Abstract
We demonstrate that a mutation-prone virus engineered to express a foreign gene is an expedient means for generating novel mutant nonviral proteins in mammalian cells. Using vesicular stomatitis virus to express a gene coding for a fluorescent DsRed protein, a number of green mutant variants including a new variant not previously described were rapidly isolated from infected cells, sequenced, and cloned. Similar methods may be useful in the development of physiologically sensitive fluorescent reporter proteins and directed evolution or mutagenesis of proteins in general.
Directed protein evolution often employs random mutagenesis to generate mutant sequences of genes of interest that can be expressed and screened to identify new proteins with altered characteristics. Early random mutagenesis strategies using radiation and chemical mutagens such as ethyl methanesulfonate have today been replaced by the use of error-prone DNA polymerases (17). The development of new fluorescent proteins (20) has made use of error-prone PCR (4) to generate mutant variants that can be screened by examining plates of Escherichia coli colonies under fluorescent illumination (3, 16) or by fluorescence-activated cell sorting (2, 18). Whereas these procedures have proven effective, they can also be labor-intensive, with plate-based screening at times requiring the examination of hundreds of plates and over 100,000 E. coli colonies for the identification of a single fluorescently novel variant (3). An additional drawback is that bacterium-based screening of mutants is of limited use in the development of fluorescent reporter proteins that alter their fluorescence in response to changing physiological conditions within a cell. For these, a mammalian cell-based method to screen for fluorescent mutants would be beneficial. Here we report the use of a recombinant vesicular stomatitis virus (VSV) to generate mutated nonviral genes coding for new proteins. We focused on a gene coding for wild-type DsRed that, due to spontaneous viral mutations, generated mutant green fluorescent variants that were easily visualized and isolated from infected cultures of mammalian cells.
Unlike most eukaryotic and prokaryotic cells, which have very low mutation rates, some viruses, particularly RNA viruses, have high mutation rates due to a lack of proofreading activity of the RNA polymerase. One such virus is VSV, a single-stranded, negative-sense RNA virus of the Rhabdoviridae family with a wild-type genome approximately 11.2 kb in length that can be engineered to accept an additional 4.5-kb gene insert (21). Genomic replication of VSV is error prone, with approximately 1 nucleotide mutation per viral genome per replication cycle (7). Using recombinant versions of VSV with a gene insert coding for a fluorescent protein, one can perform a plaque assay to examine fluorescent plaques formed by VSV-1′DsRed (21; also called VSV-p1-RFP [24]), a recombinant VSV that expresses a codon-optimized DsRed gene (DsRed1) inserted at the first position (1′) of the viral genome (Fig. 1A).
FIG. 1.
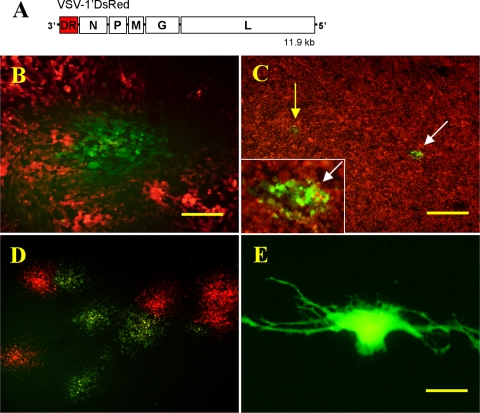
VSV mutagenesis of DsRed1. (A) Diagram of the recombinant VSV-1′DsRed RNA genome showing position of the DsRed1 gene (DR) and N, P, M, G, and L viral genes. (B) Example of mutant green fluorescent VSV-1′DsRed plaque amid a field of red fluorescent plaques grown on BHK-21 cells. Bar, 200 μm. (C) A 35-mm culture well of BHK-21 cells inoculated with 100,000 PFU of VSV-1′DsRed. Arrows highlight the green fluorescent plaques that were still identifiable despite the high number of PFU used to inoculate the culture. Inset is magnification of plaque (white arrow). (D) A mixture of red and green fluorescent plaques that developed from an agarose punch extract of a single green plaque that arose in a culture infected with 25,000 PFU. Bar, 500 μm (C and D). (E) A U-373 human glioblastoma cell transfected with green mutant variant gene DsRed-F91L. Bar, 25 μm.
Fluorescent plaque analysis.
To investigate the possibility that viral mutagenesis might lead to functional mutations of foreign nonviral genes, we examined over 80 BHK-21 cell cultures infected with VSV-1′DsRed containing hundreds of thousands of red fluorescent plaques and consistently found a small number of green fluorescent plaques (Fig. 1B) 24 h postinoculation, suggesting the presence of mutant DsRed genes. Recombinant VSV-1′DsRed virus (21, 24) expressing the DsRed1 gene (Clontech, Palo Alto, CA) in the first gene position was used (from J. Rose and I. Simon, Yale University). Prior to screening for fluorescent variants, plaque-purified viral stocks were prepared using single viral plaques to inoculate BHK-21 cells and virus was harvested 24 to 30 h later. All cells were propagated using minimum essential medium (MEM) growth medium (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin solution (Gibco) and maintained at 37°C with 5% CO2. Microscopic fluorescent screening and imaging of the DsRed plaques, which normally fluoresced red, were performed using an Olympus IX71 inverted microscope fitted with DsRed and enhanced green fluorescent protein (EGFP) filter sets. Quantification of the green variant plaques found in a series of duplicate 35-mm culture wells infected with 3 × 103, 10 × 103, 30 × 103, and 100 × 103 PFU per well resulted in the following numbers of green plaques: 3 × 103 PFU = 0 and 0, 10 × 103 PFU = 0 and 1, 30 × 103 PFU = 2 and 3, and 100 × 103 PFU = 10 and 12, respectively. Thus, we find a mean of 1 green variant plaque per 10,700 red ones. Individual green fluorescent plaques were identifiable against a background of red fluorescent plaques, even at high concentrations of virus, up to 100,000 PFU/well (Fig. 1C).
Isolation and sequencing of fluorescent mutants.
As a proof-of-principle test, we used a single 6-well culture dish infected with 10,000 to 25,000 PFU/well VSV-1′DsRed, and from the thousands of red plaques, we harvested six green mutant plaques by using a pipette to punch through the overlay of semisolid agarose. It took only about 5 min to identify the six green plaques. Since an “agarose punch” may capture not only the intended green infected cells but also some of the red, replating the extract yielded a mixture of red and green plaques (Fig. 1D) that were spatially isolated from one another, allowing individual plaques to be extracted in pure form. Six green and two red control single plaque extracts were then grown on BHK-21 cells without an agarose overlay and harvested to yield genomic viral RNA template suitable for reverse transcription-PCR (RT-PCR).
VSV genomic RNA was isolated using the QIAamp viral RNA minikit (Qiagen, Valencia, CA). Reverse transcription of VSV genomic RNA was performed using the Superscript III RT kit (Invitrogen, Carlsbad, CA) at a reaction temperature of 53°C and an RT oligonucleotide with the sequence 5′-ACG AAG ACA AAC AAA CCA TTA TTA TC-3′, designed to anneal to the initial 26 bases of the 3′ end of the VSV genome. Genomic VSV cDNA was then used as template in a PCR utilizing primers flanking the inserted fluorescent protein gene. PCR was performed using the RT oligonucleotide above as the forward primer and a reverse primer designed to anneal within the N-terminal region of the N protein gene, the sequence of which was 5′-GGA ACT ACG ACT GTG TTG TC-3′. Sequencing was performed in both the sense and antisense directions for each product.
Sequencing of the DsRed gene from each of the six green variants revealed a single nonsynonymous amino acid-altering mutation at the positions N42H, K83E, F91L, Y120H, and F124L. In contrast, no mutations were found in either of the two red control isolates. The DsRed gene used here (DsRed1) is a mammalian codon-optimized version of DsRed that produces a protein identical to the initially identified wild-type protein except for a valine that has been inserted after the start codon (3). Consistent with the literature, DsRed mutations are numbered in accordance with the original (non-valine-inserted) wild-type protein sequence. Two of the green isolates contained the same mutation (K83E), and one isolate (N42H) also carried an additional silent mutation. Four of these mutations (N42H [3], K83E [1], Y120H [1], and F124L [5]) have been previously described. The F91L mutant appears to be a previously unreported mutant of wild-type DsRed. Loss-of-function mutations were also detected, as indicated by the presence of nonfluorescent plaques. These loss-of-function variants could be due to a mutation in the fluorescent protein gene or mutation in the upstream transcription regulation sequences or a frameshift (14). Thus, we successfully identified 5 different functional mutations in a single dish. This demonstrates a remarkable saving of time and effort in generating and detecting novel mutations compared with other approaches.
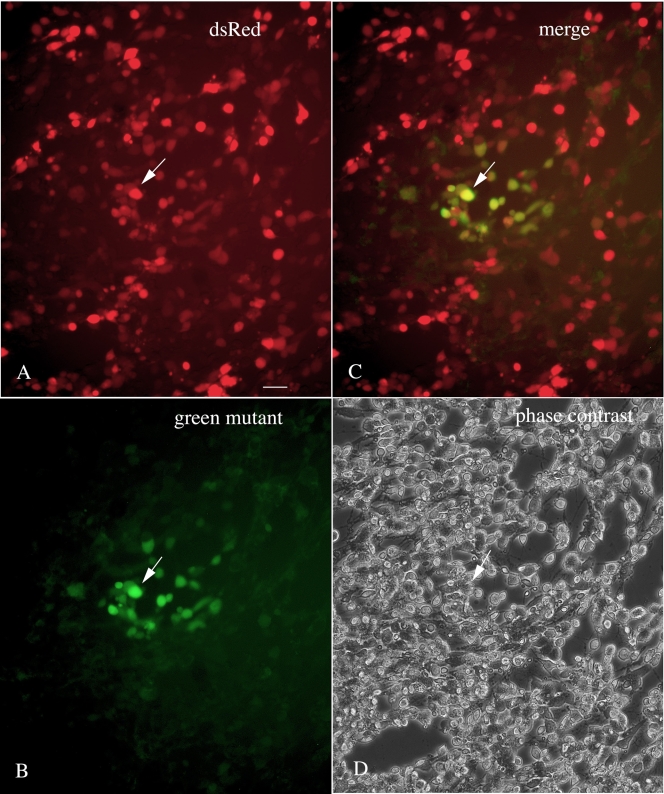
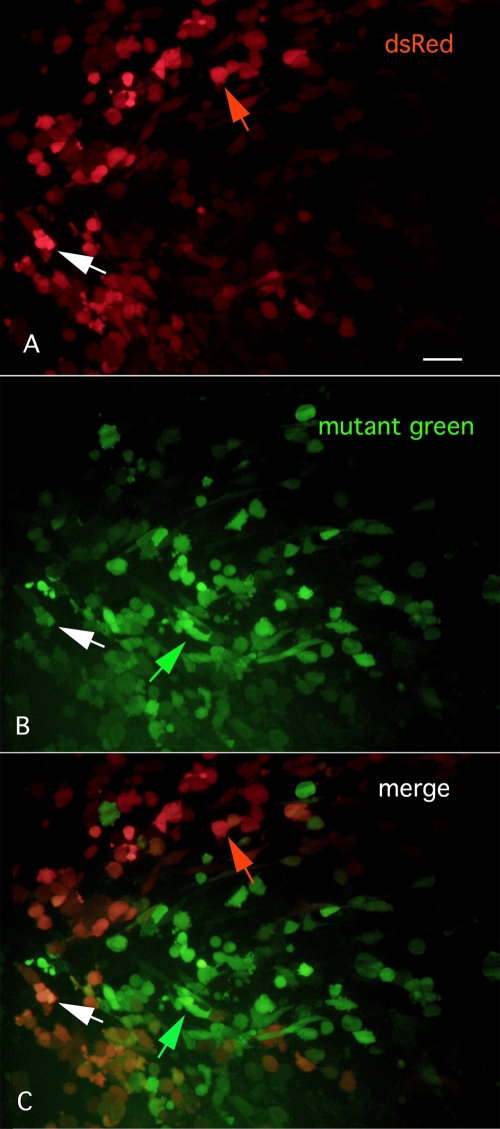
Even in the absence of an agarose overlay of the culture, one can identify and harvest VSVs that generate mutant fluorescent proteins. For instance, in Fig. 2A to D, cultures of BHK cells were infected with VSV-1′DsRed, and the following day cells or groups of cells could be found showing a green fluorescence in a background of red fluorescent cells. This approach would enable a potentially greater number of mutants to be detected per culture, as even a single cell showing a variant fluorescence could be detected and harvested. Interestingly, in some cases we find that single cells show both red and green color. This could be due to expression of fluorescent proteins encoded by two viruses, the parent virus expressing DsRed and a progeny mutant virus expressing a green-shifted reporter. Alternately, it could be due to a shift in color with protein maturation (1, 3) or to a reporter gene mutation that fluoresces red and green. Another permutation of expression is shown in Fig. 3A to C, where some infected cells show selectively either the parental red fluorescence (Fig. 3A) with little green (Fig. 3C) or a mutant green fluorescence (Fig. 3B) with little red and little overlap in the two colors in many cells (Fig. 3C). That some cells do show both green and red colors (Fig. 3A to C) is consistent with infection of primarily one virus type in single-color cells and multiple virus types in multiple-color cells.
FIG. 2.
VSV-1′DsRed was used to infect a culture of BHK-21 cells with no agarose overlay. Twenty-four hours later, among a population of infected red fluorescent cells (under green light excitation) (A), infected cells that also showed a green color (under blue light excitation) were found (B). Many cells showed both red and green color (see one such cell at the arrow in all four panels) (C). A phase-contrast image of the same field is shown in panel D. Bar, 30 μm.
FIG. 3.
Selective expression of green or red fluorescence. (A) With 535-nm excitation, infected cells show a red fluorescence. Bar, 30 μm. (B) In the same field, other cells show little red fluorescence but show selective green fluorescence with 488-nm excitation. (C) The merged image combining panels A and B above shows red or green cells.
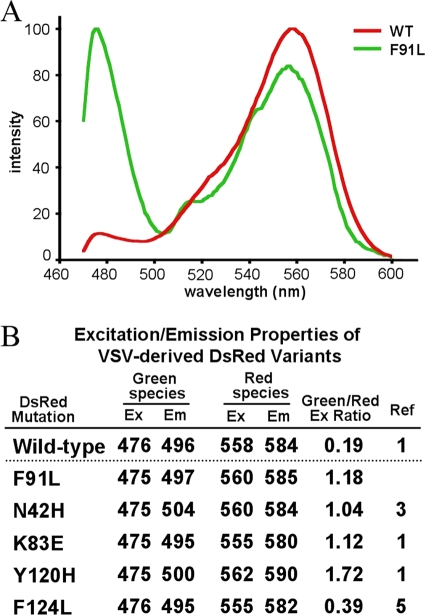
Using a TCS SP5 X Leica white-light laser confocal microscope, we collected excitation and emission spectral scans of plaques from each of the green DsRed variants and a DsRed control (Fig. 4A and B). For these measurements, BHK-21 cells were grown on glass-bottomed 35-mm culture dishes (MatTek Corp, Ashland, MA) and inoculated with virus. Excitation scan values were collected at 1-nm increments, and emission values were collected at 5-nm increments using the respective excitation peak wavelengths. Intensity values were normalized across the spectral range scanned. Overall, the fluorescent characteristics of each of the previously identified variants were consistent with reported values (Fig. 3B), demonstrating that informative measurements of fluorescence can be made from viral plaques in vitro.
FIG. 4.
(A) Excitation scans of VSV plaques expressing F91L (green) and wild-type (WT; red) DsRed. Intensity values are normalized across the spectral range. (B) Maximum excitation (Ex) and emission (Em) (nm) from the mutant plaques that we isolated. The ratios (column 5) are the green excitation peaks divided by the red excitation peaks.
Cloning and transfecting the novel F91L DsRed variant into U-373 glioma cells.
To demonstrate that the previously unreported F91L DsRed variant could be recapitulated in the absence of virus, we isolated and cloned the F91L DsRed gene into a plasmid expression vector and used this to transfect human U-373 glioma cells (Fig. 1E). Genomic VSV cDNA prepared from the F91L variant of VSV-1′DsRed was used as template in a PCR using the Phusion high-fidelity PCR kit (New England Biolabs, Ipswich, MA). Purified product was inserted into the plasmid pCR-Blunt II-TOPO (Invitrogen), and six insert-positive plasmids were then sequenced to confirm that no mutations had been introduced during the PCR amplification. All plasmids matched the consensus DsRed-F91L sequence, and one was selected for double digestion using AgeI and EcoRI and gel extracted to yield a DsRed-F91L insert. The pDsRed-Monomer-C1 plasmid (Clontech) driven by a cytomegalovirus promoter was digested using the same enzymes (thus removing the DsRed-monomer sequence), and the remaining plasmid backbone was gel extracted and ligated with the DsRed-F91L insert. The resulting plasmid (pDsRed-F91L) was sequenced to confirm the proper incorporation of insert using primers flanking the insertion site. After sequencing, pDsRed-F91L was used to transiently transfect U-373 glioblastoma cells using Lipofectamine 2000 (Invitrogen). After 48 h of incubation to allow time for fluorescent protein expression, spectral scans of individual transfected glioblastoma cells returned maximum excitation and emission wavelength values (green species: excitation, 475 nm, and emission, 491 nm; red species: excitation, 560 nm, and emission, 590 nm) comparable to those obtained from F91L DsRed viral plaques.
Conclusions.
The use of VSV to produce multiple variants of DsRed demonstrates that a mutation-prone virus can be used to generate randomly mutated fluorescent proteins amenable to fluorescent screening in mammalian cells. This is of particular interest given that similar methods may be useful to the further development of physiologically sensitive fluorescent reporter proteins (20) such as those designed to detect Ca2+ (25), glutamate (8), or cellular kinase activity (19). Multiple mutations in the gene of interest can easily be obtained simply by allowing further generations of progeny virus to infect additional cells at a rate of 4 successive rounds of replication/day. The utility of this approach is further enhanced by the fact that different viral genomes in a single cell, leading to different colors, can be detected, and the mutations can be harvested. Similar to VSV, other RNA viruses with high mutation rates such as Sindbis virus (10, 22), measles virus (12, 13), or other viruses could be used in place of VSV. For us, VSV makes an attractive choice for random mutagenesis, given its relative safety in the lab, high rate of mutation (7), routine recombinant engineering (6, 9, 15), the wide spectrum of animal cells that it can infect (11, 21, 23), and the rapid (1 to 3 h) and robust expression of reporter genes (21).
Acknowledgments
We thank J. Rose and I. Simon for recombinant VSVs, G. Wollmann for suggestions, and J. Boyd and Leica Microsystems Inc. for use of a Leica TCS SP5 X white-light laser confocal microscope.
The work was supported by NIH grants A1/NS48854 and CA124737.
Footnotes
Published ahead of print on 11 November 2009.
REFERENCES
- 1.Baird, G. S., D. A. Zacharias, and R. Y. Tsien. 2000. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl. Acad. Sci. U. S. A. 97:11984-11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bessette, P. H., and P. S. Daugherty. 2004. Flow cytometric screening of cDNA expression libraries for fluorescent proteins. Biotechnol. Prog. 20:963-967. [DOI] [PubMed] [Google Scholar]
- 3.Bevis, B. J., and B. S. Glick. 2002. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 20:83-87. [DOI] [PubMed] [Google Scholar]
- 4.Cadwell, R. C., and G. F. Joyce. 1992. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2:28-33. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalton, K. P., and J. K. Rose. 2001. Vesicular stomatitis virus glycoprotein containing the entire green fluorescent protein on its cytoplasmic domain is incorporated efficiently into virus particles. Virology 279:414-421. [DOI] [PubMed] [Google Scholar]
- 7.Drake, J. W., and J. J. Holland. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 96:13910-13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hires, S. A., Y. Zhu, and R. Y. Tsien. 2008. Optical measurements of synaptic glutamate spillover and reuptake by linker optimized glutamate-sensitive fluorescent reporters. Proc. Natl. Acad. Sci. U. S. A. 105:4411-4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawson, N. D., E. A. Stillman, M. A. Whitt, and J. K. Rose. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. U. S. A. 92:4477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu, X., and J. Silver. 2001. Transmission of replication-defective Sindbis helper vectors encoding capsid and envelope proteins. J. Virol. Methods 91:59-65. [DOI] [PubMed] [Google Scholar]
- 11.Lyles, D. S., and C. E. Rupprecht. 2007. Rhabdoviridae, p. 1363-1408. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 12.Msaouel, P., I. D. Iankov, C. Allen, J. C. Morris, V. von Messling, R. Cattaneo, M. Koutsilieris, S. J. Russell, and E. Galanis. 2009. Engineered measles virus as a novel oncolytic therapy against prostate cancer. Prostate 69:82-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paraskevakou, G., C. Allen, T. Nakamura, P. Zollman, C. D. James, K. W. Peng, M. Schroeder, S. J. Russell, and E. Galanis. 2007. Epidermal growth factor receptor (EGFR)-retargeted measles virus strains effectively target EGFR- or EGFRvIII expressing gliomas. Mol. Ther. 15:677-686. [DOI] [PubMed] [Google Scholar]
- 14.Quinones-Kochs, M. I., M. J. Schnell, L. Buonocore, and J. K. Rose. 2001. Mechanisms of loss of foreign gene expression in recombinant vesicular stomatitis viruses. Virology 287:427-435. [DOI] [PubMed] [Google Scholar]
- 15.Ramsburg, E., J. Publicover, L. Buonocore, A. Poholek, M. Robek, A. Palin, and J. K. Rose. 2005. A vesicular stomatitis virus recombinant expressing granulocyte-macrophage colony-stimulating factor induces enhanced T-cell responses and is highly attenuated for replication in animals. J. Virol. 79:15043-15053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaner, N. C., M. Z. Lin, M. R. McKeown, P. A. Steinbach, K. L. Hazelwood, M. W. Davidson, and R. Y. Tsien. 2008. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5:545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shivange, A. V., J. Marienhagen, H. Mundhada, A. Schenk, and U. Schwaneberg. 2009. Advances in generating functional diversity for directed protein evolution. Curr. Opin. Chem. Biol. 13:19-25. [DOI] [PubMed] [Google Scholar]
- 18.Subach, O. M., I. S. Gundorov, M. Yoshimura, F. V. Subach, J. Zhang, D. Gruenwald, E. A. Souslova, D. M. Chudakov, and V. V. Verkhusha. 2008. Conversion of red fluorescent protein into a bright blue probe. Chem. Biol. 15:1116-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ting, A. Y., K. H. Kain, R. L. Klemke, and R. Y. Tsien. 2001. Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc. Natl. Acad. Sci. U. S. A. 98:15003-15008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsien, R. Y. 2005. Building and breeding molecules to spy on cells and tumors. FEBS Lett. 579:927-932. [DOI] [PubMed] [Google Scholar]
- 21.van den Pol, A. N., K. Ozduman, G. Wollmann, W. S. Ho, I. Simon, Y. Yao, J. K. Rose, and P. Ghosh. 2009. Viral strategies for studying the brain, including a replication-restricted self-amplifying delta-G vesicular stomatitis virus that rapidly expresses transgenes in brain and can generate a multicolor golgi-like expression. J. Comp. Neurol. 516:456-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wahlfors, J. J., S. A. Zullo, S. Loimas, D. M. Nelson, and R. A. Morgan. 2000. Evaluation of recombinant alphaviruses as vectors in gene therapy. Gene Ther. 7:472-480. [DOI] [PubMed] [Google Scholar]
- 23.Wilkins, C., R. Dishongh, S. C. Moore, M. A. Whitt, M. Chow, and K. Machaca. 2005. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature 436:1044-1047. [DOI] [PubMed] [Google Scholar]
- 24.Wollmann, G., V. Rogulin, I. Simon, J. K. Rose, and A. N. van den Pol. 2010. Some attenuated variants of vesicular stomatitis virus show enhanced oncolytic activity against human glioblastoma cells relative to normal brain cells. J. Virol. 84:1563-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu, D., G. S. Baird, R. Y. Tsien, and R. L. Davis. 2003. Detection of calcium transients in Drosophila mushroom body neurons with camgaroo reporters. J. Neurosci. 23:64-72. [DOI] [PMC free article] [PubMed] [Google Scholar]