Abstract
For Kaposi's sarcoma-associated herpesvirus (KSHV; also called human herpesvirus 8 [HHV8]), the switch from latency to active lytic replication requires RTA, the product of open reading frame 50 (ORF50). RTA activates transcription from nearly 40 early and delayed-early viral promoters, mainly through interactions with cellular DNA binding proteins, such as CSL/RBP-Jκ, Oct-1, C/EBPα, and c-Jun. Reliance on cellular coregulators may allow KSHV to adjust its lytic program to suit different cellular contexts or interpret signals from the outside. CSL is a key component of the Notch signaling pathway and is targeted by several viruses. A search with known CSL binding sequences from cellular genes found at least 260 matches in the KSHV genome, many from regions containing known or suspected lytic promoters. Analysis of clustered sites located immediately upstream of ORF70 (thymidylate synthase), ORF19 (tegument protein), and ORF47 (glycoprotein L) uncovered RTA-responsive promoters that were validated using mRNAs isolated from KSHV-infected cells undergoing lytic reactivation. Notably, ORF19 behaves as a true late gene, indicating that RTA regulates all three phases of the lytic program. For each new promoter, the response to RTA was dependent on CSL, and 5 of the 10 candidate sites were shown to bind CSL in vitro. Analysis of individual sites highlighted the importance of a cytosine residue flanking the core CSL binding sequence. These findings broaden the role for CSL in coordinating the KSHV lytic gene expression program and help to define a signature motif for functional CSL sites within the viral genome.
Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and variant multicentric Castleman's disease are life-threatening malignancies associated with lowered cell-mediated immunity, such as in patients with AIDS (reviewed in references 15, 23, and 24). For all three tumor types, the primary etiological agent is Kaposi's sarcoma-associated herpesvirus (KSHV; also termed human herpesvirus 8 [HHV8]), a member of the γ2-herpesvirus (rhadinovirus) family. Like other herpesviruses, KSHV alternates between states of latency (temperate or semiquiescent infection) and lytic (productive) replication. The KS tumor mass is composed of elongated spindle cells that are derived from blood vessel endothelia and invariably carry latent KSHV. A fraction of infected cells will enter into lytic replication (reactivate), producing infectious virions and virus-encoded paracrine signaling factors. Reactivation is important for the growth and dissemination of the tumor, as revealed by clinical studies of ganciclovir, a potent inhibitor of lytic but not latent replication (15). What determines this low level of reactivation is not understood, but the prospect of exploiting the lytic cycle as a therapeutic target provides incentive for deeper studies of the interplay between viral lytic regulators and the host cell.
There is now ample evidence that the viral transcription factor RTA (replication and transcription activator), encoded by open reading frame 50 (ORF50), initiates the switch from latency to lytic replication (11). Ectopic RTA is sufficient to induce the lytic program in at least a proportion of latently infected PEL cells, and reactivation can be blocked using dominant-negative versions of RTA or ribozymes directed against ORF50 transcripts (18, 39, 40, 55). Recombinant viruses that lack RTA establish latency at a significant frequency but are unable to reactivate (71). During reactivation, ORF50 is expressed with immediate-early (IE) kinetics, and its promoter responds to a wide spectrum of inducing signals (55, 67, 74). The 120-kDa (691 amino acids) RTA protein localizes to the nucleus and contributes to lytic replication through at least three mechanisms: direct transcriptional activation of viral early and late genes, assembly of DNA replication complexes at lytic origins, and inhibition of antiviral defenses (11, 54, 62, 75). The N terminus contains a DNA binding domain, and a strong activation domain is located in the C terminus (reviewed in reference 64). Counterparts of RTA exist in other gammaherpesviruses, including herpesvirus saimiri (HVS), rhesus rhadinovirus (RRV), murine herpesvirus 68 (MHV68), and Epstein-Barr Virus (EBV) (10).
To date, upwards of 40 RTA-responsive elements (RREs) have been identified upstream of genes showing IE, early (E), or delayed-early (DE) kinetics (8, 13). RTA is recruited to RREs through two principal mechanisms. The simplest is by direct DNA binding, as exemplified by the core PAN and proximal K12 promoters, which share a 20-bp RTA recognition sequence (6, 52, 53). The second mechanism involves protein-protein interactions with cellular DNA binding proteins, including CSL/RBP-Jκ (33), C/EBPα (60), Oct-1 (50), and c-Jun (59). In the presence of a cellular coregulator, the DNA contacts made by RTA appear to be relatively sequence independent, although the presence of A/T-rich triplets arranged at 10-bp intervals may further stabilize the RTA-containing complex (35). Careful studies of the ORF57 promoter indicate that some coregulator binding sites are relatively weak and that binding is stabilized by RTA (3). The reliance on cellular coregulators draws obvious parallels with VP16, the initiator of herpes simplex virus lytic replication (66, 69). VP16 has weak DNA binding activity but uses a combination of protein-protein and protein-DNA interactions to redirect cellular Oct-1 onto low- or moderate-affinity binding sites present in each VP16-responsive IE promoter. By comparison, RTA uses a broader repertoire of coregulators than VP16 and is directed to a much larger set of target promoters, raising the interesting possibility that the precise mode of RTA recruitment influences the kinetics, magnitude, or context in which an individual promoter is activated.
Dependence on cellular factors provides an opportunity for the virus to integrate information about the state of the host cell into the control of its own lytic program. The RTA coregulator CSL [acronym derived from CBF-1, Su(H), and Lag2, names for the mammalian, Drosophila, and nematode orthologs, respectively; also known as RBP-Jκ or KBF2] is a major end point of the Notch signal transduction pathway, which relays signals from one cell to another and is critical for fate decisions in many tissues (1, 14). The Notch receptor resides in the plasma membrane of the responding cell, and activation leads to sequential proteolytic cleavage events that release the Notch intracellular domain (NICD) and allow it to translocate to the nucleus. There, NICD assembles a protein-DNA complex with the DNA binding protein CSL, already present on Notch-responsive elements, and subsequently forms a ternary complex with several coactivators, including MAML-1 (mastermind in Drosophila), GCN5, P/CAF, and CBP/p300 (31). Like NICD, RTA forms a complex with CSL but presumably recruits coactivators through its own C-terminal activation domain (21, 33). Profiling studies indicate that a subset of KSHV lytic genes are induced by NICD, and for K2 (vIL6) and possibly K5 (MIR2), this occurs through NICD-CSL interactions (4). Early studies of the adenovirus pIX gene revealed that CSL can also act as a repressor, and this observation has been extended to many other contexts (2, 12, 30). In the absence of an activating factor such as NICD, CSL can repress some, but not all, Notch-responsive promoters by recruiting corepressor complexes. In addition to facilitating RTA-mediated activation of lytic genes, CSL may therefore also help to limit expression during latency or render promoters responsive to Notch signals independent of the rest of the lytic program.
To better understand the contribution of CSL to KSHV persistence and reactivation, we set out to discover additional regulatory elements that contain functional CSL binding sites. A search of the KSHV genome sequence found in excess of 260 potential binding sites, many in positions that might correspond to promoters. Four regions with a cluster of three or more candidate sites were selected for further analysis, and the regions immediately upstream of ORF70, ORF19, and ORF47 were found to contain strong RTA-responsive promoters. Authentic lytic transcripts were shown to initiate at each promoter, and at least one CSL binding site from each cluster was validated in vitro. These findings support the idea that CSL contributes to activation of many, but clearly not all, KSHV lytic genes. Alignments were used to establish a sequence signature for functional sites within the viral genome that will help to delineate the full complement of promoters that require CSL for their response to RTA.
MATERIALS AND METHODS
Plasmids.
Fragments of KSHV genomic DNA containing candidate promoter regions were amplified with Expand high-fidelity PCR (Roche), using custom primers, and subcloned into the unique XhoI and HindIII sites of a modified promoterless luciferase vector, pGL3-Basic (Promega). Mutagenesis was performed using QuikChange II XL site-directed mutagenesis (Stratagene) with custom oligonucleotide primers that changed 3 or 4 nucleotides within the site of interest. Control reporters LTi-luc (pLT4-luc) and LTc-luc (pLT6-luc) have been described previously (42). PANp-luc (pSEW-PP1) (61) was a kind gift of Gary Hayward, Johns Hopkins School of Medicine. Plasmid expression vectors encoding full-length RTA have been described before (42), and SG5-Flag-CSL (pJH282) (26), encoding full-length CSL, was a kind gift of Diane Hayward, Johns Hopkins School of Medicine.
Cell lines and transfections.
HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Cellgro) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and antibiotics. SLK cells were grown in RPMI 1640 medium (HyClone) with 10% Fetalplex, 2 mM l-glutamine, and antibiotics. For luciferase reporter assays, 2 × 104 HeLa or SLK cells were seeded in 24-well plates 24 h before transfection and then transfected using Polyfect (Qiagen) with 250 ng of luciferase reporter together with 25 ng pCGFlag-RTA (encoding full-length KSHV RTA) or an empty pCGFlag expression vector. The DG75-derived CSL knockout line SM244.9 (a kind gift of B. Kempkes, Helmholtz Center Munich) was cultured in RPMI 1640 medium (HyClone) supplemented with 20% fetal bovine serum, 2 mM l-glutamine, and antibiotics. A total of 0.5 × 107 cells were electroporated with 12.5 μg luciferase reporter and 1.25 μg pCGFlag-RTA or empty vector, using a Gene Pulser II instrument (Bio-Rad) set at 250 V and 975 μF. Ectopic CSL was supplied by cotransfection with 10 μg of SG5-Flag-CSL. Luciferase activity was measured 24 h after all transfections. Vero-rKSHV.219 cells (58) (a kind gift of Jeff Vieira, University of Washington) were maintained in DMEM (Cellgro) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, antibiotics, and 5 μg/μl puromycin to maintain the recombinant KSHV genome.
Immunoblotting.
Cells were lysed in extraction buffer (250 mM KCl, 20 mM Tris, pH 7.9, 10% glycerol, 0.25% NP-40, and protease inhibitors), and the soluble proteins were separated by SDS-10% PAGE, transferred to a Protran nitrocellulose membrane (Whatman), and blocked in Tris-borate-EDTA buffer containing 5% nonfat milk. Membranes were probed with anti-Flag (M2; Sigma) (diluted 1:1,000) and anti-Rho GDIα (A-20/sc-360; Santa Cruz) (diluted 1:5,000) primary antibodies and detected using horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit secondary antibodies (Amersham) (diluted 1:5,000). Chemiluminescence detection was performed with a SuperSignal West Pico kit (Thermo Scientific).
Real-time qRT-PCR analysis.
BC3 cells (2 × 105 cells/well) were treated with 300 μg/ml phosphonoacetic acid (PAA; Sigma-Aldrich) for 1 h before the addition of 40 ng/ml phorbol-12-myristate-13-acetate (TPA; Sigma-Aldrich). After 72 h, cells were harvested, and total RNA was isolated using RNeasy (Qiagen) and reverse transcribed with SuperScript III (Invitrogen) according to the manufacturer's instructions. Transcript levels were analyzed by real-time quantitative reverse transcription-PCR (qRT-PCR) with SYBR green detection. Samples were prepared in triplicate and amplified using a MyiQ single-color real-time PCR detection system (Bio-Rad). 18S rRNA was used for sample normalization. Primers to detect KSHV transcripts were as follows: for ORF19, 5′-GGCGAAAAAGTCAGCGGTGGT-3′ and 5′-CGGCGCGTCTTCCCTAAAGA-3′; for ORF47, 5′-GCTGCCGATGCCTGAATTGC-3′ and 5′-CGCTATTTGCCGTCCTGTGGA-3′; for ORF70, 5′-CAAATCCCCCTCCCTTCTGTGC-3′ and 5′-TTTGGCGAGGCGTAGTGCAA-3′; and for ORF50, 5′-GCACTAAGGCCAAACAGGGCGCAGG-3′ and 5′-TCGCCGCTAGGAAACATAGTTGTGC-3′.
5′-Rapid amplification of cDNA ends (5′-RACE) and primer extension analysis.
Vero-rKSHV.219 cells were induced to enter lytic replication by infection with BAC50 in the presence of 3 mM sodium butyrate (58). This recombinant KSHV was engineered to express two fluorescent proteins, namely, enhanced green fluorescent protein (EGFP), expressed from the constitutive EF1-α promoter, and red fluorescent protein (RFP), expressed from the RTA-dependent PAN promoter. In the absence of ectopic RTA, the vast majority of cells in the cultures expressed EGFP, reflecting the ubiquitous presence of the rKSHV.219 episome, but few, if any, expressed RFP. In the presence of BAC50 and sodium butyrate, ≥60% of cells became strongly RFP positive after 18 to 20 h, indicative of efficient activation of the PAN lytic promoter. Total RNA was isolated after 20 h by use of RNeasy reagent (Qiagen), and the poly(A)+ RNA fraction was selected on Oligotex beads (Qiagen). cDNA was synthesized by incubating the template RNA with Moloney murine leukemia virus (MMLV) reverse transcriptase, Smart II A oligonucleotide, and 5′ CDS primer A from a Smart RACE cDNA amplification kit (Clontech) at 42°C for 1.5 h. Gene-specific amplification products were generated using a universal primer mix and the following oligonucleotides: ORF47-5′RACE, 5′-GCAGCTTGGCTATACAGACCCCGTTGC-3′; ORF70-5′RACE, 5′-CGGCACAAAATTTCCCTCAACTGCC-3′; and ORF19-5′RACE, 5′-GCAGGTGTGCCTTTGTCTTTCGTTCTCG-3′. Amplification products were subcloned into pCR2.1-TOPO vector (Invitrogen), and the inserts were sequenced.
For primer extension analysis, the ORF-specific primers were labeled with [γ-32P]ATP by use of T4 polynucleotide kinase (Promega). For each primer extension reaction, 1 ng of labeled primer was mixed with 0.4 μg poly(A)+ RNA in denaturing hybridization solution [40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 6.8, 1 mM EDTA, 0.4 M NaCl, and 80% formamide] and incubated at 45°C for 1 h. Nucleic acids were recovered by ethanol precipitation and resuspended in avian myeloblastosis virus (AMV) primer extension buffer (Promega) with 2.8 mM sodium pyrophosphate and 1 U AMV reverse transcriptase (Promega). Extension reaction mixtures were incubated at 42°C for 30 min, the reaction was stopped by ethanol precipitation, and products were resolved in an 8% acrylamide-7 M urea denaturing gel. Gels were fixed and dried, and the labeled products were visualized by autoradiography. The same labeled primers were used to generate sequencing ladders that were run alongside the primer extension samples.
Gel shift analysis.
The following oligonucleotides (only one strand shown) containing known or potential CSL binding sites (underlined) were subcloned into the unique BamHI and XbaI sites (lowercase) of pCR2.1-TOPO (Invitrogen): EBVCp, 5′-gatccAAACACGCCGTGGGAAAAAATTTGGt-3′; ORF57RRE, 5′-gatccAGTGTAACAATAATGTTCCCACGGCt-3′; ORF70.1, 5′-gatccGGGGACACGCTGGGAGCGTCCTGGCt-3′; ORF70.2, 5′-gatccGTACCCCACCTCCCAACTGTGCAGCt-3′; ORF70.3, 5′-gatccGAGTGGGATCTCCCAATTACAGGAGt-3′; ORF70.4, 5′-gatccGGCGTCACCGTGGGAACTGTGAGTAt-3′; ORF19.1, 5′-gatccACCAACACCGTGAGAACGTCACAGCt-3′; ORF19.1-C, 5′-gatccACCAACACAGTGAGAACGTCACAGCt-3′; ORF19.1/3, 5′-gatccACCAACACCGTGAGAAAGGCGCACAt-3′; ORF19.2, 5′-gatccCCGGGGCGTTTCCCACGCCGGTGCAt-3′; ORF19.3, 5′-gatccTGTGCGCCTTTCTCACTGCTAGAGAt-3′; ORF19.3+C, 5′-gatccTGTGCGCCTTTCTCACGGCTAGAGAt-3′; ORF19.3/1, 5′-gatccTCTCTAGCAGTGAGAACGTCACAGCt-3′; ORF47.1, 5′-gatccTTTTAGAAAATCTCAGCCAGACGTGt-3′; ORF47.2, 5′-gatccCCCATCGTCTTCCCACGGAAATTAAt-3′; and ORF47.3, 5′-gatccAAGTATGTCGTGGGAAATAGACCTCt-3′.
Radiolabeled double-stranded DNA probes were prepared by PCR amplification, using 32P-labeled M13 forward and reverse primers. The resulting 130-bp amplification products were gel purified to remove single-stranded DNA, plasmid template, and unincorporated primers. This method ensures that all probes have identical specific activities. Extracts were prepared by incubating cells in 250 mM KCl, 20 mM Tris-HCl, pH 7.9, 10% glycerol, 0.25% NP-40, and 0.5 mM phenylmethylsulfonyl fluoride (PMSF), followed by centrifugation to remove nuclei and other nonsoluble components. Binding reaction mixtures containing probe, 10 μg protein extract, 1 μg sonicated poly(dI-dC), 0.5 μl fetal bovine serum, 10 mM Tris-HCl, pH 7.9, 90 mM KCl, 10 mM dithiothreitol (DTT), 1 mM EDTA, 0.1% NP-40, 1% glycerol, and 2% Ficoll-400 were incubated at 30°C for 30 min and loaded on a 4% native PAGE gel with 1× Tris-glycine-EDTA buffer. Electrophoresis was carried out at room temperature and run at a constant 170 V. Gels were fixed in methanol-acetic acid and dried, and the probes were visualized by autoradiography.
RESULTS
Enumeration of potential CSL binding sites in the KSHV genome.
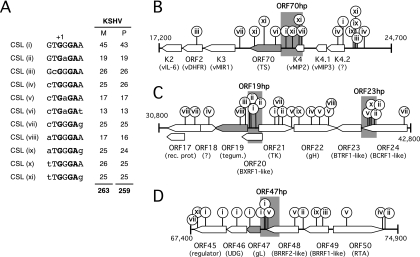
At least six KSHV promoters have been shown to contain binding sites for the host transcription factor CSL (RBP-Jκ/CBF-1) that contribute significantly to transcriptional activation by RTA. Other characterized RTA-responsive promoters lack identifiable CSL sites and rely on other cellular coregulators and/or direct binding by RTA (see the introduction). Whether there are any regulatory distinctions between CSL-dependent and -independent promoters is not yet known but may become apparent with a more complete catalogue of these two classes. To begin to address this, we searched the KSHV genomic sequence for matches to known CSL binding sequences. The canonical recognition sequence for CSL was originally defined as 5′-GTGGGAA-3′, but functional variants of this sequence have since been described, and it is likely that sequence context is also important (44, 46, 65). Using the literature, we derived a set of 11 core sequences from sites identified in mammalian Notch-responsive promoters and enhancers (Fig. 1A, sequences i through xi) and used these to search the KSHV genome sequence. By this approach, we identified 263 candidates in the prototype M-type genome (BC1 PEL; NCBI accession no. NC_003409) (49) and 259 candidates in the P-type genome (classic KS tumor biopsy GK18; NCBI accession no. NC_009333) (17). Candidate sites are scattered widely across the entire unique portion of the genome, with more than half of the matches located in intergenic regions or within a few hundred base pairs of the start of an ORF, which are favored locations for transcriptional promoters and other regulatory elements (data not shown).
FIG. 1.
The KSHV genome contains many matches to known CSL binding sites. (A) Published studies of mammalian Notch-regulated genes describe 11 variants (i through xi) of the canonical CSL recognition sequence. The numbers of matches to each variant present in the M-type (49) (NCBI accession no. NC_003409) and P-type (17) (NCBI accession no. NC_009333) KSHV genome sequences are indicated on the right. (B) Schematic showing 7.5-kbp region (positions 17,200 to 24,700 in the M-type reference sequence) between ORF K2 and the intergenic region containing the lytic origin of replication. The positions and type of potential CSL sites are indicated by lollipops. The region immediately upstream of ORF70 (ORF70hp; shaded), containing four candidate sites, was selected for functional analysis. (C) Map of the 12-kbp region (positions 30,800 to 42,800) between ORF17 and ORF24, containing ORF19hp and ORF23hp. (D) Region (7.5 kbp) between ORF45 and ORF50 (positions 67,400 to 74,900), including ORF47hp.
Identification of three new RTA-responsive promoters.
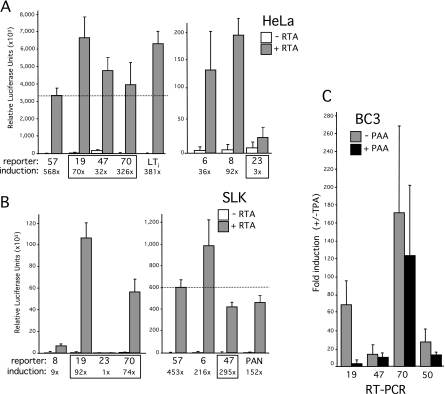
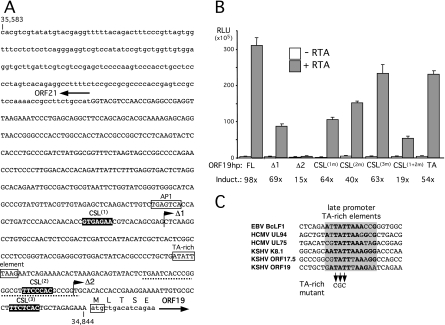
In view of the large number of potential CSL sites, we selected four clusters (M type) (Fig. 1B to D) for further testing; specifically, we chose the clusters immediately upstream of ORF70 (encoding thymidylate synthase), ORF19 (encoding tegument protein), ORF23 (encoding an EBV BTRF1 homolog), and ORF47 (encoding glycoprotein L). These were chosen on the basis of the following three criteria: (i) they include three or four candidate CSL sites; (ii) they include additional motifs, such as TATA boxes or other transcription factor binding sites; and (iii) they have not been studied in depth before. Transcriptional start sites (TSSs) have not been determined for any of these ORFs, so we arbitrarily selected the first 750 bp upstream of the translational initiation codon (ATG) as hypothetical promoters (hp). Each hp fragment included either three (ORF19hp and ORF47hp) or four (ORF70hp and ORF23hp) of the candidate CSL sites. Fragments were generated by PCR amplification and inserted into a promoterless luciferase reporter construct in the same orientation as the aforementioned open reading frames. As controls, we used the same strategy to select fragments from three previously demonstrated RTA-regulated genes, i.e., ORF6 (single-stranded binding protein), ORF8 (glycoprotein B), and ORF57 (Mta), each of which contains one or more functional CSL sites (33, 63, 77). The response of each luciferase reporter was assayed in HeLa and SLK cells by transient cotransfection with an empty expression vector or one encoding full-length RTA (Fig. 2A and B). SLK cells are an immortalized but KSHV-negative spindle cell line derived from a classical Kaposi's sarcoma tumor (25). The RTA-responsive LTi and PAN reporters served as further positive controls in the experiment (42). With the exception of the ORF23 fragment, all of the test and control fragments functioned as strong RTA-responsive promoters. The extent of induction was clearly influenced by cell type. For example, the ORF57 reporter was induced to similar extents in both cell types, whereas ORF70hp was induced 326-fold in HeLa cells but only 74-fold in SLK cells. Conversely, ORF47 was induced more strongly in SLK cells (295-fold) than in HeLa cells (32-fold). ORF19hp resembled ORF57 in showing similar degrees of activation in either context. Presumably, these differences reflect the altered transcriptional milieu of each cell line and differences in the baseline (constitutive) activity of the promoters in the absence of RTA. Regardless of these quantitative differences, it is clear that the test fragments chosen by the presence of potential CSL binding sites were capable, with one exception (ORF23hp), of directing high levels of RTA-dependent transcription.
FIG. 2.
Identification of novel RTA-responsive promoters in the intergenic regions upstream of ORF70, ORF19, and ORF47. Reporter constructs were generated by subcloning the following fragments into a promoterless luciferase reporter: ORF6 (M-type reference sequence nucleotides [nt] 2,472 to 3,209), ORF8 (nt 7,959 to 8,698), ORF70hp (nt 21,105 to 21,842), ORF23 (nt 40,517 to 41,287), ORF19hp (nt 34,844 to 35,583), ORF47 (nt 69,916 to 70,677), and ORF57 (nt 81,331 to 82,086). Activity was assayed by transient transfection of HeLa (A) and SLK (B) cells by cotransfection with a full-length RTA (pCGFlag-RTAFL) expression plasmid or the empty equivalent. Each assay was performed in triplicate, and relative luciferase activity was measured after 24 h. For each combination, the mean and standard deviation are shown, together with the calculated difference between the presence and absence of RTA (fold induction). Two previously characterized RTA-responsive reporters, LTi-luc (nucleotides 127,610 to 127,807) and PAN-luc (nucleotides 28,455 to 28,681), were included as positive controls (42). ORF57-luc showed similar levels of induction in the two cell types, and its activity is used as a point of comparison (dotted line). (C) Latently infected BC3 cells were induced with TPA in the presence or absence of PAA. After 72 h, total RNA was isolated and analyzed by RT-PCR, using primers specific to ORF19, ORF47, ORF70, and ORF50/RTA. Values represent the mean and standard deviations obtained from assaying three independent cultures. Differences in RNA handling and cDNA synthesis were normalized using 18S rRNA.
Previous profiling studies have categorized ORF70 and ORF47 as delayed-early genes and ORF19 as either a delayed-early or late gene (28, 38, 73). To confirm these assignments, latently infected BC3 cells were induced to enter lytic replication by use of the phorbol ester TPA in either the presence or absence of the viral DNA polymerase inhibitor PAA. RNA was collected after 72 h and analyzed by real-time qRT-PCR, using primer pairs located within each of the open reading frames (Fig. 2C). All three genes were strongly induced by TPA treatment, but ORF19 showed a much greater sensitivity to PAA (93% reduction), characteristic of a true late gene that relies on replication of the viral genome for activation. ORF50 served as a positive control for lytic induction. Based on these observations, we chose to pursue all three uncharacterized RTA-responsive promoters to determine their individual requirements for CSL.
Induction by RTA is dependent on CSL.
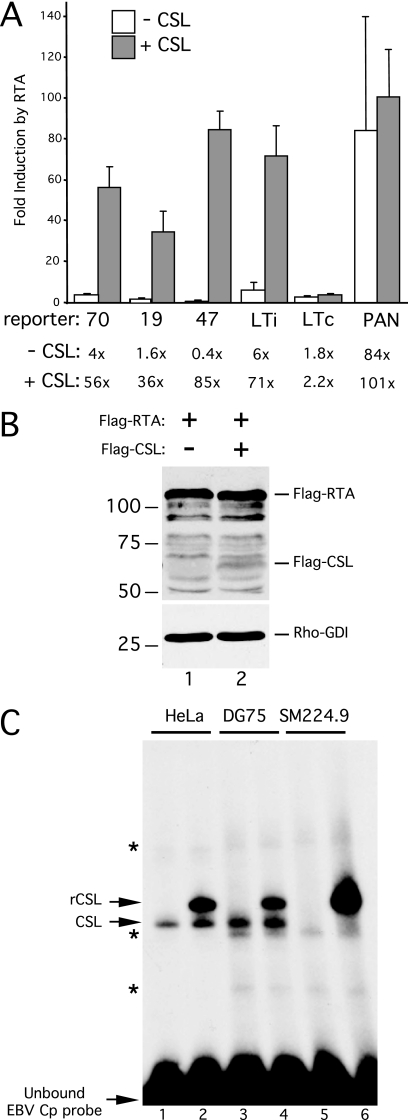
To determine whether CSL was required for the response to RTA, we assayed each reporter in CSL-negative SM224.9 cells. This line is derived from the KSHV- and EBV-negative DG75 human Burkitt's lymphoma line, and the CSL locus has been inactivated by targeted homologous recombination (41). The ORF70, ORF19, and ORF47 reporters were essentially unresponsive to RTA in these cells (Fig. 3A, open bars). As a control, we also tested the LTi promoter, which contains two CSL binding sites that are known to be essential for the response to RTA (42). Again, induction of LTi was severely compromised (5.6-fold compared to 381-fold [Fig. 2A]) in the absence of CSL. As a positive control for RTA function, we also tested the core PAN promoter that can be bound directly by RTA, and this construct was highly responsive (84-fold induction).
FIG. 3.
CSL is required for response to RTA. (A) Analysis of induction by RTA in CSL-null SM224.9 cells, a derivative of the DG75 cell line carrying a somatic knockout of the CSL gene (41). Plasmid DNA was introduced into 0.5 × 107 SM224.9 cells by electroporation, and luciferase activity was determined after 24 h. Each reporter plasmid, ORF70hp-luc, ORF19hp-luc, ORF47hp-luc, LTi-luc, LTc-luc, and PAN-luc, was cotransfected with combinations of empty vector, RTA vector, or CSL vector. Fold induction by RTA was calculated for each reporter, in either the presence (+) or absence (−) of cotransfected CSL. The mean and standard deviation of three separate assays are shown. (B) Immunoblot of whole-cell extracts prepared from SM224.9 cells transfected with plasmids encoding Flag-RTA (lane 1) or Flag-RTA and Flag-CSL (lane 2). Extracts were separated in an SDS-10% PAGE gel and probed with anti-FLAG or anti-Rho-GDI antibodies. (C) Gel shift assay using whole-cell extracts prepared from HeLa (lanes 1 and 2), DG75 (lanes 3 and 4), and SM224.9 (lanes 5 and 6) cells that were either mock transfected (lanes 1, 3, and 5) or transfected with a plasmid encoding epitope-tagged CSL (lanes 2, 4, and 6). Each extract was mixed with a 32P-labeled probe containing the canonical CSL binding site from the EBV Cp promoter, incubated for 30 min at 30°C, and resolved in a 4% native PAGE gel. Positions of the unbound (free) probe and shifted complexes corresponding to endogenous (CSL) and transfected (rCSL) CSL are indicated. Nonspecific complexes are indicated with asterisks.
Next, we complemented the CSL deficiency by transient transfection of an expression plasmid encoding human CSL in either the presence or absence of RTA. This had no effect on the constitutive LTC promoter, which is unresponsive to RTA, and had only a modest effect on induction of the PAN promoter (101-fold compared to 84-fold). However, cotransfection of CSL effectively restored RTA-mediated induction of all three test promoters (ORF70, 56-fold; ORF19, 36-fold; and ORF47, 85-fold). Immunoblotting confirmed that this increase in transcriptional activity was not due to altered expression of RTA (Fig. 3B). Finally, to verify that the SM224.9 cells lack CSL site binding activity, we prepared whole-cell extracts and performed a gel mobility shift assay using the prototype binding site from the Epstein-Barr virus Cp promoter (Fig. 3C). Extracts from the parental DG75 cells and from HeLa cells were prepared and assayed in parallel. A discrete complex corresponding to bound CSL was detected in the HeLa and DG75 cell extracts (lanes 1 and 3, respectively) but was absent in the SM224.9 cell extract (lane 5). Transfection of a CSL expression plasmid into each cell line prior to extract preparation resulted in a novel, slower-migrating shift that represents the larger recombinant protein (rCSL), which includes a 33-amino-acid extension at the N terminus added during plasmid construction (26). From these results, we concluded that transcriptional activation of the ORF70hp, ORF19hp, and ORF47hp reporters by RTA is dependent on CSL, consistent with their selection on the basis of a cluster of potential binding sites.
One or more CSL sites are required for the response to RTA.
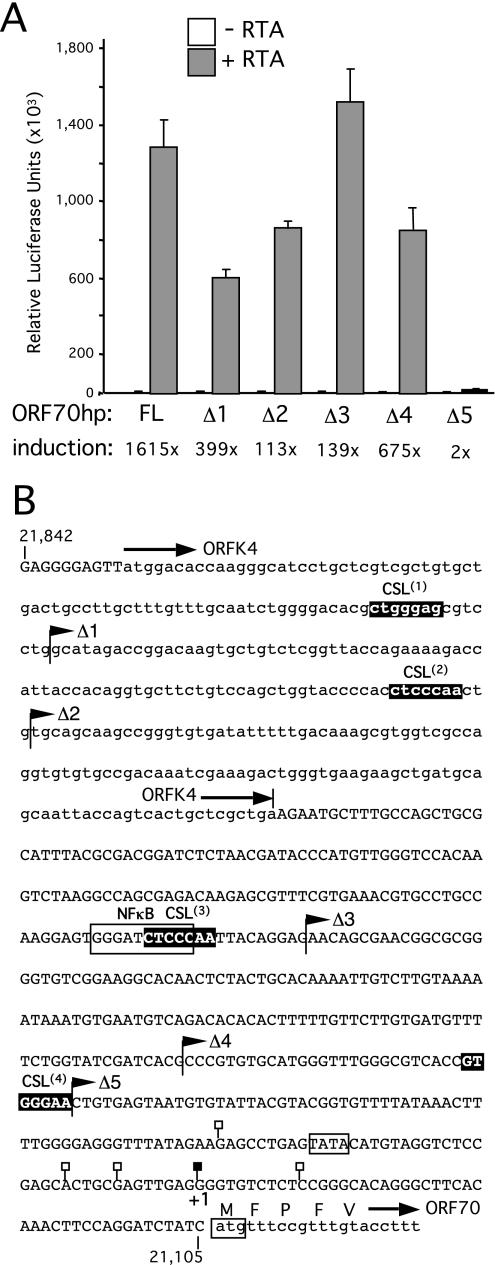
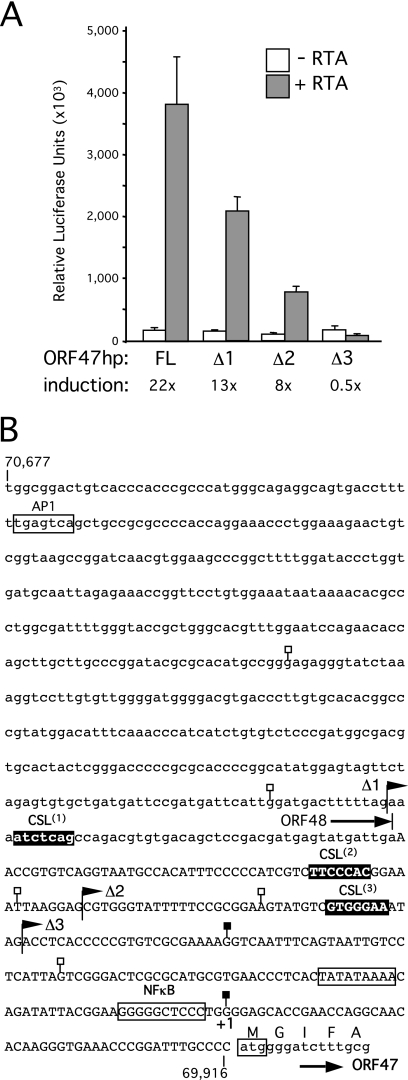
A series of truncations were prepared for ORF70hp (Fig. 4), ORF47hp (Fig. 5), and ORF19hp (Fig. 6), sequentially removing the candidate CSL sites. For ORF70hp, activity remained high until the removal of sequences that included the most proximal CSL site, at which point the response to RTA was effectively abolished (compare OR70hpΔ4 to ORF70hpΔ5). Interestingly, CSL site 3 overlaps with a consensus sequence for NF-κB (5′-GGGRNNYCCC-3′), a cellular factor that is upregulated in latently infected cells and implicated in suppression of lytic gene expression (27). Deletion of the potential NF-κB site correlated with an increase in overall reporter activity in the presence of RTA (compare ORF70hpΔ2 to ORF70hpΔ3). Enhanced activity was maintained over a 50-fold range in RTA expression, suggesting that the truncated promoter was inherently more active than the full-length version (data not shown). Analysis of three truncations in ORF47hp (Fig. 5) resulted in a stepwise decrease in the response to RTA, and all promoter activity was lost when all three potential CSL sites were removed (ORF47hpΔ3). Thus, for both promoters, sequences containing at least one of the candidate CSL sites are critical for the response to RTA.
FIG. 4.
Analysis of ORF70-K4 intergenic region. (A) Full-length (FL) and truncated (Δ1, Δ2, Δ3, Δ4, and Δ5) versions of ORF70hp-luc were tested for a response to RTA in HeLa cells by transient transfection. Assays were performed in triplicate and the luciferase values expressed as the mean and standard deviation. For each reporter, fold induction was calculated from luciferase activity measured in the presence and absence of the RTA expression plasmid. (B) Annotated sequence of the ORF70hp fragment (nucleotides 21,105 to 21,842 in the M-type reference sequence). Overlap with ORFK4 (encoding vMIP2) is shown in lowercase, as are the first few codons of ORF70, encoding thymidylate synthase. Also shown are the end points of the truncations used in panel A (filled flags), the four candidate CSL sites, ORF70.1, ORF70.2, ORF70.3, and ORF70.4 (filled boxes), and TSSs mapped by 5′-RACE (filled or open lollipops). The principal TSS (filled lollipop) is labeled +1 and corresponds to position 21,150. A putative TATA box and consensus NF-κB site are indicated with an open box.
FIG. 5.
Truncation analysis of the ORF47 promoter region. (A) Analysis of full-length (FL) and truncated (Δ1, Δ2, and Δ3) versions of ORF47hp-luc in HeLa cells. (B) Sequence of the ORF47hp fragment (nucleotides 69,916 to 70,677). The region of overlap with ORF48 (EBV BRRF2 homolog) is shown in lowercase, as are the first five codons of ORF47. Also shown are the end points of truncations used in panel A (filled flags), the three candidate CSL sites, ORF47.1, ORF47.2, and ORF47.3 (filled boxes), and TSSs mapped by 5′-RACE (filled or open lollipops). The preferred TSS (filled lollipop) is labeled +1 and corresponds to position 69,963 in the KSHV prototype genome. Consensus AP1 and NF-κB binding sequences and a putative TATA box are also indicated.
FIG. 6.
Analysis of ORF19-ORF21 intergenic region. (A) Sequence of the ORF19hp fragment (nucleotides 34,844 to 35,583). The 5′ end of ORF21 (thymidine kinase), encoded on the opposite strand from ORF19, is shown in lowercase, as are the first five codons of ORF19. Also shown are the end points of truncations used in panel B (filled flags) and the three candidate CSL sites, ORF19.1, ORF19.2, and ORF19.3 (filled boxes). Primer extension product end points were mapped to a region around the second CSL site (dotted line). A consensus AP1 site is indicated with an open box. (B) Full-length (FL) ORF19hp-luc and a series of truncations (Δ1 and Δ2) and targeted mutations were tested for a response to RTA in HeLa cells by transient transfection. Assays were performed in triplicate and luciferase values expressed as the mean and standard deviation. For each reporter, fold induction was calculated from luciferase activities measured in the presence and absence of the RTA expression plasmid. (C) Alignment of the ORF19 TA-rich element with similar sequences found in the Epstein-Barr virus (EBV) BcLF1 (51), human cytomegalovirus (HCMV) UL94 (68), HCMV UL75 (43), KSHV K8.1 (56), and KSHV ORF17.5 (5) true late promoters. Shading indicates the 12-bp core element defined by studies of the K8.1 promoter, with conserved nucleotides shown in bold.
We performed a similar truncation analysis on ORF19hp (Fig. 6), where a large deletion that included the distal CSL site (ORF19hpΔ1) significantly reduced promoter output. A further truncation (ORF19hpΔ1) that removed site 2 essentially abolished promoter activity. Mutating the putative CSL sites individually reduced the overall activity of the promoter and the extent of induction. Mutations of sites 1 (CSL1m) and 2 (CSL2m) had the greatest effects, and when combined together (CSL1+2m), they substantially reduced the response to RTA and the overall promoter activity. This result suggests that the presence of two or more CSL sites is required for a strong response to RTA. The PAA sensitivity of ORF19 mRNA accumulation in latently infected cells is indicative of a true late gene. Studies of other examples have identified a TA-rich element that is important for promoter activity (5, 43, 51, 56, 68). Examination of the ORF19hp sequence identified an 8 of 12 bp match (Fig. 6C). Substitution of three residues (TA mutant, changing TAT to CGC) reduced induction by RTA from 98-fold to 54-fold, indicating a functional element (Fig. 6B). Taken out of the context of the viral episome, late promoters can show a reduced dependence on DNA replication, and it is conceivable that the TA mutation would have a greater impact on a replicating template.
Transcript start site mapping.
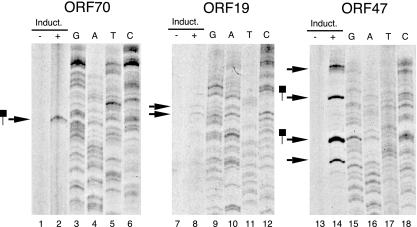
Although each test fragment displayed robust activity in the context of a reporter assay, it was important to establish that the fragments coincided with lytic promoters in the viral episome. To do this, we mapped the TSSs for mRNAs that encode ORF70, ORF19, and ORF47 by 5′-RACE and by primer extension. Vero cells carrying a recombinant version of KSHV (rKSHV.219) served as a source of lytic transcripts (58). The virus is maintained in a latent state and can be induced to reactivate relatively efficiently by the expression of ectopic RTA in the presence of a histone deacetylase (HDAC) inhibitor or other inducer. Poly(A)+ RNA was isolated after 48 h and used for 5′-RACE with gene-specific primers located within ORF70 and ORF47. Sequencing of the cloned amplification products identified multiple initiation sites within each promoter. For ORF70, 6 of 10 RACE products terminated at a guanine at position 21,150, approximately 30 bp downstream from a TATA-like sequence (Fig. 4B). Likewise, multiple potential start sites were identified for ORF47, with three clones ending at a guanine at position 69,693 (Fig. 5B). This residue lies 34 to 36 bp downstream of a TATA-like sequence, again suggestive of a valid TSS. Although ORF19 mRNA could readily be detected by qRT-PCR at this time point, no 5′-RACE products were captured (data not shown).
Because we observed heterogeneity in the end points of the RACE products, we also used primer extension analysis to identify the most commonly utilized start sites. The same ORF-specific primers were end labeled, annealed to mRNA, and extended with AMV reverse transcriptase. Products were resolved in 8% denaturing polyacrylamide gels and detected by autoradiography (Fig. 7). The results were in general agreement with those of the 5′-RACE analysis. A single extension product was detected with the ORF70 primer (Fig. 7, lane 2), and by its size, this corresponds to the most frequently occurring 5′-RACE clone, terminating at nucleotide 21,150 (Fig. 4B). For ORF47, we detected four major extension products (lane 14), two of which also correspond to 5′-RACE clones (nucleotides 69,963 and 70,054) (Fig. 5B). Faint but reproducible products were obtained with the ORF19 primer (lane 8), and assuming that the template was not spliced, these tentatively place the TSSs in a region between nucleotides 34,890 and 34,920 (dotted line in Fig. 6A). This area is on the boundary of the Δ2 truncation that abolished promoter activity, suggesting that this is the location of the core promoter. For all three primers, extension products were observed only with mRNAs from cells induced with BAC50 (Fig. 7), consistent with the lytic transcription profile of these genes (Fig. 2C). The fact that we can map initiation sites for lytic transcripts within each of the proposed promoter regions clearly indicates that our candidate approach was effective.
FIG. 7.
Mapping the 5′ ends of RTA-inducible transcripts. Poly(A)+ RNAs from induced (+) and uninduced (−) rKSHV.219 Vero cells were annealed to 32P-labeled antisense oligonucleotides specific to ORF70 (lanes 1 and 2), ORF19 (lanes 7 and 8), or ORF47 (lanes 13 and 14). Primers were extended using reverse transcriptase, and the denatured products were resolved in an 8% sequencing gel. The sizes of the extension products were determined using radiolabeled molecular weight markers and a sequencing ladder generated with the same labeled primer and a subcloned fragment of KSHV genomic DNA as the sequencing template. Extension products that coincide with end points detected by 5′-RACE are indicated with lollipops.
Validation of individual CSL binding sites.
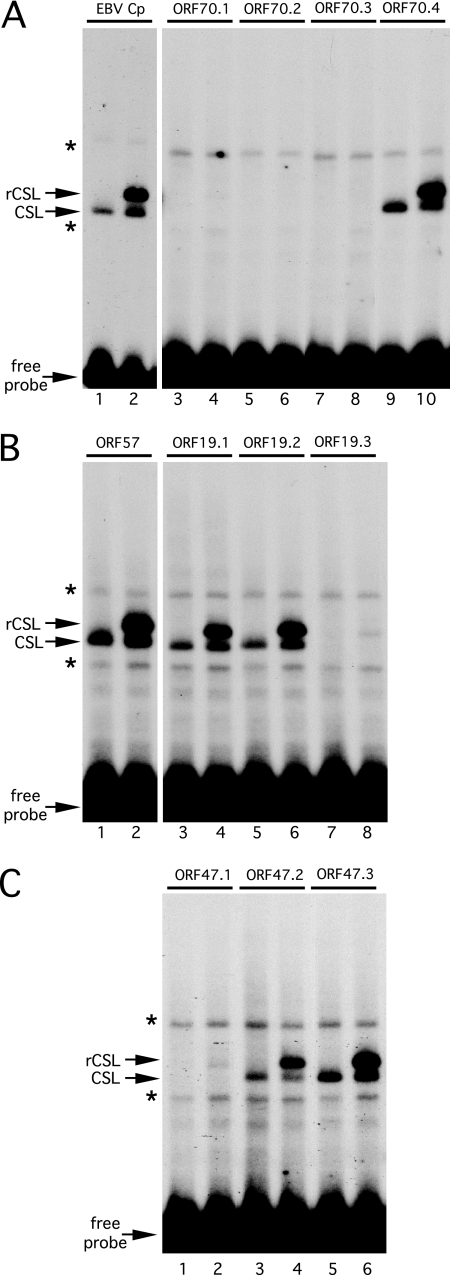
We next evaluated the ability of CSL to bind to each candidate sequence by performing a gel mobility shift assay (Fig. 8). As in Fig. 3C, incubating the canonical CSL site from the EBV Cp promoter with HeLa cell extract resulted in a robust shift indicative of a CSL-DNA complex (labeled CSL; Fig. 8A, lane 1). An additional, slower-moving complex was observed using an extract from HeLa cells that expressed recombinant CSL (labeled rCSL; Fig. 8A, lane 2). Equivalent complexes (Fig. 8B, lanes 1 and 2) were detected using a probe containing the demonstrated CSL site from the ORF57 RTA-responsive element (33). Of the four candidate sites in the ORF70 promoter, only the most proximal (site ORF70.4) was bound under these conditions (Fig. 8A), consistent with the results of the truncation analysis (Fig. 4). Robust binding was detected with the distal and middle sites from the ORF19 promoter (Fig. 8B, ORF19.1 [lanes 3 and 4] and ORF19.2 [lanes 5 and 6]) and the middle and proximal sites from the ORF47 promoter (Fig. 8C, ORF47.2 [lanes 3 and 4] and ORF47.3 [lanes 5 and 6]). We note that a very weak complex was formed by recombinant CSL with the ORF19.3 (Fig. 8B, lane 8) and ORF47.1 (Fig. 8C, lane 2) probes, and these presumably represent weak sites that may or may not be physiologically significant. The addition of RTA, transfected either alone or together with CSL, did not alter the binding profile or the mobility of the complexes (data not shown). In summary, this in vitro analysis establishes that all three of the previously uncharacterized promoters contain one or more functional CSL binding sites. Moreover, the locations of the validated sites agree with the truncation analyses, indicating that these CSL binding sites are likely to form part of the RTA response element(s) of each promoter. No strong conclusions can be drawn for the five sequences that did not form a robust complex in this assay. It is possible that they represent low-affinity sites that nonetheless are functional in vivo. This is supported by the fact that mutation of ORF19.3 reduced promoter activity by approximately one-third (Fig. 6B).
FIG. 8.
In vitro binding analysis of candidate CSL binding sites. Gel mobility shift assays were performed using 32P-labeled probes containing candidate CSL binding sites from the ORF70 (A), ORF19 (B), and ORF47 (C) promoters. Individual sites are numbered in accordance with Fig. 4, 5, and 6. Characterized binding sites from the EBV Cp promoter (panel A, lanes 1 and 2) and the ORF57 promoter (panel B, lanes 1 and 2) were included as positive controls. All probes were identical in length and were mixed with HeLa whole-cell extract from mock-transfected cells (odd-numbered lanes) or cells expressing tagged CSL (even-numbered lanes). Binding reaction mixtures were incubated at 30°C for 30 min prior to being loaded on a 4% native PAGE gel. Positions of the unbound probe and complexes corresponding to endogenous (CSL) or transfected CSL (rCSL) are indicated. Nonspecific complexes are marked with asterisks.
KSHV uses a subset of possible CSL binding sequences.
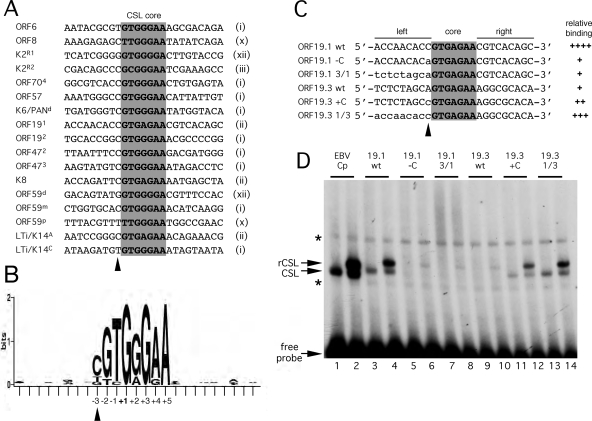
The sequences of the five functional CSL binding sites were compared to those already known from characterized RTA response elements (Fig. 9A). Only 4 of the original 11 core sequences (i, ii, iii, and x) are represented, with type i being the most frequent (9 of 17 sequences). The absence of significant sequence conservation beyond the core positions is very evident from the sequence logo representation (Fig. 9B), with the one exception being a cytosine (C) at position −3 (indicated with an arrowhead), present in 12 of the 17 sequences. It should be noted that the so-called R1 site from the K2/vIL6 promoter (4) and the distal site from the ORF59 promoter (36) represent a sequence variant (5′-GTGGGGA-3′; dubbed type xii) that was not included in our original search. There are 26 versions of sequence type xii in the M- and P-type KSHV genomes, bringing the total number of candidate sites in each strain to 289 and 285, respectively.
FIG. 9.
Defining a signature for CSL binding sites in the KSHV genome. (A) Alignment of 17 confirmed CSL binding sites from the KSHV genome. The core sequence is shaded, and the core motif type, as defined in Fig. 1A, is noted on the right. Labeling of individual sites from K2/vIL6, K6/PAN, ORF59, and LTi/K14 follows published naming schemes (4, 33, 34, 36). (B) A positional nucleotide frequency pattern (sequence logo) for the sites listed in panel A was generated using WebLogo (9). Only positions +1, +3, and +5 are invariant. This visual representation emphasizes the strong preference for a cytosine at position −3 and the striking absence of sequence conservation beyond the eight central positions (−3 to +5). (C) Sequences of probes used to test the contributions of flanking residues to the differential binding of CSL to ORF19 sites 1 and 3. The wild-type sequence is shown in uppercase, and the altered sequence is shown in lowercase. (D) Gel mobility shift analysis of the probes shown in panel C. Each was labeled to an identical specific activity and then incubated with HeLa whole-cell extract from mock-transfected cells (odd-numbered lanes) or cells expressing tagged CSL (even-numbered lanes). The EBV Cp site (lanes 1 and 2) was included as a reference. Positions of the unbound probe and complexes corresponding to endogenous (CSL) or transfected (rCSL) CSL are indicated. Nonspecific complexes are marked with asterisks. The ability of each probe to form a stable complex with CSL is summarized in panel C.
Within the ORF19 promoter, there are two examples of the type ii sequence. Interestingly, CSL formed a stable complex with the distal site ORF19.1 and not the proximal site ORF19.3. This argues that sequences outside the core also determine binding affinity. Of special note is the presence of a C residue at position −3 in ORF19.1. To test the relevance of the flanking sequences, and this residue in particular, we made additional gel mobility shift assay probes that swapped the C and adenosine (A) residues at position −3 (Fig. 9C, ORF19.1 −C and ORF19.3 +C) or exchanged the entire left flanking sequence (ORF19.1 3/1 and ORF19.3 1/3). Changing the C to A in ORF19.1 virtually abolished binding, whereas replacement of the A in ORF19.3 with C conferred increased binding, a clear demonstration that this position is critical. Finally, a chimera corresponding to the left flanking sequence of ORF19.1 and the right flank of ORF19.3 resulted in a complex similar to that seen with wild-type ORF19.1, indicating that an additional residue contributes to the overall affinity of the site.
DISCUSSION
Viruses often forge an intimate relationship with their host cell, intermeshing their regulatory machineries to create a suitable niche for virus propagation and to circumvent antiviral defenses. Nowhere is this more apparent than in the herpesviruses, which need to persist for long periods as latent viruses but must also respond to external stimuli by entering a productive (lytic) phase that produces new infectious virus. For KSHV, the transition from latency to lytic replication is initiated by RTA, an IE protein that functions as a transcription factor to directly or indirectly stimulate the transcription of 80 or more lytic genes. RTA is capable of binding to a specific sequence found in a few viral promoters; however, for most, it is reliant on cellular coregulators for recruitment. In this study, we focused on CSL, a cellular transcription factor involved in Notch signaling and a coregulator for several viral proteins. Ganem and colleagues first uncovered a physical interaction between RTA and CSL and identified binding sites in the ORF6 (single-stranded binding protein) and ORF57 (Mta) promoters that are critical for the response to RTA (33). They also described a site well upstream of the RTA-responsive PAN promoter that acts much like an enhancer element, boosting activity without being strictly essential for induction. Subsequent work by this group and several others pinpointed additional CSL sites upstream of ORF8 (glycoprotein B) (77), K2 (vIL6) (4), K6 (MIP1) (7), K8 (K-bZIP) (63), ORF59 (DNA replication processivity factor) (36), and the bidirectional LTi/K14 promoter (34, 42). It is likely that the K5 promoter will be added to this list. It has been shown to respond to ectopic NICD, implying that it also contains one or more functional CSL binding sites, but involvement in the response to RTA still needs to be addressed (4).
ORF70 and ORF47 behave as delayed-early genes and thus resemble other known targets of direct RTA transactivation (28). In contrast, ORF19 mRNA accumulates with late kinetics in TPA-treated PELs, and the addition of viral DNA polymerase inhibitors phosphonoacetic acid (this study) and cidofovir (38) leads to a marked reduction in ORF19 transcript levels, indicating a strong requirement for DNA replication, the hallmark of a true late (γ2) gene. Close examination of the ORF19 promoter sequence also revealed a TA-rich sequence resembling a sequence found in true late promoters, including K8.1 and ORF17.5 from KSHV (5, 43, 51, 56, 68). These observations indicate that ORF19 is most likely a true late gene that is regulated by RTA through CSL. A similar claim has been made for ORF8 (encoding glycoprotein B), but the authors of that study noted that ORF8 expression is relatively insensitive to the DNA replication inhibitor ganciclovir and that ORF8 might be classified better as a delayed-early or leaky-late gene (38, 77).
KSHV uses a subset of possible CSL binding sequences.
In this study, we deliberately inverted the discovery process, using a motif search to predict the locations of uncharacterized RTA-responsive promoters. Of the four candidate regions selected for detailed analysis, three were found to contain a strong CSL/RTA-dependent promoter and to coincide with the 5′ ends of lytic transcripts. Removal of the predicted CSL binding sites correlated with the loss of RTA responsiveness, and for half of the candidate sites, we were able to demonstrate binding by CSL in vitro. Closer examination of the sequences of these validated sites reveals some clear preferences with respect to the core sequence. Of the initial 11 unique variants, only types i, ii, iii, and x are represented as experimentally validated sites. Alignment of these sequences clearly highlights the conservation of only one residue outside the core motif, an additional cytosine (C) residue at position −3, consistent with an earlier in vitro binding site selection assay using purified murine CSL that also indicated a strong preference for a cytosine at this position (57). Presumably, this residue contributes to a higher-affinity binding site and explains the contrasting behaviors of sites ORF19.1 and ORF19.3 in the ORF19 promoter. The core sequences of the two sites are identical, yet only ORF19.1 bound CSL in the in vitro assay. The contribution of the flanking cytosine present in ORF19.1 (5′-CGTGAGAA-3′) but absent in ORF19.3 (5′-AGTGAGAA-3′) was demonstrated by making exchanges that swapped either the entire flanking sequence or this single position. Genome-wide, there are 47 candidate CSL binding sites in the M-type genome (44 in the P-type genome) that have a cytosine at the −3 position.
Independent studies of the K2/vIL6 and ORF59 promoters revealed one additional variant (5′-GTGGGGA-3′; subsequently termed type xii) that was not included in our original motif search and that extends the number of candidate binding sites spread across the KSHV genome. This sequence does not occur in the ORF70, ORF19, and ORF47 promoter regions, but paradoxically, there is a copy within the unresponsive ORF23hp fragment, with another immediately upstream (data not shown). The failure of the ORF23hp construct to respond to RTA is puzzling. Not only does the test fragment contain five potential CSL binding sites, two of which include the extra cytosine at position −3, but Izumiya and colleagues identified a strong RTA-responsive promoter within the proximal 500 bp of ORF23hp (13). Whether the lack of activity of our reporter construct reflects a negative element in the upstream portion of the fragment used or some more trivial explanation remains to be determined.
We contend that the same candidate-driven approach can be applied to other gammaherpesviruses. For example, a similar number of candidate sites are present in the murine herpesvirus 68 genome sequence, and this may reflect the conservation of coregulator usage across the rhadinovirus RTA proteins (47, 54). More than 330 candidate sites are present in the EBV genome (data not shown); this is an unexpected observation because the prototype RTA transactivator encoded by EBV is not known to use CSL as a coregulator. Instead, the EBNA2 and EBNA3A to -C proteins, regulators of latent gene expression, interact with CSL at the viral Cp, LMP-1, LMP-2A, and TP1 promoters (22, 29). All of the known CSL binding sites belong to core sequence type i, and with the exception of a site in the LMP-1 promoter, they all include the flanking C at the −3 position. It will be interesting to determine whether any of the additional sites in the EBV genome, especially those associated with lytic genes, are functional during latency or even facilitate the response to EBV RTA. Some of these sites may also contribute to cross talk between EBV and KSHV in PELs, which are often colonized by both viruses simultaneously (70).
Organization of KSHV transcription units.
A recurring question during these studies is how to meaningfully define the boundaries of individual promoters in the context of gene-dense viruses. KSHV, like all other herpesviruses, often uses bidirectional promoters that share regulatory elements, an economy afforded by having genes that can be transcribed coordinately. Likewise, regulatory elements may be embedded within the coding sequence of an adjacent gene, such that genetic units are quite literally layered on top of one another. Whether long-range elements similar to cellular enhancer sequences operate in herpesvirus genomes is unclear. A previous report described four potential CSL binding sites within a 3-kb segment upstream of ORF50, and binding of CSL to the most distal of these sites (position −1461 with respect to the major ORF50 TSS) was confirmed by gel shift assay (32). This corresponds to site ORF47.2 of the ORF47 promoter described here, and its relevance to ORF50 transcription is open to debate. In transient assays, all four CSL binding sites could be deleted from the parental 3-kb ORF50 promoter fragment without diminishing either the baseline activity of the promoter or its response to RTA (32). Moreover, other studies have mapped key elements required for the response to various inducers to a region much closer to the TSS (37, 72), again suggesting that the important promoter elements are within a few hundred base pairs, at most. Recently, a second, more distal ORF50 promoter was described, first in murine herpesvirus 68 and subsequently in KSHV and EBV (19). Although the limits of this promoter have not been defined yet, it may overlap with (and conceivably share) the ORF47 elements described here.
The realization that KSHV uses several mechanisms to bring RTA to target promoters raises the possibility that lytic genes can be subdivided on the basis of how they are activated rather than when they are activated. As others have pointed out, this versatility would enable the virus to customize the lytic program to better fit the environment of a given host cell or allow for selective expression of part of the lytic repertoire. Determining functional relationships between sets of coregulated genes is complicated by the presence of polycistronic transcription units, weak polyadenylation sites, alternative splicing, and so on. Primary transcripts arising from the ORF70 promoter extend to a polyadenylation site downstream of K3, and possibly farther (48). In addition to a 1.5-kb spliced IE mRNA, there are at least three different E mRNAs, of which only the 2.5-kb unspliced version retains the complete ORF70 open reading frame. The protein encoded by ORF70 is a functional thymidylate synthase, one of four virus-encoded enzymes involved in nucleoside and nucleotide biosynthesis (16, 45, 49). Interestingly, another enzyme in this network, thymidine kinase (TK), is encoded by ORF21, which is transcribed in the opposite direction from ORF19, from what is very likely a bidirectional promoter (76). Continuing with this theme, we noticed that ORF2, encoding a third enzyme in the network, dihydrofolate reductase (vDHFR), lies immediately downstream of K3. Sequences immediately upstream of ORF2 are unresponsive to RTA (13), and it may be that ORF2 mRNAs originate from the ORF70 promoter, located a relatively short distance upstream (Fig. 1B). Lastly, the two subunits of ribonucleotide reductase (ORF61 and ORF62), a fourth nucleotide biosynthesis enzyme encoded by KSHV, are believed to initiate at an RTA-responsive promoter immediately upstream of ORF64, and the sequence includes a potential CSL binding site (13; our unpublished data). These correlations are intriguing because they hint at the possibility that the expression of functionally related genes may be coupled through the use of coregulators such as CSL. Counterparts to these enzymes are found in other herpesviruses, but in every case, they appear to be dispensable for replication in tissue culture. This is not true for infections of quiescent primary cells, where the levels of cellular nucleotide biosynthesis enzymes are generally very low and the virus must either upregulate the cellular genes, as shown for murine cytomegalovirus (20), or provide its own versions, as in the case of KSHV.
Based on this survey, it is very likely that several RTA-responsive promoters containing functional CSL binding sites remain to be discovered, either through analysis of individual regions or by alternative approaches such as chromatin immunoprecipitation. Knowing whether individual sites are occupied in the absence of RTA will be especially interesting given the ability of CSL to act as a repressor. So far, the emphasis has been on establishing a positive role for CSL in mediating promoter recruitment and transcriptional activation by RTA, but its role in maintaining the same genes in a repressed state may turn out to be important in the context of natural long-term infections.
Acknowledgments
We are indebted to a number of colleagues for valuable reagents and advice. In particular, we thank Gary Hayward and Diane Hayward for plasmids, Bettina Kempkes and Ursula Holter for SM224.9 cells, Jeff Vieira for Vero-rKSHV.219 cells, and Carolina Arias for production of a high-titer stock of BAC50. Insightful suggestions from various members of the Wilson and Mohr labs helped to shape these studies. Naoko Tanese and Ian Mohr provided constructive comments that strengthened the project and the manuscript.
This work was supported by grants from the NIH (GM61139 and S10 RR017970) and by a pilot project grant from the NYU CFAR (NIH/NIAID P30 AI027742) for HIV-associated malignancy research.
Footnotes
Published ahead of print on 11 November 2009.
REFERENCES
- 1.Borggrefe, T., and F. Oswald. 2009. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol. Life Sci. 66:1631-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray, S. J. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678-689. [DOI] [PubMed] [Google Scholar]
- 3.Carroll, K. D., F. Khadim, S. Spadavecchia, D. Palmeri, and D. M. Lukac. 2007. Direct interactions of KSHV/HHV-8 ORF50/Rta protein with the cellular protein octamer-1 and DNA are critical for specifying transactivation of a delayed-early promoter and stimulating viral reactivation. J. Virol. 81:8451-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, H., D. P. Dittmer, S. Y. Chul, Y. Hong, and J. U. Jung. 2005. Role of Notch signal transduction in Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 79:14371-14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, J., and D. Ganem. 2000. On the control of late gene expression in Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8). J. Gen. Virol. 81:2039-2047. [DOI] [PubMed] [Google Scholar]
- 6.Chang, P. J., D. Shedd, L. Gradoville, M. S. Cho, L. W. Chen, J. Chang, and G. Miller. 2002. Open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J. Virol. 76:3168-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, P. J., D. Shedd, and G. Miller. 2005. Two subclasses of Kaposi's sarcoma-associated herpesvirus lytic cycle promoters distinguished by open reading frame 50 mutant proteins that are deficient in binding to DNA. J. Virol. 79:8750-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J., F. Ye, J. Xie, K. Kuhne, and S. J. Gao. 2009. Genome-wide identification of binding sites for Kaposi's sarcoma-associated herpesvirus lytic switch protein, RTA. Virology 386:290-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damania, B., J. H. Jeong, B. S. Bowser, S. M. DeWire, M. R. Staudt, and D. P. Dittmer. 2004. Comparison of the Rta/Orf50 transactivator proteins of gamma-2-herpesviruses. J. Virol. 78:5491-5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng, H., Y. Liang, and R. Sun. 2007. Regulation of KSHV lytic gene expression. Curr. Top. Microbiol. Immunol. 312:157-183. [DOI] [PubMed] [Google Scholar]
- 12.Dou, S., X. Zeng, P. Cortes, H. Erdjument-Bromage, P. Tempst, T. Honjo, and L. D. Vales. 1994. The recombination signal sequence-binding protein RBP-2N functions as a transcriptional repressor. Mol. Cell. Biol. 14:3310-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellison, T. J., Y. Izumiya, C. Izumiya, P. A. Luciw, and H. J. Kung. 2009. A comprehensive analysis of recruitment and transactivation potential of K-Rta and K-bZIP during reactivation of Kaposi's sarcoma-associated herpesvirus. Virology 387:76-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fortini, M. E. 2009. Notch signaling: the core pathway and its posttranslational regulation. Dev. Cell 16:633-647. [DOI] [PubMed] [Google Scholar]
- 15.Ganem, D. 2006. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu. Rev. Pathol. 1:273-296. [DOI] [PubMed] [Google Scholar]
- 16.Gaspar, G., E. De Clercq, and J. Neyts. 2002. Human herpesvirus 8 gene encodes a functional thymidylate synthase. J. Virol. 76:10530-10532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glenn, M., L. Rainbow, F. Aurade, A. Davison, and T. F. Schulz. 1999. Identification of a spliced gene from Kaposi's sarcoma-associated herpesvirus encoding a protein with similarities to latent membrane proteins 1 and 2A of Epstein-Barr virus. J. Virol. 73:6953-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray, K. S., R. D. Allen, M. L. Farrell, J. C. Forrest, and S. H. Speck. 2008. Alternatively initiated gene 50/RTA transcripts expressed during murine and human gammaherpesvirus reactivation from latency. J. Virol. 83:14-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gribaudo, G., L. Riera, T. L. Rudge, P. Caposio, L. F. Johnson, and S. Landolfo. 2002. Human cytomegalovirus infection induces cellular thymidylate synthase gene expression in quiescent fibroblasts. J. Gen. Virol. 83:2983-2993. [DOI] [PubMed] [Google Scholar]
- 21.Gwack, Y., H. J. Baek, H. Nakamura, S. H. Lee, M. Meisterernst, R. G. Roeder, and J. U. Jung. 2003. Principal role of TRAP/mediator and SWI/SNF complexes in Kaposi's sarcoma-associated herpesvirus RTA-mediated lytic reactivation. Mol. Cell. Biol. 23:2055-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayward, S. D. 2004. Viral interactions with the Notch pathway. Semin. Cancer Biol. 14:387-396. [DOI] [PubMed] [Google Scholar]
- 23.Hengge, U. R., T. Ruzicka, S. K. Tyring, M. Stuschke, M. Roggendorf, R. A. Schwartz, and S. Seeber. 2002. Update on Kaposi's sarcoma and other HHV8 associated diseases. 1. Epidemiology, environmental predispositions, clinical manifestations, and therapy. Lancet Infect. Dis. 2:281-292. [DOI] [PubMed] [Google Scholar]
- 24.Hengge, U. R., T. Ruzicka, S. K. Tyring, M. Stuschke, M. Roggendorf, R. A. Schwartz, and S. Seeber. 2002. Update on Kaposi's sarcoma and other HHV8 associated diseases. 2. Pathogenesis, Castleman's disease, and pleural effusion lymphoma. Lancet Infect. Dis. 2:344-352. [DOI] [PubMed] [Google Scholar]
- 25.Herndier, B. G., A. Werner, P. Arnstein, N. W. Abbey, F. Demartis, R. L. Cohen, M. A. Shuman, and J. A. Levy. 1994. Characterization of a human Kaposi's sarcoma cell line that induces angiogenic tumors in animals. AIDS 8:575-581. [DOI] [PubMed] [Google Scholar]
- 26.Hsieh, J. J., S. Zhou, L. Chen, D. B. Young, and S. D. Hayward. 1999. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 96:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izumiya, Y., C. Izumiya, D. Hsia, T. J. Ellison, P. A. Luciw, and H. J. Kung. 2009. NF-kappaB serves as a cellular sensor of KSHV latency and negatively regulates K-Rta by antagonizing the RBP-Jκ coactivator. J. Virol. 83:4435-4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johannsen, E., E. Koh, G. Mosialos, X. Tong, E. Kieff, and S. R. Grossman. 1995. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jkappa and PU.1. J. Virol. 69:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovall, R. A. 2008. More complicated than it looks: assembly of Notch pathway transcription complexes. Oncogene 27:5099-5109. [DOI] [PubMed] [Google Scholar]
- 31.Kovall, R. A. 2006. Structures of CSL, Notch and Mastermind proteins: piecing together an active transcription complex. Curr. Opin. Struct. Biol. 17:117-127. [DOI] [PubMed] [Google Scholar]
- 32.Lan, K., D. A. Kuppers, and E. S. Robertson. 2005. Kaposi's sarcoma-associated herpesvirus reactivation is regulated by interaction of latency-associated nuclear antigen with recombination signal sequence-binding protein Jkappa, the major downstream effector of the Notch signaling pathway. J. Virol. 79:3468-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang, Y., and D. Ganem. 2004. RBP-J (CSL) is essential for activation of the K14/vGPCR promoter of Kaposi's sarcoma-associated herpesvirus by the lytic switch protein RTA. J. Virol. 78:6818-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao, W., Y. Tang, Y. L. Kuo, B. Y. Liu, C. J. Xu, and C. Z. Giam. 2003. Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 transcriptional activator Rta is an oligomeric DNA-binding protein that interacts with tandem arrays of phased A/T-trinucleotide motifs. J. Virol. 77:9399-9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, Y., Y. Cao, D. Liang, Y. Gao, T. Xia, E. S. Robertson, and K. Lan. 2008. Kaposi's sarcoma-associated herpesvirus RTA activates the processivity factor ORF59 through interaction with RBP-Jkappa and a cis-acting RTA responsive element. Virology 380:264-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu, F., J. Zhou, A. Wiedmer, K. Madden, Y. Yuan, and P. M. Lieberman. 2003. Chromatin remodeling of the Kaposi's sarcoma-associated herpesvirus ORF50 promoter correlates with reactivation from latency. J. Virol. 77:11425-11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu, M., J. Suen, C. Frias, R. Pfeiffer, M. H. Tsai, E. Chuang, and S. L. Zeichner. 2004. Dissection of the Kaposi's sarcoma-associated herpesvirus gene expression program by using the viral DNA replication inhibitor cidofovir. J. Virol. 78:13637-13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 41.Maier, S., M. Santak, A. Mantik, K. Grabusic, E. Kremmer, W. Hammerschmidt, and B. Kempkes. 2005. A somatic knockout of CBF1 in a human B-cell line reveals that induction of CD21 and CCR7 by EBNA-2 is strictly CBF1 dependent and that downregulation of immunoglobulin M is partially CBF1 independent. J. Virol. 79:8784-8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsumura, S., Y. Fujita, E. Gomez, N. Tanese, and A. C. Wilson. 2005. Activation of the Kaposi's sarcoma-associated herpesvirus major latency locus by the lytic switch protein RTA (ORF50). J. Virol. 79:8493-8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McWatters, B. J., R. M. Stenberg, and J. A. Kerry. 2002. Characterization of the human cytomegalovirus UL75 (glycoprotein H) late gene promoter. Virology 303:309-316. [DOI] [PubMed] [Google Scholar]
- 44.Nellesen, D. T., E. C. Lai, and J. W. Posakony. 1999. Discrete enhancer elements mediate selective responsiveness of enhancer of split complex genes to common transcriptional activators. Dev. Biol. 213:33-53. [DOI] [PubMed] [Google Scholar]
- 45.Nicholas, J., V. Ruvolo, J. Zong, D. Ciufo, H. G. Guo, M. S. Reitz, and G. S. Hayward. 1997. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J. Virol. 71:1963-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ong, C. T., H. T. Cheng, L. W. Chang, T. Ohtsuka, R. Kageyama, G. D. Stormo, and R. Kopan. 2006. Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J. Biol. Chem. 281:5106-5119. [DOI] [PubMed] [Google Scholar]
- 47.Pavlova, I., C. Y. Lin, and S. H. Speck. 2005. Murine gammaherpesvirus 68 Rta-dependent activation of the gene 57 promoter. Virology 333:169-179. [DOI] [PubMed] [Google Scholar]
- 48.Rimessi, P., A. Bonaccorsi, M. Sturzl, M. Fabris, E. Brocca-Cofano, A. Caputo, G. Melucci-Vigo, M. Falchi, A. Cafaro, E. Cassai, B. Ensoli, and P. Monini. 2001. Transcription pattern of human herpesvirus 8 open reading frame K3 in primary effusion lymphoma and Kaposi's sarcoma. J. Virol. 75:7161-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serio, T. R., N. Cahill, M. E. Prout, and G. Miller. 1998. A functionally distinct TATA box required for late progression through the Epstein-Barr virus life cycle. J. Virol. 72:8338-8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song, M. J., H. J. Brown, T. T. Wu, and R. Sun. 2001. Transcription activation of polyadenylated nuclear RNA by RTA in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:3129-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song, M. J., X. Li, H. J. Brown, and R. Sun. 2002. Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 76:5000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Staudt, M. R., and D. P. Dittmer. 2007. The Rta/Orf50 transactivator proteins of the gamma-herpesviridae. Curr. Top. Microbiol. Immunol. 312:71-100. [DOI] [PubMed] [Google Scholar]
- 55.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang, S., K. Yamanegi, and Z. M. Zheng. 2004. Requirement of a 12-base-pair TATT-containing sequence and viral lytic DNA replication in activation of the Kaposi's sarcoma-associated herpesvirus K8.1 late promoter. J. Virol. 78:2609-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tun, T., Y. Hamaguchi, N. Matsunami, T. Furukawa, T. Honjo, and M. Kawaichi. 1994. Recognition sequence of a highly conserved DNA binding protein RBP-Jkappa. Nucleic Acids Res. 22:965-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vieira, J., and P. M. O'Hearn. 2004. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology 325:225-240. [DOI] [PubMed] [Google Scholar]
- 59.Wang, S. E., F. Y. Wu, H. Chen, M. Shamay, Q. Zheng, and G. S. Hayward. 2004. Early activation of the Kaposi's sarcoma-associated herpesvirus RTA, RAP, and MTA promoters by the tetradecanoyl phorbol acetate-induced AP1 pathway. J. Virol. 78:4248-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, S. E., F. Y. Wu, M. Fujimuro, J. Zong, S. D. Hayward, and G. S. Hayward. 2003. Role of CCAAT/enhancer-binding protein alpha (C/EBPalpha) in activation of the Kaposi's sarcoma-associated herpesvirus (KSHV) lytic-cycle replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J. Virol. 77:600-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, S. E., F. Y. Wu, Y. Yu, and G. S. Hayward. 2003. CCAAT/enhancer-binding protein-alpha is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J. Virol. 77:9590-9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang, Y., Q. Tang, G. G. Maul, and Y. Yuan. 2006. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: dual role of replication and transcription activator. J. Virol. 80:12171-12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, Y., and Y. Yuan. 2007. Essential role of RBP-Jkappa in activation of the K8 delayed-early promoter of Kaposi's sarcoma-associated herpesvirus by ORF50/RTA. Virology 359:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.West, J. T., and C. Wood. 2003. The role of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 regulator of transcription activation (RTA) in control of gene expression. Oncogene 22:5150-5163. [DOI] [PubMed] [Google Scholar]
- 65.Whelan, J. T., S. L. Forbes, and F. E. Bertrand. 2007. CBF-1 (RBP-Jkappa) binds to the PTEN promoter and regulates PTEN gene expression. Cell Cycle 6:80-84. [DOI] [PubMed] [Google Scholar]
- 66.Wilson, A. C., M. A. Cleary, J. S. Lai, K. LaMarco, M. G. Peterson, and W. Herr. 1993. Combinatorial control of transcription: the herpes simplex virus VP16-induced complex. Cold Spring Harbor Symp. Quant. Biol. 58:167-178. [DOI] [PubMed] [Google Scholar]
- 67.Wilson, S. J., E. H. Tsao, B. L. Webb, H. Ye, L. Dalton-Griffin, C. Tsantoulas, C. V. Gale, M. Q. Du, A. Whitehouse, and P. Kellam. 2007. XBP-1s transactivates the KSHV ORF50 promoter, linking plasma cell differentiation to KSHV reactivation from latency. J. Virol. 81:13578-13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wing, B. A., R. A. Johnson, and E. S. Huang. 1998. Identification of positive and negative regulatory regions involved in regulating expression of the human cytomegalovirus UL94 late promoter: role of IE2-86 and cellular p53 in mediating negative regulatory function. J. Virol. 72:1814-1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wysocka, J., and W. Herr. 2003. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem. Sci. 28:294-304. [DOI] [PubMed] [Google Scholar]
- 70.Xu, D., T. Coleman, J. Zhang, A. Fagot, C. Kotalik, L. Zhao, P. Trivedi, C. Jones, and L. Zhang. 2007. Epstein-Barr virus inhibits Kaposi's sarcoma-associated herpesvirus lytic replication in primary effusion lymphomas. J. Virol. 81:6068-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu, Y., D. P. Aucoin, A. R. Huete, S. A. Cei, L. J. Hanson, and G. S. Pari. 2005. A Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50 deletion mutant is defective for reactivation of latent virus and DNA replication. J. Virol. 79:3479-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ye, J., D. Shedd, and G. Miller. 2005. An Sp1 response element in the Kaposi's sarcoma-associated herpesvirus open reading frame 50 promoter mediates lytic cycle induction by butyrate. J. Virol. 79:1397-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoo, S. M., F. C. Zhou, F. C. Ye, H. Y. Pan, and S. J. Gao. 2005. Early and sustained expression of latent and host modulating genes in coordinated transcriptional program of KSHV productive primary infection of human primary endothelial cells. Virology 343:47-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu, F., J. Feng, J. N. Harada, S. K. Chanda, S. C. Kenney, and R. Sun. 2007. B cell terminal differentiation factor XBP-1 induces reactivation of Kaposi's sarcoma-associated herpesvirus. FEBS Lett. 581:3485-3488. [DOI] [PubMed] [Google Scholar]
- 75.Yu, Y., S. E. Wang, and G. S. Hayward. 2005. The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity 22:59-70. [DOI] [PubMed] [Google Scholar]
- 76.Zhang, L., J. Chiu, and J. C. Lin. 1998. Activation of human herpesvirus 8 (HHV-8) thymidine kinase (TK) TATAA-less promoter by HHV-8 ORF50 gene product is SP1 dependent. DNA Cell Biol. 17:735-742. [DOI] [PubMed] [Google Scholar]
- 77.Ziegelbauer, J., A. Grundhoff, and D. Ganem. 2006. Exploring the DNA binding interactions of the Kaposi's sarcoma-associated herpesvirus lytic switch protein by selective amplification of bound sequences in vitro. J. Virol. 80:2958-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]