Abstract
We characterized the cellular immune response to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in 12- to 14-month-old BALB/c mice, a model that mimics features of the human disease. Following intranasal administration, the virus replicated in the lungs, with peak titers on day 2 postinfection. Enhanced production of cytokines (tumor necrosis factor alpha [TNF-α] and interleukin-6 [IL-6]) and chemokines (CXCL10, CCL2, CCL3, and CCL5) correlated with migration of NK cells, macrophages, and plasmacytoid dendritic cells (pDC) into the lungs. By day 7, histopathologic evidence of pneumonitis was seen in the lungs when viral clearance occurred. At this time, a second wave of enhanced production of cytokines (TNF-α, IL-6, gamma interferon [IFN-γ], IL-2, and IL-5), chemokines (CXCL9, CXCL10, CCL2, CCL3, and CCL5), and receptors (CXCR3, CCR2, and CCR5), was detected in the lungs, associated with an influx of T lymphocytes. Depletion of CD8+ T cells at the time of infection did not affect viral replication or clearance. However, depletion of CD4+ T cells resulted in an enhanced immune-mediated interstitial pneumonitis and delayed clearance of SARS-CoV from the lungs, which was associated with reduced neutralizing antibody and cytokine production and reduced pulmonary recruitment of lymphocytes. Innate defense mechanisms are able to control SARS-CoV infection in the absence of CD4+ and CD8+ T cells and antibodies. Our findings provide new insights into the pathogenesis of SARS, demonstrating the important role of CD4+ but not CD8+ T cells in primary SARS-CoV infection in this model.
The global outbreak of severe acute respiratory syndrome (SARS) in 2003 that infected more than 8,000 people in 29 countries across five continents, with 774 deaths reported by the World Health Organization (54), was caused by a highly contagious coronavirus designated SARS-CoV (33). The elderly were more likely to die from SARS-CoV infection than younger people (7), with a case-fatality rate of 50% in people older than 65 years (14, 53). Disease pathogenesis in SARS is complex, with multiple factors leading to severe pulmonary injury and dissemination of the virus to other organs. High viral load; systemic infection; a cytokine storm with high levels of CXCL10/IP-10, CCL3/MIP-1α, and CCL2/MCP-1; massive lung infiltration by monocytes and macrophages; and rapid depletion of T cells are hallmarks of SARS (5, 13, 15, 21, 28, 35). The role of neutralizing antibodies (Abs) in protection from SARS-CoV infection has been well documented. Virus-specific neutralizing Abs reduce viral load, protect against weight loss, and reduce histopathology in animal models (42, 47, 48). Although the role of type I interferons (IFNs) in the natural history of SARS is controversial (5, 9, 59), the innate defense system appears to be critical for controlling SARS-CoV replication in mice (23, 41). Mice lacking normal innate signaling due to STAT1 or MyD88 deficiency are highly susceptible to SARS-CoV infection. Virus-specific T-cell responses are present in convalescent patients with SARS (27, 55). However, little is known about the role of T cells in the acute phase of SARS.
Several mouse models have been developed for the in vivo study of SARS pathogenesis. However, no single model accurately reproduces all aspects of the human disease. SARS-CoV replicates in the upper and lower respiratory tracts of 4- to 8-week-old mice and is cleared rapidly; infection is associated with transient mild pneumonitis, and cytokines are not detectable in the lungs (20, 42, 49). A SARS-CoV isolate that was adapted by serial passage in mice (MA-15) replicates to a higher titer and for a longer duration in the lungs than the unadapted (Urbani) virus and is associated with viremia and mortality in young mice (36), but the histologic changes in the lungs are caused by high titers of virus and cell death without significant infiltrates of inflammatory cells. The heightened susceptibility of elderly patients to SARS led us to develop a pneumonia model in 12- to 14-month-old (mo) BALB/c mice using the Urbani virus. In this model, pulmonary replication of virus was associated with signs of clinical illness and histopathological evidence of disease characterized by bronchiolitis, interstitial pneumonitis, diffuse alveolar damage, and fibrotic scarring (3), thus resembling SARS in the elderly. We evaluated the host response to SARS-CoV infection by examining the gene expression profile in the senescent mouse model and found a robust response to virus infection, with an increased expression of several immune response and cell-to-cell signaling genes, including those for tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), CCL2, CCL3, CXCL10, and IFN-γ (1).
In this study, we characterize the cellular immune response to SARS-CoV infection in 12- to 14-mo BALB/c mice in terms of the protein and gene expression of inflammatory mediators, migration of inflammatory cells, and virus-specific T-cell responses in the lungs during the course of disease. We evaluated the role of T cells in disease pathogenesis and viral clearance by depleting T-cell subsets at the time of infection and found an important role for CD4+ T cells (but not CD8+ T cells) in primary infection with SARS-CoV in this model.
MATERIALS AND METHODS
Virus.
SARS-CoV (Urbani strain), a generous gift from L. J. Anderson and T. G. Ksiazek (Centers for Disease Control and Prevention, Atlanta, GA), was propagated in Vero cells, with a titer of 106.5 50% tissue culture infective doses (TCID50)/ml. Vero cells were maintained in OptiPro SFM (Invitrogen, CA). All work with infectious virus was performed inside a biosafety cabinet in a biosafety level 3 facility.
Mouse model of SARS.
The animal studies were approved by the National Institutes of Health Animal Care and Use Committee. Female BALB/c mice, 12 to 14 mo, were purchased from Taconic (Germantown, NY). We administered 105 TCID50 of SARS-CoV intranasally (i.n.) to mice as previously described (42). Leibovitz 15 medium was used to mock infect control groups of mice. Lungs were harvested at serial time points without perfusion for virus titration, cytokine assays, histopathology, and flow cytometry.
Depletion of T-cell subsets.
Monoclonal antibodies (MAb) Gk1.5 specific for mouse CD4, 2.43 specific for mouse CD8, and SFR3-DR5 specific for human leukocyte antigen as an isotype control were used for in vivo depletion as described previously (17). All MAbs are rat IgG2b and were prepared as ascites fluid and delipified by the National Cell Culture Center (Biovest International). Mice received 1 mg/ml of MAb for each dose, given intraperitoneally (i.p.) 3 days before and 3, 7, and 10 days after infection.
Passive transfer of serum Ab.
Postinfection hyperimmune SARS (HIS) antiserum was generated in BALB/c mice following SARS-CoV infection. Mice treated with anti-CD4 or anti-CD4 and anti-CD8 MAbs were given an i.p. injection of 500 μl of serum on day 7 postinfection (p.i.). The control group received normal (nonimmune) BALB/c mouse serum (Harlan, Indianapolis, IA); the experimental groups received either undiluted HIS or a 1:4 dilution (in phosphate-buffered saline [PBS]) of HIS. Lungs were harvested on day 9 p.i.
Virus titration.
Supernatants of 10% (wt/vol) lung homogenates were prepared and titrated on Vero cell monolayers in 24- and 96-well plates as previously described (42). Virus titers are expressed as TCID50 per g of tissue. The lower limit of detection was 101.5 TCID50/g.
Histopathology and immunohistochemistry (IHC).
Mice were euthanized by cervical dislocation on days 7, 9, and 12 p.i. The lungs were inflated with 10% formalin and embedded in paraffin wax. Lung tissue sections were prepared for histopathological examination and evaluated by using conventional hematoxylin and eosin (H&E) staining and an immunoalkaline phosphatase staining technique. The primary Ab was a polyclonal rabbit Ab against SARS-CoV nucleocapsid used at a dilution of 1/1,000 as described previously (37). The scoring system was developed prior to the experiment and allowed for blinded review. Mild multifocal interstitial inflammation was scored as 1 to 2, moderate disease as 3 to 4, and severe disease as 5.
BD CBA and ELISA for protein expression.
Supernatants of 10 or 20% (wt/vol) lung homogenates were used for detection of cytokine and chemokine protein expression using BD cytometric bead array (CBA) kits (BD Biosciences), the Bio-plex Protein Array system (Bio-Rad, CA), Quantikine immunoassay kits (R&D, MN), and IFN-α and IFN-β enzyme-linked immunosorbent assay (ELISA) kits (PBL, NJ). Data are expressed as pg or ng of protein per g of tissue. The lower limit of detection for each protein is included in the kit protocol.
Quantitative RT-PCR.
Total RNA (0.5 μg) was isolated from lung samples using the RNeasy mini kit (Qiagen) and reverse transcribed into cDNA using StrataScript first-strand synthesis (Stratagene). The following primers were used: 18S forward (5′-GGTACAGTGAAACTGCGAAT-3′) and 18S reverse (5′-CAGTTATCCAAGTAGGAGAG-3′), CCR2 forward (5′-GTTACCTCAGTTCATCCA-3′) and CCR2 reverse (5′-CAAGGCTCACCATCATCGTAGTC-3′), CCR5 forward (5′-TTGCAAACGGTGTTCAATTTTC-3′) and CCR5 reverse (5′-TCTCCTGTGGATCGGGTATAGAC-3′), CXCR3 forward (5′-GCTAGATGCCTCGGACTTTGC-3′) and CXCR3 reverse (5′-CGCTCTCGTTTTCCCCATAA-3′), CCL2 forward (5′-GCTGGAGCATCCACGTGTT-3′) and CCL2 reverse (5′-ATCTTGCTGGTGAATGAGTAGCA-3′), CCL3 forward (5′-CCAAGTCTTCTCAGCGCCAT-3′) and CCL3 reverse (5′-GAATCTTCCGGCTGTAGGAGAAG-3′), CCL5 forward (5′-GTGCCCACGTCAAGGAGTAT-3′) and CCL5 reverse (5′-GGGAAGCTATACAGGGTCA-3′), CXCL9 forward (5′-TGCACGATGCTCCTGCA-3′) and CXCL9 reverse (5′-AGGTCTTTGAGGGATTTGTAGTGG-3′), and CXCL10 forward (5′-GACGGTCCGCTGCAACTG-3′) and CXCL10 reverse (5′-GCTTCCCTATGGCCCTCATT-3′). Quantitative reverse transcription-PCR (RT-PCR) was performed by heating cDNA samples mixed with primers and Brilliant SYBR green QPCR master mix (Stratagene) at 95°C for 10 min, followed by 40 cycles of amplification at 95°C for 30 s, 55 to 65°C for 1 min, and 72°C for 1 min using an Mx4000 multiplex quantitative PCR system (Stratagene). All infected samples and mock controls were tested at the same time to enable comparisons of 18S rRNA-normalized values. Negative control samples without RT or RNA were included. Each data point was examined for integrity by analysis of the amplification plot and dissociation curves. For data analysis, the amount of reaction product was determined using the comparative threshold cycle method, as described elsewhere (30). mRNA levels for each primer pair were normalized to the 18S rRNA housekeeping gene, and fold changes were calculated against mock-infected controls.
Preparation of lung single-cell suspensions.
To obtain single-cell suspensions, lungs were incubated in RPMI 1640 containing 10% fetal bovine serum (FBS) and 2 mg/ml collagenase A (Sigma) at 37°C for 30 to 45 min. A single-cell suspension was prepared after red blood cell (RBC) lysis, filtered through a 40-nm cell strainer (BD), and used for fluorescence-activated cell sorter (FACS) analysis.
Flow cytometry.
All Abs used in this study were purchased from BD Biosciences, except PCDA-1, which was obtained from Miltenyi (Auburn, CA), and F4/80, which was from eBioscience (San Diego, CA). Leukocytes isolated from lungs were cultured in the presence of 1 μM peptide (KCYGVSATKL, an H-2d-restricted CD8+ T-cell epitope) (24) and 25 IU per ml of IL-2 at 37°C for 5 h, incubated with anti-CD107a-fluorescein isothiocyanate (FITC) and anti-CD107b-FITC MAbs (0.1 and 0.5 μg per well), respectively. After treatment with an Fc block for 15 min on ice, cells were surface stained with anti-CD8-allophycocyanin (ALPC) for 30 min on ice and were fixed and permeabilized using a Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer's instructions. For intracellular staining, phycoerythrin (PE)-conjugated anti-mouse IFN-γ MAb (clone XMG1.2) was used. To evaluate the CD4+ T-cell response, lung cells were stimulated with anti-CD28 and anti-CD49d MAbs (0.1 μg per well) and SARS-CoV spike (S) protein (0.5 μg per well; Protein Sciences Corporation, Meriden, CT) for 16 h, with Golgi-plug, Golgi-stop, and anti-CD107a/b MAbs added for the last 8 h. Cells were then stained with anti-CD4-peridinin chlorophyll protein (PerCP)- and PE-conjugated rat anti-mouse IFN-γ MAbs (clone XMG1.2) using the Cytofix/Cytoperm kit for the detection of IFN-γ-positive CD4+ lymphocytes. The final analysis and graphical output were performed using FlowJo software (Tree Star, Inc.). Cell populations were identified as CD11c+ PDCA1+ for plasmacytoid dendritic cells (pDC), CD3− DX5+ for NK cells, CD3+ DX5+ for NK T cells, CD11b+ F4/80+ for macrophages, and CD11c− Gr1+ for neutrophils.
Neutralizing Ab assay.
Twofold dilutions of heat-inactivated serum were tested in a microneutralization assay using 100 TCID50 of SARS-CoV as previously described (42). The presence of viral cytopathic effect was read on days 3 and 4.
ELISA for Ab.
Microtiter plates were coated with 100 ng per well of recombinant truncated SARS-CoV S protein (Protein Sciences Corp., Meriden, CT). Serially diluted heat-inactivated mouse sera were incubated overnight. Bound Abs were detected with alkaline phosphatase-conjugated goat anti-mouse IgG and p-nitrophenyl phosphate (Sigma, MO) substrate. An optical absorbance of >0.2 at 405 nm was considered positive.
Statistical analysis.
Statistically significant differences between groups of mice were determined by Student's t test and the Mann-Whitney U test. P values of <0.05 were considered significant.
RESULTS
SARS-CoV infection is associated with pneumonitis in 12- to 14-mo BALB/c mice.
Pulmonary viral infection was induced in senescent BALB/c mice by i.n. administration of 105 TCID50 SARS-CoV. High titers of virus (108.6 TCID50/g) were detected in the lungs as early as day 2 p.i. Titers remained high for several days (∼105 TCID50/g on day 7), after which titers decreased significantly and were just above the limit of detection (101.5 TCID50/g) by day 9 p.i. (data not shown).
Histologic examination of the lungs on day 2 p.i. showed focal perivascular infiltrates comprised predominantly of macrophages and some lymphocytes and airway lesions characterized by bronchiolar epithelial necrosis and luminal necrotic debris (data not shown). On day 7 p.i., multifocal interstitial infiltrates comprised predominantly of mononuclear cells were identified, while high titers of virus remained in the lungs. Perivascular infiltrates persisted on day 9. Diffuse alveolar damage and fibrotic scarring were seen on day 9 and day 12 p.i., as described previously (references 37 and 38 and data not shown).
SARS-CoV infection induces a biphasic pattern of increased expression of inflammatory mediators in the lungs.
To identify the inflammatory mediators involved in the pathogenesis of SARS-CoV-induced pneumonitis, we measured the protein levels of cytokines and chemokines in lung homogenates (Fig. 1A and B). We describe events in the first 1 to 5 days p.i. as early and those during 6 to 13 days p.i. as late events and noted four patterns, with early, late, biphasic, or no increases, respectively. First, a modest increase in the levels of IFN-α and IFN-β was detected in the lungs early (on day 3) following SARS-CoV infection, but no statistical difference was found compared to mock-infected control samples (P > 0.05). Second, a marked increase in inflammatory chemokines CCL5/RANTES and CXCL9/MIG, accompanied by an increase of inflammatory cytokines IFN-γ, IL-2, and IL-5, was detected in the lungs predominantly at day 7, the late stage of infection (P < 0.05). Third, a biphasic pattern of significantly increased production of proinflammatory cytokines TNF-α and IL-6 and chemokines CCL2/MCP-1, CCL3/MIP-1α, and CXCL10/IP-10 was seen both early and late following infection (P < 0.05). The first wave of these cytokines and chemokines was detected as early as day 2 p.i. High levels of these cytokines and chemokines remained on day 3 and decreased dramatically on day 5 p.i. By day 7, when pneumonitis was observed, we detected a second peak of production of these proteins. Fourth, IL-4, IL-10, and IL-12 were not detectable throughout the course of disease (data not shown).
FIG. 1.
Kinetics of inflammatory mediators in the lungs following SARS-CoV infection. Protein levels of cytokines (A) and chemokines (B) and fold increase in gene transcripts of chemokines and the receptors (C) were measured in the lungs at the indicated time points following i.n. inoculation of 105 TCID50 SARS-CoV. Data are shown as mean ± standard error of the mean (SEM) for four mice at each time point, and the units are reported next to the name of the protein. *, P < 0.05 for the comparison between mock- and SARS-infected groups by Student's t test.
Consistent with the findings of chemokine protein levels, a biphasic pattern of increased expression of chemokine mRNA was observed in the lungs early and late in infection (Fig. 1C). An increase in mRNA transcripts for CCL2/MCP-1, CCL3/MIP-1α, and CXCL10/IP-10 was detected in the lung early on days 2 and 3 p.i., when peak viral replication occurred. Compared to those in mock-infected animals, these mRNA transcripts in SARS-CoV-infected lungs significantly increased at day 2 p.i., declined thereafter, and then showed a moderate increase on day 7. In contrast, mRNA transcripts for CCL5/RANTES and CXCL9/MIG increased predominantly late in infection on days 5 and 7, when histopathologic evidence of pneumonitis was seen in the infected lungs.
An increase in mRNA transcripts for CCR2, CCR5, and CXCR3, which are receptors for the upregulated chemokines (CCR2 for CCL2/MCP-1, CCR5 for CCL3/MIP-1a and CCL5/RANTES, and CXCR3 for CXCL9/MIG and CXCL10/IP-10), was also found in the lungs after infection with SARS-CoV (Fig. 1C). Compared to those in mock-infected lungs, a 4-fold increase in CCR2 mRNA and a >9-fold increase in CCR5 or CXCR3 mRNA were detected around days 7 to 9 p.i. The profile of increased expression of these receptor transcripts correlated closely with the upregulation of their ligands in the SARS-CoV-infected lung in the late phase of infection.
SARS-CoV infection induces two waves of recruitment of inflammatory cells into the lungs.
To analyze the cellular responses to SARS-CoV infection, we evaluated cell migration into the lungs. FACS analyses revealed a local accumulation of leukocytes in the lungs following infection (Fig. 2A and B). We observed a significant increase in the number of CD45+ leukocytes in the lungs of SARS-CoV-infected mice 7 days p.i. (P = 0.0015). The number of CD45+ leukocytes slightly increased as early as day 2 p.i., reaching a peak on day 7 and progressively declining by days 9 and 12 p.i. Three patterns of cell migration were detected in the lungs following SARS-CoV infection. First, plasmacytoid DC (pDC) migrated into the lungs at the early phase of infection. The number of pDC peaked on day 2 (P = 0.001), followed by a sharp decline by day 7 and day 9 p.i. (P < 0.001). Second, the pulmonary migration of inflammatory cells (such as NK T cells, NK cells, macrophages, and CD4+ T cells) began on day 2, increased rapidly to reach a peak on day 7 and/or 9 (P < 0.01), and declined thereafter. Third, neutrophils and CD8+ T cells migrated into the lungs at the late phase of infection. A large number of neutrophils were detected in the lungs of mock-infected senescent mice, and these numbers increased modestly on day 7 p.i. (P < 0.05). The number of CD8+ T cells increased sharply on days 7 and 9 (P < 0.05), remaining at a high level or declining by day 12 p.i. The two waves of inflammatory cells recruited into the lungs at the early and late phases of infection correlated well with the biphasic expression of chemokines and the receptors in the lungs of SARS-CoV-infected mice (Fig. 1B and C).
FIG. 2.
Presence of inflammatory cells in the lungs of mice following SARS-CoV infection. (A) FACS analysis of inflammatory cells (CD11c+ PDCA1+ for pDC, CD3− DX5+ for NK, CD3+ DX5+ for NKT, CD11b+ F4/80+ for macrophages, and CD11c− Gr1+ for neutrophils) in mock-infected and SARS-CoV-infected lungs. Cells for analysis are gated on a CD45+ population. (B) Total number of inflammatory cells isolated from the lungs on the indicated day p.i. Data are the means ± SEMs for three to five mice analyzed at each time point. Results from one representative experiment out of three are shown. *, P < 0.05; **, P < 0.01 (for the comparison between mock-and SARS-CoV-infected groups) by Student's t test. (C) FACS analysis of IFN-γ-producing CD8+ and CD4+ T cells isolated from the lungs of SARS-CoV-infected mice on day 7 p.i., with or without ex vivo stimulation with PMA/ionomycin or SARS-CoV S or N peptides. Cells for analysis are gated on a CD3+ population. (D) Total number of CD8+ and CD4+ T cells producing IFN-γ in the lungs of mock- or SARS-CoV-infected mice, with or without ex vivo stimulation with PMA/ionomycin or SARS-CoV S or N peptides. Data are from pools of cells from four mice analyzed at day 7 p.i. Results from a representative experiment out of two is shown.
SARS-CoV infection induces an enhanced virus-specific T-cell response in the lungs.
To determine the function of T cells in response to SARS-CoV infection, we measured the number and frequency of IFN-γ-producing CD8+ and CD4+ T cells isolated from the lungs of mock- and SARS-CoV-infected mice on day 7 p.i. by intracellular staining, with and without exogenous stimulation with phorbol myristate acetate (PMA)/ionomycin or viral peptides (Fig. 2C and D). FACS analysis revealed an increased frequency of CD8+ IFN-γ+ and CD4+ IFN-γ+ cells seen in the lungs of SARS-CoV-infected mice following exogenous stimulation with SARS-CoV S protein and to a lesser extent nucleocapsid (N) proteins and PMA/ionomycin. Compared to that in mock-infected mice, the number of CD8+ IFN-γ+ cells increased from 1.6 × 104 to 4.3 × 104 cells/lung in SARS-CoV-infected mice following stimulation with PMA/ionomycin and from 0 to 1 × 105 cells/lung when stimulated with a pool of overlapping peptides from the SARS-CoV S protein. The number of CD4+ IFN-γ+ cells increased from 1.1 × 104 cells/lung in mock-infected mice to 3.7 × 104 cells/lung in SARS-CoV-infected mice when stimulated with PMA/ionomycin and from 0 to 1.9 × 105 cells/lung when stimulated with SARS-CoV S peptides. Stimulation with a pool of overlapping peptides from the SARS-CoV N protein did not significantly increase IFN-γ production by T cells. This result indicates that antigen-specific T-cell responses occurred to the SARS-CoV S protein but not to the N protein. CD4+ T cells exhibited a higher IFN-γ response to S peptides than CD8+ T cells.
Taking the above data together, our studies showed an enhanced virus-specific T-cell response in the lungs that coincided with the development of pneumonitis and was accompanied by viral clearance (see Fig. 7 and data not shown). These data indicate that T cells may play an important role in SARS-CoV infection.
FIG. 7.
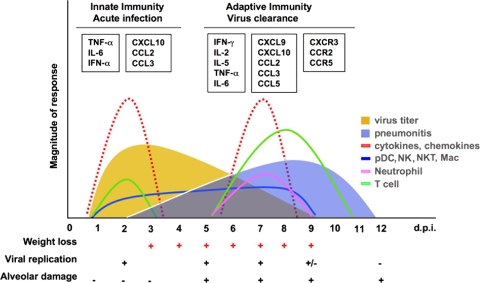
Time course of host responses to primary SARS-CoV infection in senescent mice. A biphasic expression of inflammatory mediators associated with cellular infiltration into the lungs of infected mice, coincident with peaks in viral replication and clearance, respectively, is seen. Clinical illness such as weight loss was observed coincident with pulmonary viral replication, while lung pathology (pneumonitis) was associated with T-cell infiltration when virus was cleared from the lung.
Depletion of CD4+ T cells (but not CD8+ T cells) results in delayed pulmonary viral clearance.
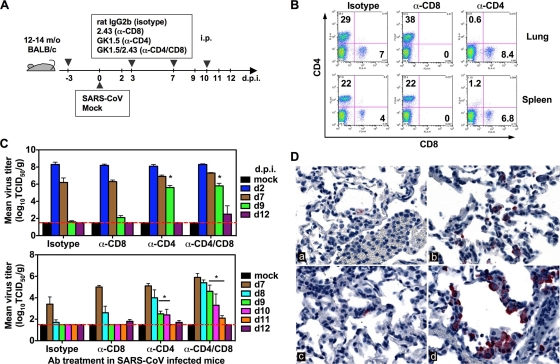
To evaluate the role of T cells in disease pathogenesis and clearance of SARS-CoV, we depleted T-cell subsets before and after infection by intraperitoneal (i.p.) injection of MAbs against CD4+ and/or CD8+ T cells (Fig. 3A). To determine the completeness of the depletion on days 8, 9, 10, 11, and 12 p.i., we used clones RM4-5 (anti-CD4) and 53-6.7 (anti-CD8) for cell staining, which are different from the anti-CD4 (GK1.5) and anti-CD8 (2.43) clones used for T-cell depletion. FACS analysis revealed that treatment with the MAbs at the time of infection led to a nearly complete depletion of T-cell subsets from the lungs and spleen (Fig. 3B).
FIG. 3.
Delayed viral clearance in the lungs of SARS-CoV-infected mice depleted of CD4+ T cells. (A) Depletion of T-cell subsets in senescent mice infected with SARS-CoV. Mice were treated with MAbs i.p. at 3 days before and 3 and 7 days after SARS-CoV infection. (B) FACS analysis of CD4 and CD8 expression on lymphocytes isolated from lungs and spleen on day 9 p.i. The recorded numbers denote the percentages of CD4+ or CD8+ cells in the total lymphocyte population. (C) Virus titers in the lungs were determined on the indicated day p.i. The limit of detection was 101.5 TCID50/g of tissue (dashed line). Data are means ± SEMs for four mice at each time point. (D) IHC staining of SARS-CoV antigen in the lungs of mice depleted of T-cell subsets. By day 9 p.i., SARS-CoV antigens (red) are extremely sparse or entirely absent from the lungs of infected mice (a) and are rare in the lungs of mice depleted of CD8+ T cells (c); however, abundant viral antigens remain in the lungs of mice depleted of CD4+ T cells (b) or both CD4+ and CD8+ T cells (d) at day 9 (immunoalkaline phosphatase stain with naphthol-fast red and hematoxylin counterstain).
SARS-CoV replicated efficiently in the lungs of mice that received the isotype control Ab and reached a peak titer on day 2 p.i. Levels of virus remained high through day 7 and declined below the limit of detection of 101.5 TCID50/g by day 9 p.i. (Fig. 3C, top). The mice depleted of CD8+ T cells cleared the virus from the lungs as well as the isotype control-treated animals did and exhibited a near complete clearance of virus by day 9 p.i. However, depletion of CD4+ T cells resulted in delayed viral clearance from the lungs. High titers of virus were detected in the lungs of mice depleted of CD4+ or CD4+ and CD8+ cells on day 9 p.i. (P = 0.00001 compared to isotype control-treated animals by Student's t test). By day 12, one of four mice depleted of both CD4+ and CD8+ T cells still had virus detectable in the lungs. The kinetics of viral clearance was further evaluated by daily examination from day 7 to day 12 p.i. (Fig. 3C, bottom). Complete clearance of virus was observed by day 9 in isotype control-treated animals and in mice depleted of CD8+ T cells. Prolonged viral replication was detected in mice depleted of CD4+ or CD4+ and CD8+ T cells, with a decline of virus titer below the limit of detection by day 11 and day 12 p.i., respectively. IHC staining for SARS-CoV antigen on day 9 p.i. confirmed the presence of viral antigen in the lungs of mice depleted of CD4+ T cells or both CD4+ and CD8+ T cells (Fig. 3D).
Depletion of CD4+ T cells results in a diminished virus-specific Ab response.
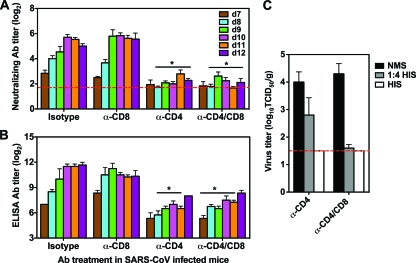
In order to identify the mechanism(s) by which CD4+ T cells control SARS-CoV infection, we measured neutralizing Ab titers in the sera of mice infected with SARS-CoV following depletion of T-cell subsets (Fig. 4A). Titers of SARS-CoV-specific neutralizing Abs were detected in isotype control-treated mice on day 7 p.i., reached a peak titer of 25.7 on day 10, and plateaued. Depletion of CD8+ T cells did not affect production of neutralizing Abs, and the virus-specific neutralizing Ab response seen in both isotype control-treated and CD8-depleted mice coincided with viral clearance on day 9 p.i. However, depletion of CD4+ cells resulted in a diminished neutralizing Ab response along with delayed viral clearance from the lungs. Compared to isotype control-treated animals, a significantly lower neutralizing Ab titer was detected from day 8 to day 12 p.i. in mice depleted of CD4+ or both CD4+ and CD8+ cells (P < 0.05 by Student's t test). In addition, mice depleted of CD4+ or both CD4+ and CD8+ cells exhibited reduced SARS-CoV S protein-specific ELISA IgG Ab production (Fig. 4B) from day 8 to day 11 p.i. (P < 0.05 by Student's t test), while a high titer of S protein-specific ELISA IgG Ab was detected in isotype control-treated and CD8-depleted mice. These data show a close relationship between the CD4-dependent virus-specific Ab response and viral clearance, indicating the important role that Abs play in the control of SARS-CoV infection.
FIG. 4.
Reduced Ab production in mice depleted of CD4+ T cells. Mice were treated with isotype control and MAbs against T-cell subsets as described in Materials and Methods. (A) Neutralizing Ab titers in the serum were determined on the indicated day p.i. Geometric mean neutralizing Ab titers from four mice per group are expressed as log2. (B) Virus-specific IgG titers in the serum were determined by ELISA on the indicated day p.i. Data are means ± SEMs for four mice at each time point. (C) HIS or NMS (500 μl per mouse) was given i.p. to mice depleted of CD4+ or CD4+ and CD8+ T cells on day 7 p.i., and virus titers in the lungs were determined on day 9 p.i. The limit of detection was 101.5 TCID50/g of tissue (dashed line). Data are means ± SEMs for four mice at each time point.
Passive transfer of SARS-specific Abs restores viral clearance in mice depleted of CD4+ T cells.
We have previously shown that passive transfer of HIS can protect senescent mice from SARS-CoV infection (48). We passively transferred HIS into mice depleted of CD4+ or both CD4+ and CD8+ cells on day 7 p.i. and assessed virus titers in the lungs 2 days later to determine whether Abs could clear the virus in the absence of T cells. On day 9 p.i., high titers of virus were detected in the lungs of mice that received normal mouse serum (NMS) that did not contain neutralizing Abs. Passive transfer of HIS dramatically reduced pulmonary virus titers in mice depleted of CD4+ or both CD4+ and CD8+ cells (Fig. 4C). Thus, passive transfer of neutralizing Abs can control virus infection and restore viral clearance in mice depleted of CD4+ T cells.
Depletion of CD4+ T cells reduces production of both Th1 and Th2 cytokines.
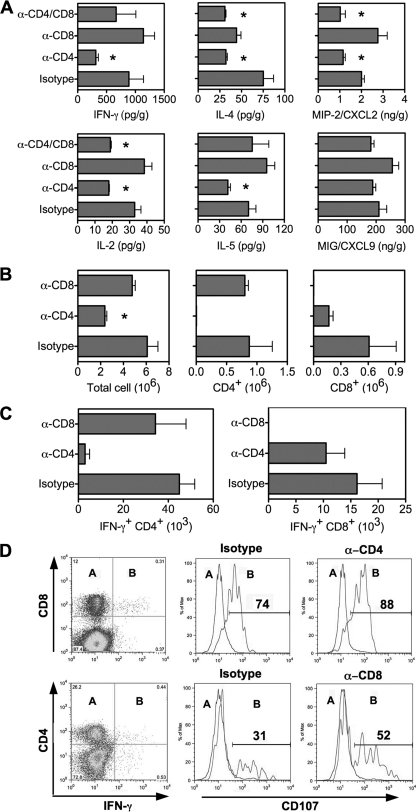
To determine whether depletion of CD4+ T cells affects control of virus infection through cytokine production, protein levels of cytokines and chemokines were measured in lung homogenates on day 7 p.i. (Fig. 5A). Depletion of CD8+ cells did not affect cytokine production, but depletion of CD4+ cells resulted in a decrease in the production of both Th1 and Th2 cytokines. Compared to the isotype-treated controls, levels of cytokines IFN-γ, IL-2, IL-4, and IL-5 and chemokine MIP-2/CXCL2 were lower in mice depleted of CD4+ T cells (P < 0.05 by Student's t test), and IL-2, IL-4, and MIP-2/CXCL2 levels were lower in mice depleted of both CD4+ and CD8+ T cells (P < 0.05 by Student's t test).
FIG. 5.
Reduced T-cell responses in the lungs of SARS-CoV-infected mice depleted of CD4+ T cells. Mice were treated with isotype control and MAbs against T-cell subsets as described in Materials and Methods. (A) Cytokines and chemokines were determined in lung homogenates by Cytokine Beads Array and Bioplex on day 7 p.i. Data are shown as means ± SEMs for four mice. (B) Number of cells isolated from the lungs of mice treated with MAbs for FACS analysis by day 9 p.i. Data are presented as means ± SEMs for four mice from a typical experiment. (C) Intracellular staining for IFN-γ on CD4+ and CD8+ T cells isolated from the lungs by day 9 p.i., with ex vivo stimulation with SARS-CoV antigen. Data are presented as means ± SEMs for four mice. (D) FACS analysis of CD107 expression on IFN-γ-negative (subset A) and IFN-γ-positive (subset B) T cells isolated from the lungs on day 9 p.i., with ex vivo stimulation with viral antigen. The recorded numbers denote the percentages of IFN-γ-positive CD4+ or CD8+ T cells expressing CD107.
Depletion of CD4+ T cells reduces cell recruitment to the lungs.
FACS analysis revealed that depletion of CD4+ T cells resulted in decreased number of leukocytes infiltrating into the lungs on day 9 p.i. (Fig. 5B), consistent with the decreased chemokine production. Total cell counts in the lungs were significantly lower in mice depleted of CD4+ T cells than in isotype control-treated animals (P < 0.05 by Student's t test). Depletion of CD4+ T cells resulted in reduced CD8+ cell counts in the lungs (not significant compared to the isotype control-treated animals), while depletion of CD8+ T cells did not lead to reduced cell recruitment into the lungs. FACS analysis also revealed that depletion of CD4+ T cells resulted in fewer IFN-γ-producing CD8+ T cells infiltrating into the lungs on day 9 p.i., while depletion of CD8+ T cells did not noticeably affect pulmonary recruitment of virus-specific IFN-γ-producing CD4+ T cells (Fig. 5C).
Depletion of CD4+ T cells does not affect the cytolytic activity of CD8+ CTLs.
To determine whether depletion of T cells affected cytotoxic T lymphocyte (CTL) activity, in a novel assay for CTL activity (58) we measured the surface expression of CD107 on virus-specific T cells isolated from the lungs on day 9 p.i. (Fig. 5D). CD107 was expressed predominantly on IFN-γ-producing T cells, exhibiting 74% on CD8+ cells and less (31%) on CD4+ cells specific to the virus. Depletion of CD4+ T cells did not dramatically alter CD107 expression on virus-specific CD8+ T cells (82%), and depletion of CD8+ T cells moderately increased CD107 expression by virus-specific CD4+ T cells (52%). These data indicate that CD8+, but not CD4+, T cells are the major population of CTLs in SARS-CoV infection and that CD8+ cytolytic activity reflected by CD107 staining is CD4 independent. The fact that viral clearance is delayed in mice depleted of CD4+ T cells further indicates that CD8+ CTLs alone are not sufficient to control SARS-CoV infection.
Depletion of CD4+ T cells results in enhanced interstitial pneumonitis.
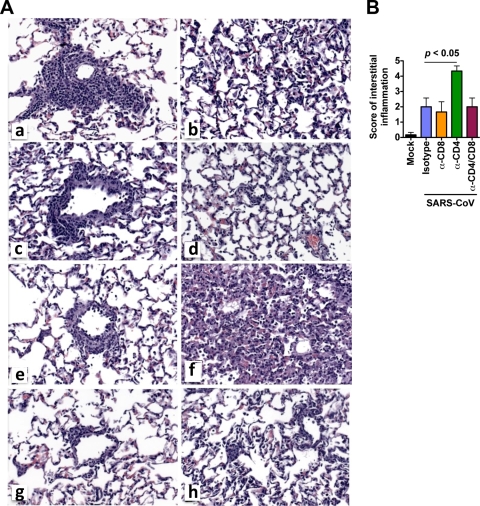
Histologic examination of the lungs was carried out to determine whether depletion of T-cell subsets affected disease severity. Lung histopathology on day 9 p.i. revealed reduced perivascular lymphocytic infiltrates in mice depleted of T-cell subsets compared with isotype control-treated animals (Fig. 6A, panels a, c, e, and g). These data are consistent with our findings on FACS analysis (Fig. 3B and 5B). Interestingly, enhanced multifocal interstitial inflammation was seen in mice depleted of CD4+ cells (Fig. 6A, panel f, and B), and this was associated with high titers of virus in the lungs (Fig. 3C). However, although virus titers were also high at day 9 p.i. in mice depleted of both CD4+ and CD8+ T cells, the severity of interstitial pneumonitis was noticeably less than that in mice lacking CD4+ T cells alone (Fig. 6A, panels h and f, respectively). The correlation between enhanced disease severity and depletion of CD4+ T cells alone suggests that CD8-mediated virus-specific lung immunopathology occurs in the absence of CD4+ T cells in senescent mice.
FIG. 6.
Histopathologic changes in the lungs of SARS-CoV-infected mice at day 9 p.i. (A) Mice were treated with isotype control Ab (a and b), anti-CD8 (c and d), anti-CD4 (e and f), and anti-CD4 and CD8 Ab (g and h) as described in Materials and Methods. Compared to isotype control-treated mice (a), perivascular lymphocytic infiltrates are diminished in mice by treatment with anti-CD8 (c), anti-CD4 (e), and anti-CD4 and CD8 Ab (g) MAbs. Multifocal interstitial infiltrates are identified in the lungs of all infected mice (b, d, f, and h) but are most prominent in the mice depleted of CD4+ T cells (f). (B) Pathology scores of pulmonary inflammation in the lungs were assessed on day 9 p.i. Data represent means ± SEMs for four mice at the indicated time point.
Clearance of SARS-CoV in the absence of both T cells and Abs.
Mice depleted of both CD4+ and CD8+ T cells, which therefore lacked both T-cell and Ab responses to the virus, were able to control SARS-CoV replication in the lungs by day 12 p.i., suggesting an immune mechanism independent of Abs and T cells. We studied the cytokine and chemokine profiles in lung homogenates at day 10 and day 12 p.i. and detected a more intense cytokine response in mice depleted of both CD4+ and CD8+ cells than in isotype control-treated animals. Compared to isotype control-treated animals, we detected a trend toward increased expression of cytokines IL-1α and IL-1β on day 10 p.i., in association with increased expression of chemokines MCP-1/CCL2 and MIP-2/CXCL2 (data not shown), which are critical for tissue recruitment of monocytes and neutrophils (2, 39). MIP-1b/CCL4, RANTES/CCL5, and MIG/CXCL9 were not increased in mice depleted of both CD4+ and CD8+ cells (data not shown). Although the total number of cells in the lungs of mice depleted of both T-cell subsets was similar to that for isotype control Ab-treated animals on day 10, complete depletion of CD4+ and CD8+ T cells was observed in these mice (data not shown). Increased numbers of pDC and fewer granular CD11b+ CD11c− Ly-6G+ Gr-1+ cells were detected in mice depleted of both CD4+ and CD8+ cells on day 10 (data not shown), and this was followed by viral clearance by day 12. Of interest, an increase in NK cells and macrophages was not seen in the lungs of mice depleted of both CD4+ and CD8+ cells. These data indicate that in the absence of both T cells and Abs, innate defense mechanisms are able to control SARS-CoV infection. Consistent with our findings in mice, high neutrophil and low lymphocyte counts were reported in the peripheral blood of SARS patients (29). The cellular mechanisms underlying Ab- and T-cell-independent control of SARS-CoV infection in this model require further investigation.
DISCUSSION
In a pneumonia model of SARS in senescent mice, we have identified a biphasic cellular immune response to SARS-CoV and found that CD4+ T cells (but not CD8+ T cells) are important in the control of disease pathogenesis and SARS-CoV replication.
The first important finding of our study is the biphasic expression of inflammatory mediators in the lungs of SARS-CoV-infected mice, associated with two waves of influx of inflammatory cells. The cytokines TNF-α and IL-6, associated with chemokines CCL2/MCP-1, CCL3/MIP-1α, and CXCL10/IP-10, were preferentially induced on days 2 to 3, when virus titers peaked in the lungs, and were likely produced by infected cells, such as airway epithelial cells and alveolar macrophages (26, 43, 56). An early accumulation of pDC, a major source of IFN-α in SARS-CoV infection (6), was accompanied by a modest type I IFN response early after infection. Early recruitment of other inflammatory cells, such as CD4+, NK T cells, NK cells, and macrophages, was also detected in the lungs following the first wave of increased expression of chemokines. The activation of the innate defense system at the early phase of infection appears to play an important role in the control of SARS-CoV replication. A second wave of inflammatory mediators noted on day 7 p.i. involved an increase of the cytokines TNF-α and IL-6 and chemokines CCL2/MCP-1, CCL3/MIP-1α, CCL5/RANTES, CXCL9/MIG, and CXCL10/IP-10 and an increase of T-cell-mediated inflammatory cytokines IFN-γ, IL-2, and IL-5. The peak of the second wave of cytokines, chemokines, and their receptors correlated with infiltration of the lungs by T cells and neutrophils. By day 7 after infection, an intense virus-specific T-cell response (particularly CD4+ T cells), in association with the second wave of inflammatory mediators, coincides with the onset of pneumonitis and is followed by complete viral clearance by day 9. Weight loss ceases by the time the virus is cleared from the lungs (Fig. 7). These findings indicate that leukocyte-mediated antiviral responses may contribute to the clearance of SARS-CoV, but they can contribute to pneumonitis.
Although relatively little is known about the early events in human SARS, local and systemic elevations of the inflammatory mediators reported in SARS patients are consistent with our findings in the senescent mouse model of SARS pneumonia. Very high levels of cytokines and chemokines in the blood and lungs of SARS patients were reported on autopsy. Protein or RNA expression of IL-6, CCL2/MCP-1, and CXCL10/IP-10 was detected by IHC and RT-PCR in the lungs of patients with fatal SARS cases (26). Elevation of IL-6 and IL-8 during the acute phase of disease and persistent expression of CCL2/MCP-1, CXCL9/MIG, and CXCL10/IP-10 in both acute and fatal cases were consistently detected in the blood of SARS patients (25, 26, 34, 43, 51). Moreover, increased serum levels of CXCL10/IP-10, IL-2, and IL-6 correlated significantly with SARS pneumonitis in patients (10), and this observation parallels our finding of the second wave of increased expression of these cytokines/chemokines coinciding with the onset of pneumonitis in mice.
Chemokine receptors play a crucial role in directing inflammatory cells to the sites of infection (8, 40). Previous studies have documented the importance of chemokine receptors in protection from SARS-associated disease. CCR1-, CCR2-, and CCR5-deficient mice developed severe disease and mortality without inflammatory cell recruitment to the lungs (41). In contrast, upregulation of chemokines MCP-1/CCL2 (CCR1), MIP-1α/CCL3 (CCR2), and RANTES/CCL5 (CCR5) in the lungs of wild-type mice coincided with pulmonary recruitment of inflammatory cells and protected the mice from mortality. The protective role of CXCR3 (CXCL9/MIG and CXCL10/IP-10) in cell-mediated viral clearance of West Nile virus (WNV) infection has been documented, where CXCR3-deficient mice exhibited significantly enhanced mortality (57). In our experiments with SARS-CoV infection in senescent mice, the biphasic increased expression of chemokines (MCP-1/CCL2, MIP-1α/CCL3, RANTES/CCL5, MIG/CXCL9, and IP-10/CXCL10) and their receptors (CCR2, CCR5, and CXCR3) is coincident with the appearance of inflammatory cells in the lungs. Taken together, these data suggest an important role for immune-mediated inflammation and chemokine recruitment of inflammatory cells in severe SARS-CoV disease.
The second novel finding of our study is that CD8+ T cells are not required but CD4+ T cells are important for the control of viral replication in primary SARS-CoV infection in senescent mice. CD8+ CTLs alone are not sufficient to clear SARS-CoV in the absence of both CD4+ T cells and Abs. Depletion of CD8+ T cells alone does not affect clearance of the virus. These findings are rather surprising because CD8+ T cells are required for the control of influenza virus and other respiratory viruses (3, 16, 45). Depletion of CD8+ T cells results in prolonged replication of influenza virus and is associated with 100% mortality in mice (3, 16). Transfer of virus-specific CD8+ T cells can provide protection and independently mediate viral clearance (45). CD4+ T cells are less efficient than CD8+ T cells at clearing influenza virus (19), and CD4+ T-cell-mediated protection occurs through Ab-dependent mechanisms (46) and Ab-independent direct killing mechanisms that require perforin (4). In the senescent mouse model for SARS, depletion of CD4+ T cells results in reduced production of neutralizing Ab and Th1 and Th2 cytokines and pulmonary recruitment of inflammatory cells. Passive transfer of neutralizing Ab against SARS-CoV can control viral replication in mice depleted of CD4+ T cells. These findings imply that the CD4-mediated control of SARS-CoV infection most likely operates through Ab and cytokine-dependent mechanisms.
It is of interest that mice depleted of both CD4+ and CD8+ T-cell subsets were able to clear the virus from the lungs by day 12 p.i. in the absence of neutralizing Ab and T-cell responses. High levels of cytokine IL-1α and chemokine MCP-1/CCL2 and MIP-2b/CXCL2 were detected in the lungs, coincident with a local accumulation of neutrophils and DCs, and this was followed by viral clearance by day 12 p.i. MCP-1/CCL2 and MIP-2b/CXCL2 are known to play a critical role in the tissue migration of neutrophils, mononuclear phagocytes, and monocytes (2, 31, 39). Recent studies using MAbs to deplete neutrophils suggest that neutrophils can play a protective role in influenza virus infection in vivo by limiting influenza virus replication and controlling disease severity (18, 31, 44). Although the cellular mechanism remains unclear, our data suggest that an extended innate antiviral response may be responsible for the clearance of SARS-CoV in the senescent mouse in the absence of both T-cell and neutralizing Ab responses.
Another important insight revealed by our study is the importance of CD4+ T cells in reducing the severity of SARS-associated pneumonitis. Depletion of CD4+ T cells alone led to enhanced interstitial pneumonitis associated with high titers of SARS-CoV in the lungs. Thus, in the absence of CD4+ T cells and virus-specific Abs, CD8+ T cells mediate virus-specific lung pathology. CD8+ CTLs have also been implicated in the exacerbation of lung pathology in mice infected with influenza virus (32) and in IFN-γ-deficient mice depleted of CD4+ T cells following infection with lymphocytic choriomeningitis virus and influenza virus (11, 12, 50). Moreover, consistent with our findings in mice, the rapid development of lymphopenia in SARS patients during acute infection involved a greater reduction in CD4+ than CD8+ T cells and was associated with adverse disease outcome (22, 52). Although the mechanism for SARS-mediated lymphopenia remains unclear, the observations in mice and humans of a correlation between a reduction in CD4+ T cells and increased severity of disease indicates that CD4+ T cells are important for the control of SARS-associated disease.
In summary, we describe the biphasic expression of inflammatory mediators (cytokine, chemokine, and receptor) at protein and mRNA levels in the lungs of 12- to 14-mo SARS-CoV-infected BALB/c mice, in association with two waves of recruitment of inflammatory cells into the lungs. We demonstrate that CD4+ T-cell (but not CD8+ T-cell)-mediated immunity plays an important role in controlling viral replication and disease severity in primary SARS-CoV infection in senescent mice, possibly through Ab- and/or cytokine-dependent mechanisms. Our findings provide new insights into the cell-mediated immune response to SARS-CoV infection and highlight the role of CD4+ T cells in the clearance of SARS-CoV.
Acknowledgments
We thank Suzanne Epstein and Julia Misplon for useful discussions and advice; Leatrice Vogel for technical assistance in the T-cell-depletion study; and Jadon Jackson and the staff of the Building 50 Shared Animal Facility, NIAID, for assistance with the animal studies.
This research was supported by the Intramural Research Program of NIAID, NIH.
Footnotes
Published ahead of print on 11 November 2009.
REFERENCES
- 1.Baas, T., A. Roberts, T. H. Teal, L. Vogel, J. Chen, T. M. Tumpey, M. G. Katze, and K. Subbarao. 2008. Genomic analysis reveals age-dependent innate immune responses to severe acute respiratory syndrome coronavirus. J. Virol. 82:9465-9476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belperio, J. A., M. P. Keane, M. D. Burdick, J. P. Lynch III, Y. Y. Xue, A. Berlin, D. J. Ross, S. L. Kunkel, I. F. Charo, and R. M. Strieter. 2001. Critical role for the chemokine MCP-1/CCR2 in the pathogenesis of bronchiolitis obliterans syndrome. J. Clin. Invest. 108:547-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, B. S., T. Croghan, L. Zhang, and P. A. Small, Jr. 1992. Transgenic mice lacking class I major histocompatibility complex-restricted T cells have delayed viral clearance and increased mortality after influenza virus challenge. J. Exp. Med. 175:1143-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, D. M., E. Roman, and S. L. Swain. 2004. CD4 T cell responses to influenza infection. Semin. Immunol. 16:171-177. [DOI] [PubMed] [Google Scholar]
- 5.Cameron, M. J., L. Ran, L. Xu, A. Danesh, J. F. Bermejo-Martin, C. M. Cameron, M. P. Muller, W. L. Gold, S. E. Richardson, S. M. Poutanen, B. M. Willey, M. E. DeVries, Y. Fang, C. Seneviratne, S. E. Bosinger, D. Persad, P. Wilkinson, L. D. Greller, R. Somogyi, A. Humar, S. Keshavjee, M. Louie, M. B. Loeb, J. Brunton, A. J. McGeer, and D. J. Kelvin. 2007. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J. Virol. 81:8692-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cervantes-Barragan, L., R. Zust, F. Weber, M. Spiegel, K. S. Lang, S. Akira, V. Thiel, and B. Ludewig. 2007. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 109:1131-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan-Yeung, M., and R. H. Xu. 2003. SARS: epidemiology. Respirology 8(Suppl.):S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charo, I. F., and R. M. Ransohoff. 2006. The many roles of chemokines and chemokine receptors in inflammation. N. Engl. J. Med. 354:610-621. [DOI] [PubMed] [Google Scholar]
- 9.Cheung, C. Y., L. L. Poon, I. H. Ng, W. Luk, S. F. Sia, M. H. Wu, K. H. Chan, K. Y. Yuen, S. Gordon, Y. Guan, and J. S. Peiris. 2005. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 79:7819-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chien, J. Y., P. R. Hsueh, W. C. Cheng, C. J. Yu, and P. C. Yang. 2006. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology 11:715-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen, J. P., C. Bartholdy, D. Wodarz, and A. R. Thomsen. 2001. Depletion of CD4+ T cells precipitates immunopathology in immunodeficient mice infected with a noncytocidal virus. J. Immunol. 166:3384-3391. [DOI] [PubMed] [Google Scholar]
- 12.Cousens, L. P., J. S. Orange, and C. A. Biron. 1995. Endogenous IL-2 contributes to T cell expansion and IFN-gamma production during lymphocytic choriomeningitis virus infection. J. Immunol. 155:5690-5699. [PubMed] [Google Scholar]
- 13.Ding, Y., L. He, Q. Zhang, Z. Huang, X. Che, J. Hou, H. Wang, H. Shen, L. Qiu, Z. Li, J. Geng, J. Cai, H. Han, X. Li, W. Kang, D. Weng, P. Liang, and S. Jiang. 2004. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J. Pathol. 203:622-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donnelly, C. A., M. C. Fisher, C. Fraser, A. C. Ghani, S. Riley, N. M. Ferguson, and R. M. Anderson. 2004. Epidemiological and genetic analysis of severe acute respiratory syndrome. Lancet Infect. Dis. 4:672-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 16.Eichelberger, M., W. Allan, M. Zijlstra, R. Jaenisch, and P. C. Doherty. 1991. Clearance of influenza virus respiratory infection in mice lacking class I major histocompatibility complex-restricted CD8+ T cells. J. Exp. Med. 174:875-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epstein, S. L., C. Y. Lo, J. A. Misplon, C. M. Lawson, B. A. Hendrickson, E. E. Max, and K. Subbarao. 1997. Mechanisms of heterosubtypic immunity to lethal influenza A virus infection in fully immunocompetent, T cell-depleted, beta2-microglobulin-deficient, and J chain-deficient mice. J. Immunol. 158:1222-1230. [PubMed] [Google Scholar]
- 18.Fujisawa, H. 2008. Neutrophils play an essential role in cooperation with antibody in both protection against and recovery from pulmonary infection with influenza virus in mice. J. Virol. 82:2772-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerhard, W., A. M. Haberman, P. A. Scherle, A. H. Taylor, G. Palladino, and A. J. Caton. 1991. Identification of eight determinants in the hemagglutinin molecule of influenza virus A/PR/8/34 (H1N1) which are recognized by class II-restricted T cells from BALB/c mice. J. Virol. 65:364-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glass, W. G., K. Subbarao, B. Murphy, and P. M. Murphy. 2004. Mechanisms of host defense following severe acute respiratory syndrome-coronavirus (SARS-CoV) pulmonary infection of mice. J. Immunol. 173:4030-4039. [DOI] [PubMed] [Google Scholar]
- 21.Gu, J., E. Gong, B. Zhang, J. Zheng, Z. Gao, Y. Zhong, W. Zou, J. Zhan, S. Wang, Z. Xie, H. Zhuang, B. Wu, H. Zhong, H. Shao, W. Fang, D. Gao, F. Pei, X. Li, Z. He, D. Xu, X. Shi, V. M. Anderson, and A. S. Leong. 2005. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 202:415-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, Z., C. Zhao, Q. Dong, H. Zhuang, S. Song, G. Peng, and D. E. Dwyer. 2005. Effects of severe acute respiratory syndrome (SARS) coronavirus infection on peripheral blood lymphocytes and their subsets. Int. J. Infect. Dis. 9:323-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogan, R. J., G. Gao, T. Rowe, P. Bell, D. Flieder, J. Paragas, G. P. Kobinger, N. A. Wivel, R. G. Crystal, J. Boyer, H. Feldmann, T. G. Voss, and J. M. Wilson. 2004. Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J. Virol. 78:11416-11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, J., Y. Cao, J. Du, X. Bu, R. Ma, and C. Wu. 2007. Priming with SARS CoV S DNA and boosting with SARS CoV S epitopes specific for CD4+ and CD8+ T cells promote cellular immune responses. Vaccine 25:6981-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, K. J., I. J. Su, M. Theron, Y. C. Wu, S. K. Lai, C. C. Liu, and H. Y. Lei. 2005. An interferon-gamma-related cytokine storm in SARS patients. J. Med. Virol. 75:185-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang, Y., J. Xu, C. Zhou, Z. Wu, S. Zhong, J. Liu, W. Luo, T. Chen, Q. Qin, and P. Deng. 2005. Characterization of cytokine/chemokine profiles of severe acute respiratory syndrome. Am. J. Respir. Crit. Care Med. 171:850-857. [DOI] [PubMed] [Google Scholar]
- 27.Li, C. K., H. Wu, H. Yan, S. Ma, L. Wang, M. Zhang, X. Tang, N. J. Temperton, R. A. Weiss, J. M. Brenchley, D. C. Douek, J. Mongkolsapaya, B. H. Tran, C. L. Lin, G. R. Screaton, J. L. Hou, A. J. McMichael, and X. N. Xu. 2008. T cell responses to whole SARS coronavirus in humans. J. Immunol. 181:5490-5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, T., Z. Qiu, L. Zhang, Y. Han, W. He, Z. Liu, X. Ma, H. Fan, W. Lu, J. Xie, H. Wang, G. Deng, and A. Wang. 2004. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J. Infect. Dis. 189:648-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lien, T. C., C. S. Sung, C. H. Lee, H. K. Kao, Y. C. Huang, C. Y. Liu, R. P. Perng, and J. H. Wang. 2008. Characteristic features and outcomes of severe acute respiratory syndrome found in severe acute respiratory syndrome intensive care unit patients. J. Crit. Care 23:557-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 31.Matzer, S. P., T. Baumann, N. W. Lukacs, M. Rollinghoff, and H. U. Beuscher. 2001. Constitutive expression of macrophage-inflammatory protein 2 (MIP-2) mRNA in bone marrow gives rise to peripheral neutrophils with preformed MIP-2 protein. J. Immunol. 167:4635-4643. [DOI] [PubMed] [Google Scholar]
- 32.Moskophidis, D., and D. Kioussis. 1998. Contribution of virus-specific CD8+ cytotoxic T cells to virus clearance or pathologic manifestations of influenza virus infection in a T cell receptor transgenic mouse model. J. Exp. Med. 188:223-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peiris, J. S., Y. Guan, and K. Y. Yuen. 2004. Severe acute respiratory syndrome. Nat. Med. 10:S88-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peiris, J. S., W. C. Yu, C. W. Leung, C. Y. Cheung, W. F. Ng, J. M. Nicholls, T. K. Ng, K. H. Chan, S. T. Lai, W. L. Lim, K. Y. Yuen, and Y. Guan. 2004. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet 363:617-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perlman, S., and A. A. Dandekar. 2005. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 5:917-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts, A., D. Deming, C. D. Paddock, A. Cheng, B. Yount, L. Vogel, B. D. Herman, T. Sheahan, M. Heise, G. L. Genrich, S. R. Zaki, R. Baric, and K. Subbarao. 2007. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 3:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts, A., C. Paddock, L. Vogel, E. Butler, S. Zaki, and K. Subbarao. 2005. Aged BALB/c mice as a model for increased severity of severe acute respiratory syndrome in elderly humans. J. Virol. 79:5833-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts, A., L. Vogel, J. Guarner, N. Hayes, B. Murphy, S. Zaki, and K. Subbarao. 2005. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J. Virol. 79:503-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roche, J. K., T. R. Keepers, L. K. Gross, R. M. Seaner, and T. G. Obrig. 2007. CXCL1/KC and CXCL2/MIP-2 are critical effectors and potential targets for therapy of Escherichia coli O157:H7-associated renal inflammation. Am. J. Pathol. 170:526-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serbina, N. V., T. Jia, T. M. Hohl, and E. G. Pamer. 2008. Monocyte-mediated defense against microbial pathogens. Annu. Rev. Immunol. 26:421-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheahan, T., T. E. Morrison, W. Funkhouser, S. Uematsu, S. Akira, R. S. Baric, and M. T. Heise. 2008. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog. 4:e1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subbarao, K., J. McAuliffe, L. Vogel, G. Fahle, S. Fischer, K. Tatti, M. Packard, W. J. Shieh, S. Zaki, and B. Murphy. 2004. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 78:3572-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang, N. L., P. K. Chan, C. K. Wong, K. F. To, A. K. Wu, Y. M. Sung, D. S. Hui, J. J. Sung, and C. W. Lam. 2005. Early enhanced expression of interferon-inducible protein-10 (CXCL-10) and other chemokines predicts adverse outcome in severe acute respiratory syndrome. Clin. Chem. 51:2333-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tate, M. D., A. G. Brooks, and P. C. Reading. 2008. The role of neutrophils in the upper and lower respiratory tract during influenza virus infection of mice. Respir. Res. 9:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor, P. M., and B. A. Askonas. 1986. Influenza nucleoprotein-specific cytotoxic T-cell clones are protective in vivo. Immunology 58:417-420. [PMC free article] [PubMed] [Google Scholar]
- 46.Topham, D. J., and P. C. Doherty. 1998. Clearance of an influenza A virus by CD4+ T cells is inefficient in the absence of B cells. J. Virol. 72:882-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Traggiai, E., S. Becker, K. Subbarao, L. Kolesnikova, Y. Uematsu, M. R. Gismondo, B. R. Murphy, R. Rappuoli, and A. Lanzavecchia. 2004. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 10:871-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogel, L. N., A. Roberts, C. D. Paddock, G. L. Genrich, E. W. Lamirande, S. U. Kapadia, J. K. Rose, S. R. Zaki, and K. Subbarao. 2007. Utility of the aged BALB/c mouse model to demonstrate prevention and control strategies for severe acute respiratory syndrome coronavirus (SARS-CoV). Vaccine 25:2173-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wentworth, D. E., L. Gillim-Ross, N. Espina, and K. A. Bernard. 2004. Mice susceptible to SARS coronavirus. Emerg. Infect. Dis. 10:1293-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiley, J. A., A. Cerwenka, J. R. Harkema, R. W. Dutton, and A. G. Harmsen. 2001. Production of interferon-gamma by influenza hemagglutinin-specific CD8 effector T cells influences the development of pulmonary immunopathology. Am. J. Pathol. 158:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong, C. K., C. W. Lam, A. K. Wu, W. K. Ip, N. L. Lee, I. H. Chan, L. C. Lit, D. S. Hui, M. H. Chan, S. S. Chung, and J. J. Sung. 2004. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin. Exp. Immunol. 136:95-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong, R. S., A. Wu, K. F. To, N. Lee, C. W. Lam, C. K. Wong, P. K. Chan, M. H. Ng, L. M. Yu, D. S. Hui, J. S. Tam, G. Cheng, and J. J. Sung. 2003. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ 326:1358-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.World Health Organization. 2003. Update 49: SARS case fatality ratio, incubation period. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/sars/archive/2003_05_07a/en/.
- 54.World Health Organization. 2003. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. World Health Organization, Geneva, Switzerland. http://www.who.int/csr/sars/country/table2003_09_23/en/.
- 55.Yang, L., H. Peng, Z. Zhu, G. Li, Z. Huang, Z. Zhao, R. A. Koup, R. T. Bailer, and C. Wu. 2007. Persistent memory CD4+ and CD8+ T-cell responses in recovered severe acute respiratory syndrome (SARS) patients to SARS coronavirus M antigen. J. Gen. Virol. 88:2740-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yen, Y. T., F. Liao, C. H. Hsiao, C. L. Kao, Y. C. Chen, and B. A. Wu-Hsieh. 2006. Modeling the early events of severe acute respiratory syndrome coronavirus infection in vitro. J. Virol. 80:2684-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, B., Y. K. Chan, B. Lu, M. S. Diamond, and R. S. Klein. 2008. CXCR3 mediates region-specific antiviral T cell trafficking within the central nervous system during West Nile virus encephalitis. J. Immunol. 180:2641-2649. [DOI] [PubMed] [Google Scholar]
- 58.Zhou, W., M. Sharma, J. Martinez, T. Srivastava, D. J. Diamond, W. Knowles, and S. F. Lacey. 2007. Functional characterization of BK virus-specific CD4+ T cells with cytotoxic potential in seropositive adults. Viral Immunol. 20:379-388. [DOI] [PubMed] [Google Scholar]
- 59.Ziegler, T., S. Matikainen, E. Ronkko, P. Osterlund, M. Sillanpaa, J. Siren, R. Fagerlund, M. Immonen, K. Melen, and I. Julkunen. 2005. Severe acute respiratory syndrome coronavirus fails to activate cytokine-mediated innate immune responses in cultured human monocyte-derived dendritic cells. J. Virol. 79:13800-13805. [DOI] [PMC free article] [PubMed] [Google Scholar]