection, two viral mutants were constructed: A30A′ (silent alanine) and FSSM (frameshift silent motif). A30A′ contains two silent mutations, GCC AAG to GCU AAA, in codon positions 30/31 of the NS2A coding sequence, and FSSM contains two silent mutations in the slippery heptanucleotide, from U CCU UUU to U CCA UUC (Fig. 4A). The introduced mutations do not change the amino acid sequence of the NS2A protein but, if our hypothesis is correct, should abolish or significantly diminish production of NS1′: A30A′ by eliminating stem 2 of the pseudoknot (similar to KUNV A30P) and FSSM by changing the slippery heptanucleotide to a nonslippery heptanucleotide (Fig. 4A). To examine the effect of these mutations on NS1′ production, BHK21 cells were infected with A30A′, FSSM, A30P, or WT KUNV. Two days after infection, cells were lysed in RIPA lysis buffer (10 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 1% sodium deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 1 mM phenylmethylsulfonyl fluoride) and analyzed by Western blotting for the presence/absence of NS1′. Antibodies (FS ab) specific for the frameshift peptide detected NS1′ in the lysates of WT KUNV-infected cells but not in cells infected with the mutant viruses A30A′, FSSM, and A30P (Fig. 4B). 4G4 antibodies detected NS1 in all samples and NS1′ only in the lysates from cells infected with WT KUNV (Fig. 4B). These results demonstrate that, in the context of KUNV viral infection, production of NS1′ requires the presence of both a slippery heptanucleotide and a 3′-adjacent pseudoknot-forming sequence. The presence or absence of NS1′ did not seem to play a role in virus replication in the examined cell lines; all mutant viruses accumulated to similar levels in BHK cells, and all but A30P virus replicated with similar efficiencies in C6/36 cells (Fig. 4C). It is likely that decreased efficiency of A30P virus replication in C6/36 cells is caused by the effect of amino acid mutation at this position on the properties of NS2A protein in virus-host/vector interactions (see below).
FIG. 4.
Sequence requirements for the production of NS1′ and its role in virus infection. (A) Predicted structure(s) of wild-type and mutated heptanucleotide or pseudoknot-forming sequences in KUNV. Altered nucleotides are highlighted, and stimulatory elements are underlined. Predicted interactions between bases in the pseudoknot structure are shown as dashed lines. (B) Western blot detection of NS1 and NS1′ with 4G4 or FS antibodies in lysates from Vero cells infected with WT or mutant KUNV viruses. Lysates from WT-, A30P-, A30A′-, or FSSM-infected cells were separated by PAGE and transferred onto nitrocellulose membranes followed by detection with 4G4 or FS antibodies as in Fig. 2C. (C) Kinetics of replication of WT, A30P, A30A′, and FSSM viruses in BHK and C6/36 cells. Cells were infected at an MOI of 1, and viral accumulation was measured up to 96 h after inoculation using plaque assays of harvested culture fluids. Growth kinetics from a typical experiment are shown. (D) Neuroinvasiveness of mutant viruses in mice. Swiss outbred weanling mice (18 to 19 days old) were inoculated with 1,000 or 10,000 PFU of WT KUNV or mutant viruses. Survival rates were recorded daily up to day 15 after inoculation, when the experiments were terminated. Shown are compiled results from multiple (up to 4) experiments involving WT KUNV (1,000 PFU, 20 mice; 10,000 PFU, 26 mice), KUNV A30P (1,000 PFU, 10 mice; 10,000 PFU, 10 mice), KUNV A30A′ (1,000 PFU, 24 mice; 10,000 PFU, 19 mice), and KUNV FSSM (1,000 PFU, 8 mice; 10,000 PFU, 10 mice).
Absence of NS1′ correlates with reduced neuroinvasiveness in mice.
Previous work by our group showed that the A30P mutation in the context of KUNV significantly attenuated virus neuroinvasiveness in mice (18). However, as shown above, the A30P mutation also disrupted pseudoknot formation, which abolished frameshifting and hence NS1′ production. Thus, the attenuated phenotype of the A30P mutant may be due to the amino acid change in the NS2A protein (from alanine to proline) or the absence of the NS1′ product or both. To dissect the roles of NS1′ and NS2A in viral neuroinvasiveness, weanling mice were injected intraperitoneally with A30A′, FSSM, A30P, and WT KUNV and the mice were monitored for signs of encephalitis (neuroinvasiveness) for 15 days after inoculation (Fig. 4D). As expected from previous studies, KUNV A30P was highly attenuated in comparison to WT KUNV. Most of the A30P virus-injected mice survived challenge with 1,000 or 10,000 PFU (80% and 87%, respectively; Fig. 4D). Fifty percent and 60% of mice infected with A30A′ virus survived challenge with 1,000 or 10,000 PFU, respectively, while survival rates of FSSM virus-inoculated mice were 40% and 75%, respectively (Fig. 4D). Only 11.5% and 20% of mice survived inoculation with 1,000 and 10,000 PFU, respectively, of WT KUNV. Thus, viruses containing silent mutations in the frameshift motif or pseudoknot structure showed reduced neuroinvasiveness, with the effect being intermediate between the WT and A30P viruses. Sequencing of viral progeny isolated from the brain of infected animals showed the retention of the introduced mutations (data not shown), demonstrating that mortality observed in mice inoculated with any of the frameshift mutants was not due to reversions to the WT sequence. Overall, these results demonstrate that NS1′ plays some role in the neuroinvasiveness of KUNV. It is unclear why NS1′ is consistently produced by members of the JE serogroup of flaviviruses that are generally associated with invasion and disease of the central nervous system (CNS) but not by other mosquito-borne flaviviruses such as DENV or YFV that produce disease in other organs and tissues of the body. Given almost exclusive use of avian species and Culex mosquitoes in the transmission cycles of the JEV group compared to a primate-Aedes cycle for the DENV and YFV groups, it is also possible that the NS1′ phenotype could have been evolutionarily selected to provide an advantage in virus replication/transmission in the mosquito-bird cycle. Whether this is indeed the case and whether neuroinvasiveness in mammals is only a coincidental effect remain to be determined.
Our previous studies showed that the A30P mutation resulted in increased transcription of IFN-β mRNA, which was suggested to be the main reason for virus attenuation (17). In the light of our current results with the silent A30A′ mutant, it could be argued that the highly attenuated phenotype of the A30P mutant is likely to be caused by two independent effects: (i) the absence of the NS1′ product as the result of the disrupted pseudoknot structure or (ii) the effect of the A30P mutation on the properties of the NS2A protein in inhibiting host antiviral response. Current studies are focused on dissecting further the roles of NS1′ and NS2A independently in evading the host antiviral response and determining the outcome of virus infection.
In conclusion, we have provided solid experimental evidence that NS1′ is produced as the result of −1 ribosomal frameshifting and showed that mutations abolishing NS1′ production lead to partial attenuation of viral neuroinvasiveness. We do not know, however, whether NS1 and NS1′ have different functions in the virus life cycle or whether NS1′ production is simply a viral strategy to produce NS1-like protein without requiring complete NS2A translation and cleavage. JEV group viruses may also use the ribosomal slippage strategy simply to alter the amounts of viral proteins, i.e., relatively more structural and NS1 proteins are produced as the result. Now that the identity and the mechanism of NS1′ production are determined, these and other important questions on the role of NS1′ in the virus life cycle can be addressed.
Acknowledgments
We are grateful to Trish Hitchcock and Ruth Lee for assistance in mouse experiments and to Mario Lobigs for sharing results with MVEV and for helpful discussions. We are also grateful to David Clark for helpful suggestions at the initial stage of the project.
The work was funded by grants 5U01AI066321 and GM079523 from the National Institutes of Health, USA, and grants from the National Health and Medical Research Council of Australia and from Science Foundation Ireland.
Footnotes
Published ahead of print on 11 November 2009.
REFERENCES
- 1.Bekaert, M., A. E. Firth, Y. Zhang, V. N. Gladyshev, J. F. Atkins, and P. V. Baranov. 25 September 2009. Recode-2: new design, new search tools, and many more genes. Nucleic Acids Res. [Epub ahead of print.] doi: 10.1093/nar/gkp788. [DOI] [PMC free article] [PubMed]
- 2.Blitvich, B. J., J. S. Mackenzie, R. J. Coelen, M. J. Howard, and R. A. Hall. 1995. A novel complex formed between the flavivirus E and NS1 proteins: analysis of its structure and function. Arch. Virol. 140:145-156. [DOI] [PubMed] [Google Scholar]
- 3.Blitvich, B. J., D. Scanlon, B. J. Shiell, J. S. Mackenzie, and R. A. Hall. 1999. Identification and analysis of truncated and elongated species of the flavivirus NS1 protein. Virus Res. 60:67-79. [DOI] [PubMed] [Google Scholar]
- 4.Brierley, I., A. J. Jenner, and S. C. Inglis. 1992. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 227:463-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, L. K., C. L. Liao, C. G. Lin, S. C. Lai, C. I. Liu, S. H. Ma, Y. Y. Huang, and Y. L. Lin. 1996. Persistence of Japanese encephalitis virus is associated with abnormal expression of the nonstructural protein NS1 in host cells. Virology 217:220-229. [DOI] [PubMed] [Google Scholar]
- 6.Chung, K. M., G. E. Nybakken, B. S. Thompson, M. J. Engle, A. Marri, D. H. Fremont, and M. S. Diamond. 2006. Antibodies against West Nile virus nonstructural protein NS1 prevent lethal infection through Fc gamma receptor-dependent and -independent mechanisms. J. Virol. 80:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung, K. M., B. S. Thompson, D. H. Fremont, and M. S. Diamond. 2007. Antibody recognition of cell surface-associated NS1 triggers Fc-gamma receptor-mediated phagocytosis and clearance of West Nile virus-infected cells. J. Virol. 81:9551-9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark, D. C., M. Lobigs, E. Lee, M. J. Howard, K. Clark, B. J. Blitvich, and R. A. Hall. 2007. In situ reactions of monoclonal antibodies with a viable mutant of Murray Valley encephalitis virus reveal an absence of dimeric NS1 protein. J. Gen. Virol. 88:1175-1183. [DOI] [PubMed] [Google Scholar]
- 9.Coia, G., M. D. Parker, G. Speight, M. E. Byrne, and E. G. Westaway. 1988. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J. Gen. Virol. 69:1-21. [DOI] [PubMed] [Google Scholar]
- 10.Falgout, B., R. Chanock, and C. J. Lai. 1989. Proper processing of dengue virus nonstructural glycoprotein NS1 requires the N-terminal hydrophobic signal sequence and the downstream nonstructural protein NS2a. J. Virol. 63:1852-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Firth, A. E., and J. F. Atkins. 2009. A conserved predicted pseudoknot in the NS2A-encoding sequence of West Nile and Japanese encephalitis flaviviruses suggests NS1′ may derive from ribosomal frameshifting. Virol. J. 6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grentzmann, G., J. A. Ingram, P. J. Kelly, R. F. Gesteland, and J. F. Atkins. 1998. A dual-luciferase reporter system for studying recoding signals. RNA 4:479-486. [PMC free article] [PubMed] [Google Scholar]
- 13.Khromykh, A. A., P. L. Sedlak, K. J. Guyatt, R. A. Hall, and E. G. Westaway. 1999. Efficient trans-complementation of the flavivirus Kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J. Virol. 73:10272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khromykh, A. A., and E. G. Westaway. 1994. Completion of Kunjin virus RNA sequence and recovery of an infectious RNA transcribed from stably cloned full-length cDNA. J. Virol. 68:4580-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindenbach, B. D., and C. M. Rice. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608-9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, W. J., H. B. Chen, X. J. Wang, H. Huang, and A. A. Khromykh. 2004. Analysis of adaptive mutations in Kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. J. Virol. 78:12225-12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, W. J., X. J. Wang, D. C. Clark, M. Lobigs, R. A. Hall, and A. A. Khromykh. 2006. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J. Virol. 80:2396-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackenzie, J. M., M. K. Jones, and P. R. Young. 1996. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 220:232-240. [DOI] [PubMed] [Google Scholar]
- 19.Mason, P. W. 1989. Maturation of Japanese encephalitis virus glycoproteins produced by infected mammalian and mosquito cells. Virology 169:354-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muylaert, I. R., T. J. Chambers, R. Galler, and C. M. Rice. 1996. Mutagenesis of the N-linked glycosylation sites of the yellow fever virus NS1 protein: effects on virus replication and mouse neurovirulence. Virology 222:159-168. [DOI] [PubMed] [Google Scholar]
- 21.Reeder, J., P. Steffen, and R. Giegerich. 2007. pknotsRG: RNA pseudoknot folding including near-optimal structures and sliding windows. Nucleic Acids Res. 35:W320-W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, A. L., H. M. Yang, K. A. Shen, and C. C. Wang. 1993. Giardiavirus double-stranded RNA genome encodes a capsid polypeptide and a gag-pol-like fusion protein by a translation frameshift. Proc. Natl. Acad. Sci. U. S. A. 90:8595-8599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westaway, E. G., J. M. Mackenzie, and A. A. Khromykh. 2003. Kunjin RNA replication and applications of Kunjin replicons. Adv. Virus Res. 59:99-140. [DOI] [PubMed] [Google Scholar]
- 24.Wilson, J. R., P. F. de Sessions, M. A. Leon, and F. Scholle. 2008. West Nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J. Virol. 82:8262-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]