Abstract
Human papillomavirus 16 is a causative agent of most cases of cervical cancer and has also been implicated in the development of some head and neck cancers. The early viral E6 gene codes for two alternatively spliced isoforms, E6large and E6*. We have previously demonstrated the differential effects of E6large and E6* binding on the expression and stability of procaspase 8, a key mediator of the apoptotic pathway. Additionally, we have reported that E6 binds to the FADD death effector domain (DED) at a novel E6 binding domain. Sequence similarities between the FADD and procaspase 8 DEDs suggested a specific region for E6large/procaspase 8 binding, which was subsequently confirmed by mutational analysis as well as by the ability of peptides capable of blocking E6/FADD binding to also block E6large/caspase 8 binding. However, the binding of the smaller isoform, E6*, to procaspase 8 occurs at a different region, as deletion and point mutations that disrupt E6large/caspase 8 DED binding do not disrupt E6*/caspase 8 DED binding. In addition, peptide inhibitors that can block E6large/procaspase 8 binding do not affect the binding of E6* to procaspase 8. These results demonstrate that the residues that mediate E6*/procaspase 8 DED binding localize to a different region on the protein and employ a separate binding motif. This provides a molecular explanation for our initial findings that the two E6 isoforms affect procaspase 8 stability in an opposing manner.
The relationship between viruses and cancers is reflected in the observation that viral infections account for approximately 10 to 15% of the cancer burden worldwide (6, 60). This makes viral infections one of the preventable risk factors of cancer. Viruses are associated with several human malignancies, including hepatitis B and C virus-associated hepatocellular carcinomas (48), Epstein-Barr virus-associated nasopharyngeal carcinomas and lymphomas (36), and human T-cell leukemia virus-associated adult T-cell leukemia (8, 28). Although there is a correlation between infection and the onset of cancer, the frequency of infection supersedes the incidence of cancer inception, suggesting that the presence of the virus alone is not sufficient to trigger carcinogenesis. Progression from viral infection to tumor development therefore requires additional environmental and cellular factors in addition to the expression and activity of virus-encoded proteins (40).
High-risk strains of human papillomavirus (HPV) (high-risk HPV [HR-HPV]) such as HPV16 and HPV18 have been implicated in most cases of cervical cancer and also in a subset of head and neck cancers (24, 26, 39). Infection with oncogenic strains of HPV represents up to 75% of all infections. Furthermore, 1/10 of all deaths among women worldwide can be attributed to HR-HPV-related cancers (44, 45). The key players in promoting cell transformation and immortalization following HPV infection are the viral early proteins E6 and E7. These proteins are well known for their ability to interact with the tumor suppressor p53 or members of the retinoblastoma family of proteins including pRb, p107, and p130, respectively (3, 17, 41). In addition to p53, HR-HPV E6 (HR-E6) binds to a number of cellular proteins involved in various aspects of cell proliferation and virus survival (reviewed in references 34 and 53). Our laboratory has reported that E6 binds to key mediators of the apoptotic pathway including tumor necrosis factor (TNF) R1 (22), the FADD death effector domain (DED) (21), and the procaspase 8 DED (20) and, in doing so, impedes apoptosis from taking place.
As noted above, HR-E6 binds to TNF R1, blocking the adaptor molecule TRADD from binding to the membrane receptor. Similarly, the binding of HR-E6 to the FADD DED, a molecule common to the TNF-, Fas L-, and TRAIL-mediated extrinsic pathways of apoptosis, leads to the accelerated degradation of FADD and thereby inhibits the binding of additional downstream molecules necessary for programmed cell death. Additionally, we have reported that HR-E6 binds to procaspase 8, another molecule common to all three receptor-mediated pathways. The importance of procaspase 8 can be demonstrated by the many proteins produced by viruses to either inactivate or inhibit this apoptotic mediator in order to evade clearance by the host immune response. Such proteins include the herpes simplex virus R1 subunit that interferes with caspase 8 activation (31); the molluscum contagiosum virus MC159 protein that binds to the DEDs of both FADD and procaspase 8, thereby inhibiting their interaction (25); the human herpesvirus 8 FLICE protein that obstructs procaspase 8 cleavage and prevents its activation (4); and the cowpox virus serpin CrmA, which, along with the human cytomegalovirus UL136 proteins, inhibits caspase 8 activation (50, 56). In a like manner, HR-HPV16 produces the early protein E6 that binds to procaspase 8. Interestingly, however, we have found that the two splice products of the E6 gene, E6large, a protein of about 16 kDa, and E6*, a protein less than half the size of E6large, bind to and affect procaspase 8 stability differentially. While the large isoform accelerates the degradation of procaspase 8, leading to its destabilization, the short isoform leads to the stabilization of protein expression and an increase in activity. These observations suggest that the bindings of these two E6 isoforms have different functional consequences and may well localize to different regions on procaspase 8.
We have previously identified a novel E6 binding site on the FADD DED (54). Based on sequence comparisons between the DEDs of FADD and procaspase 8, we proposed that the binding motif that mediates oncoprotein binding to both proteins would be similar. To test this possibility, we performed a series of mutational and peptide competitor-based experiments and discovered that the motifs on caspase 8 and on FADD that mediate binding between E6 and its cellular partner are indeed similar. Interestingly, however, the motif by which E6* binds to procaspase 8 is located in another region of the protein. These findings provide a molecular explanation for our previously reported observations concerning the differential effects of the binding of each isoform to the procaspase 8 DED. These findings also demonstrate the ability of peptide inhibitors to successfully impair E6 binding to its cellular targets and contribute to the discovery of therapeutic agents that are effective against cervical cancer.
MATERIALS AND METHODS
Reagents.
The following reagents were used in these studies: lyophilized human recombinant TNF-α (R&D Systems, Minneapolis, MN), monoclonal antibodies directed against Fas (clone CH-11; Medical and Biological Laboratories Co., Ltd., Nagoya, Japan), mouse monoclonal antibodies against caspase 8 (BD Pharmingen, San Jose, CA), monoclonal antibodies directed against β-actin or against Flag (Sigma), monoclonal antibodies directed against the HPV16 E6 N terminus (Euromedex, France), peroxidase-coupled anti-mouse polyclonal antibodies (Thermo Fisher), polyclonal anti-FADD, monoclonal anti-green fluorescent protein (GFP), and peroxidase-coupled monoclonal antibodies against glutathione (GSH) S-transferase (GST) (Santa Cruz Biotechnology, Santa Cruz, CA), cycloheximide (Sigma), and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma, St. Louis, MO).
Cell culture.
U2OS cells derived from a human osteosarcoma were obtained from the ATCC (Manassas, VA) and cultured in McCoy's 5A medium (Invitrogen, Carlsbad, CA) supplemented to contain 10% fetal bovine serum (Invitrogen), penicillin (100 U/ml), and streptomycin (100 μg/ml) (Sigma). Cells were passaged and used at approximately 80% confluence. The production of the stably transfected U2OStet, U2OStetE6, and U2OSE6large (U2OSE6L) cell lines was described previously (18, 20).
Plasmids.
Carl Ware (La Jolla Institute for Allergy and Immunology, La Jolla, CA) kindly provided plasmid pcDNA3-FADD. To express FADD and its variants in the Escherichia coli system for subsequent protein purification, the EcoRI-XhoI fragment from pM-FADD, coding for FADD, was cloned in frame with the His6 epitope of pTriEx-4 (Novagen) by using the SmaI-XhoI sites in the multiple-cloning site to produce plasmid pHis-FADD.
To express His-tagged E6AP in E. coli for subsequent protein purification, EcoRI was used to remove the sequence of E6AP necessary for E6 binding from plasmid pOTB7 (Open Biosystems), amino acids (aa) 288 to 496. This fragment was subsequently cloned in frame with the His6 epitope of pTriEx-4 (Novagen) by using the SmaI-PvuII sites in the multiple-cloning site to produce plasmid pHis-E6AP.
To express GST-tagged E6 in E. coli cells for subsequent protein purification, we created a plasmid coding for the fusion protein GST-E6 by cloning E6 (Ecl136II-EcoRI blunt-end fragment) into the SmaI site of pGEX-2T (GE Healthcare). The creation of plasmids that express GST-tagged versions of the large (E6large) and small (E6*) isoforms of E6 in E. coli was previously described (20). Mutagenesis of GST-E6* was performed as described previously (54). To produce the pGST-E6*QA mutant, H21 was exchanged with a Q, and L22 was replaced with an A. According to the three-dimensional (3D) structure predicted previously by Nomine et al. (42), these two amino acids are positioned within a loop and have the potential to be part of a fragile site.
The sequence for the caspase 8 DED was obtained from cDNA prepared from U2OS cells by PCR amplification using primers 5′-GACTTCAGCAGAAATCTTTATGATATTGGGGAAC-3′ and 5′-GAGATTGTCATTACCCCACACA-3′ and served as the basis for our additional caspase 8 DED-expressing constructs. It produces a protein of approximately 25 kDa, while the full-length, endogenous procaspase 8 encodes a protein of approximately 50 kDa. To express wild-type (wt) caspase 8 and its variants in the E. coli system or in mammalian cells, the sequence coding for the procaspase 8 DED was cloned in frame with the His6 epitope of pTriEx-4 (Novagen) to produce pTriEx-casp 8 ded. This is the plasmid that was used for the bead-based binding assays as well as the pulldown assays, as it contains a His tag. To remove the first 16 aa from the N terminus of the caspase 8 DED, plasmid pTriEx-casp 8 ded was digested with BglII. The BglII-BglII fragment was then cloned into the pTriEx-4 vector to produce the ΔN version of the plasmid. To produce the pcDNA-3 version of the plasmid, which was used for assays examining how E6 affects caspase 8 stability, the caspase 8 DED sequence was cloned into the pcDNA-3 vector (Invitrogen); this version of the protein lacks the His tag.
Transfections and siRNA treatment.
FuGENE VI (Roche Applied Science) was used to transfect cells (Roche Applied Science) with the indicated plasmids as directed by the manufacturer and as described previously (21). siControl, siFADD, and siCaspase 8 were obtained from Santa Cruz Technology. For siRNA inhibition experiments, X-tremeGENE small interfering RNA (siRNA) transfection reagent (Roche) was used according to the manufacturer's protocol.
Treatment of cells with anti-Fas and TNF-α.
To measure cell survival following anti-Fas or TNF-α treatment, U2OS cells (1 × 104 cells/well) were seeded into 96-well plates and allowed to adhere overnight. Anti-Fas (50 ng/ml) or TNF-α (5 ng/ml) was then added in the presence of cycloheximide (5 μg/ml) to inhibit de novo protein synthesis, and the cells were incubated for 16 h prior to measuring cell viability by the MTT assay (20).
Immunoblotting.
To prepare cell lysates for immunoblotting, 5 × 105 cells were lysed in 100 μl of lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 5% glycerol, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]) for 10 min on ice. One tablet of protease inhibitor mixture (Roche Applied Science) was added per 10 ml of lysis buffer just prior to use. The protein concentration in cleared lysates was measured by using the Bio-Rad DC protein assay (Bio-Rad). Anti-caspase 8 monoclonal antibodies at a 1:3,000 dilution, anti-GST monoclonal antibodies at a 1:5,000 dilution, and horseradish peroxidase (HRP)-conjugated secondary anti-mouse antibodies at a 1:3,000 dilution were used for detection using the chemiluminescent SuperSignal West Femto or Pico maximum-sensitivity substrate (Thermo Fisher).
Expression and purification of recombinant proteins.
The purification of GST-E6large, GST-E6*, and the various His-tagged proteins was carried out as previously described (21).
In vitro pulldown assays.
In vitro pulldown assays were performed to assess the ability of bead-bound GST-E6 proteins (glutathione beads from Sigma) to bind to the caspase 8 protein as previously described (21).
Bead-based binding assay.
AlphaScreen technology was used to assess the interaction between GST-bound E6 and the various His-tagged caspase 8 DED mutants, His-tagged FADD, or His-tagged E6AP proteins as previously described (54). For experiments involving peptide A, the indicated concentrations/dilutions were added prior to the initial 1-h incubation.
qRT-PCR.
Quantitative real-time PCR (qRT-PCR) to measure the levels of the E6 isoforms was conducted by using primers designed as described previously by Hafner et al. (27) along with the Absolute QPCR Sybr green kit according to the manufacturer's protocol (ABgene). Observed E6 isoform concentrations were normalized using the level of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) expression.
Statistics.
Experiments were done in triplicate, and the error bars represent the standard deviations. Asterisks are used to indicate comparisons where the level of confidence is greater than 0.99.
RESULTS
The TNF-mediated apoptotic pathway inhibited by E6large is caspase 8 dependent.
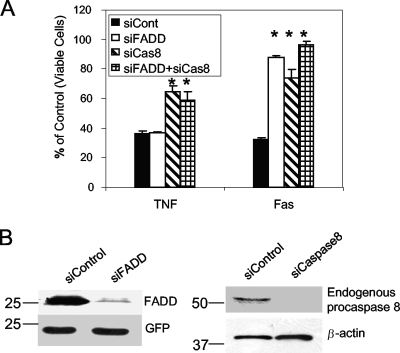
The interaction between E6large and procaspase 8 destabilizes procaspase 8 by accelerating the rate at which it is degraded and therefore impairs procaspase 8 dimerization and the procaspase 8/FADD association (19). This inactivation of procaspase 8 activity in cells expressing E6large then leads to the resultant inability of these cells to respond to treatment with TNF or Fas. To validate the importance of procaspase 8 in the apoptotic cascade, U2OS cells were transfected with siRNA targeting procaspase 8 or FADD or a control siRNA and subsequently treated with TNF-α or anti-Fas to trigger cell death. Figure 1A shows that a deficiency of functional procaspase 8 impairs both TNF- and Fas-induced apoptosis. Interestingly, cell viability measurements obtained following the treatment of cells expressing reduced FADD protein levels with apoptosis-inducing agents revealed that although FADD is necessary for the mediation of Fas-triggered apoptosis, it does not seem to be necessary for TNF-mediated apoptosis. Figure 1B illustrates the effectiveness of the respective siRNAs in inhibiting procaspase 8 and FADD protein expression.
FIG. 1.
Caspase 8 is required for apoptosis induced by both TNF and anti-Fas. (A) siRNA directed against procaspase 8 affects the cellular response to both TNF and Fas, while siRNA directed against FADD affects only the cellular response to Fas. U2OS cells were transfected with siRNA targeting procaspase 8 (siCas8) or FADD (siFADD) or a control siRNA (siCont) followed by treatment with TNF-α (5 ng/ml) or anti-Fas (50 ng/ml) in the presence of cycloheximide (5 μg/ml) for 16 h. Cell viability was then measured with the MTT assay and is presented as a percentage of viable cells, with untreated cells serving as the control. Measurements were made in triplicate, and the error bars represent the standard deviations. (B) siRNA efficiently reduces FADD and procaspase 8 protein expression. (Left) U2OS cells were transfected with control siRNA or siRNA targeting FADD and were cotransfected with pcDNA-FADD and pcDNA-GFP the next day. Forty-eight hours posttransfection, the level of expressed FADD was determined by immunoblotting using antibodies directed against FADD. The membrane was then stripped and reblotted with antibodies against GFP in order to normalize for sample loading. (Right) U2OS cells were transfected with control siRNA or siRNA targeting procaspase 8. Forty-eight hours posttransfection, cells were lysed and subjected to immunoblot analysis. The resultant blot was initially probed with antibodies directed against procaspase 8. The same membrane was then stripped and reprobed with antibodies against β-actin to normalize for loading.
HPV16 E6large binds to the caspase 8 DED and decreases its level in cells.
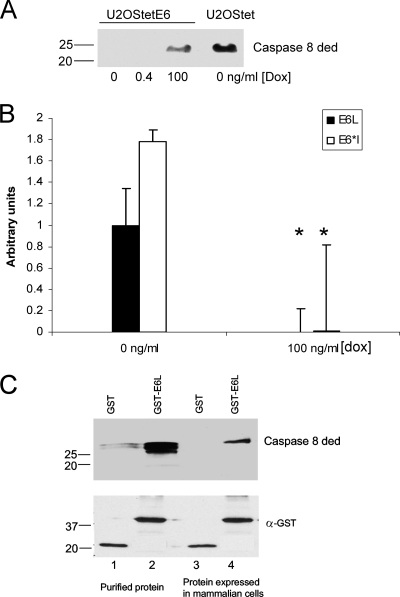
We previously reported that levels of the exogenously expressed caspase 8 DED are lower in E6-expressing cells than in cells lacking E6 (20). To further confirm this finding, U2OS cells expressing the HA-E6 wild type under the control of the tet-responsive element were maintained in the presence of different concentrations of doxycycline (0, 0.4, and 100 ng/ml) in the growth medium. In this system, high levels of doxycycline (100 ng/ml) correspond to low levels of E6, while low levels of doxycycline (0 ng/ml) correspond to high levels of E6. Cells were then transfected with plasmid pcDNA3-casp 8 ded, and 48 h posttransfection, the lysates were subjected to SDS-PAGE. As expected, the expression of the caspase 8 DED was not detected at high and moderate levels of E6 expression (0 and 0.4 ng/ml doxycycline). At lower levels of E6 expression (100 ng/ml doxycycline), however, the caspase 8 DED was detected, although the level of expression was significantly lower than that seen for control, non-E6-expressing cells (U2OStet) (Fig. 2A). qRT-PCR was used to quantify the mRNA levels of both E6 isoforms in cells grown in the presence of 0 or 100 ng/ml doxycycline (Fig. 2B), and as expected, the levels of both the full-length and shorter isoforms were much lower for cells grown in the presence of high levels of doxycycline. Interestingly, the level of expression of the shorter isoform, E6*, was higher than that of the full-length isoform. The decreased levels of procaspase 8 in the presence of E6 correlate with the ability of E6 to bind to this protein, and the binding of E6large to the caspase 8 DED is demonstrated by the pulldown assays shown in Fig. 2C. In this experiment, GST-E6large was expressed in and purified from E. coli, and the caspase 8 DED was either expressed in and purified from E. coli or expressed in U2OS cells after transient transfection.
FIG. 2.
E6 binds to the caspase 8 DED and reduces its level in cells. (A) Caspase 8 DED levels are reduced in cells expressing E6. U2OStetE6 cells grown in medium containing the indicated concentrations of doxycycline (Dox) and control U2OStet cells that do not express E6 were transfected with a plasmid expressing the caspase 8 DED (pcDNA/cas 8 ded). Cells were lysed after a 48-h incubation period at 37°C and subjected to SDS-PAGE separation. The levels of procaspase 8 were estimated by immunoblotting with antibodies directed against procaspase 8. (B) Doxycycline reduces the level of expression of both E6 isoforms. A total of 106 U2OStetE6 cells were grown in the absence (0 ng/ml) or presence (100 ng/ml) of doxycycline. RNA was extracted and purified, cDNA was synthesized, and qRT-PCR was performed using primers specific for the E6large or E6*I transcripts. Results are presented relative to the level of E6large transcripts produced in U2OStetE6 cells grown in the absence of doxycycline. (C) E6large and procaspase 8 physically bind to one another. The His-caspase 8 DED protein was expressed in E. coli cells and purified or expressed in U2OS cells. Either the purified protein (lanes 1 and 2) or the cell lysate (lanes 3 and 4) was then incubated with GST (lanes 1 and 3) or GST-E6large (lanes 2 and 4) bound to glutathione beads. Proteins bound to GST or to GST-E6large were then subjected to immunoblot analysis. The detection of the caspase 8 DED was performed by using antibodies directed against caspase 8, and the GST and GST-E6large proteins were detected by using antibodies directed against GST.
Overexpression of the caspase 8 DED resensitizes E6large-expressing cells to TNF.
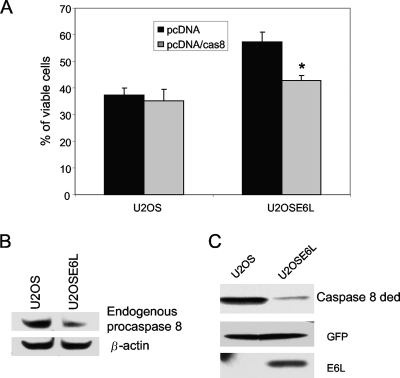
Since it is E6large that is responsible for the degradation of procaspase 8 and since our results shown in Fig. 1 demonstrate that reduced procaspase 8 levels impair apoptosis, it seemed plausible that increasing the cellular level of procaspase 8 by transfection could lead to the resensitization of E6large-expressing cells to TNF-induced apoptosis. To test this possibility, cells expressing the 16-kDa isoform of E6, U2OSE6large (U2OSE6L), were transfected with a plasmid coding for the caspase 8 DED and then tested for their response to TNF. As expected, the levels of both endogenous procaspase 8 and the exogenously expressed caspase 8 DED were lower in cells expressing E6large than in the parental U2OS cells (Fig. 3B and C). The results following TNF treatment (Fig. 3A) demonstrate that the overexpression of procaspase 8 did indeed restore the sensitivity of E6large-expressing cells to TNF.
FIG. 3.
Overexpression of the caspase 8 DED in E6-expressing cells resensitizes the cells to TNF-induced apoptosis. (A) E6-expressing cells regain sensitivity to TNF following the expression of the exogenous caspase 8 DED. Control U2OS and E6large-expressing U2OSE6L cells were transfected with either the empty pcDNA vector or with pcDNA/cas8 ded. Forty-eight hours posttransfection, cells were treated with TNF-α (5 ng/ml) in the presence of cycloheximide (5 μg/ml) and incubated for an additional 16 h. Viability was determined by using the MTT assay. Measurements were made in triplicate, and the error bars represent the standard deviations. (B and C) E6 expression reduces the level of both endogenous procaspase 8 and the exogenously expressed caspase 8 DED. (B) Lysates of U2OS and U2OSE6L cells were subjected to SDS-PAGE, and the level of endogenous procaspase 8 was estimated by immunoblotting using anti-caspase 8 antibodies (top). Normalization for loading using antibodies directed against β-actin is shown (bottom). (C) U2OS and U2OSE6large cells were transfected with plasmids coding for the caspase 8 DED and GFP. Forty-eight hours posttransfection, cell lysates were prepared, and proteins were separated by SDS-PAGE and transferred onto a membrane. The membrane was then probed with antibodies against caspase 8 (top), GFP (middle), and E6 (bottom).
The E6large binding sites on the FADD DED and procaspase 8 DED are similar.
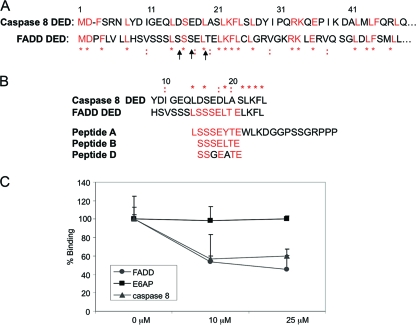
Apoptosis can proceed via the mitochondrion-mediated intrinsic pathway or by way of the cell membrane receptor-mediated extrinsic pathway (46, 55, 58, 59). The engagement of a receptor with its appropriate ligand at the cell surface initiates the extrinsic arm of the apoptotic cascade and leads to the recruitment of an adaptor molecule such as FADD or TRADD and the ensuing interaction between these molecules and procaspase 8. The binding of procaspase 8 to FADD is mediated by amino acids in the death effector domain (DED) regions of both proteins. We previously identified a novel binding site for E6 on the FADD DED (54) that differs from the previously reported binding domains for HPV16 E6 (2, 15, 23, 32, 35). Since E6 binds to both FADD and procaspase 8, we performed a sequence alignment of the two proteins to look for possible sequence similarities. If regions of conserved residues exist, this may indicate that E6 utilizes a similar binding domain to bind to procaspase 8. An alignment conducted with Biology Workbench indicates that there are indeed a number of conserved residues between the two proteins, with a few of these residues being in the region which aligns with the novel E6 binding domain on the FADD DED (Fig. 4A).
FIG. 4.
The residues that comprise the DEDs of caspase 8 and FADD share homology, and a peptide inhibitor of the E6/FADD interaction also impedes the E6/caspase 8 DED interaction. (A) Sequence alignment of the DEDs of both caspase 8 and FADD utilizing Biology Workbench (http://workbench.sdsc.edu). Arrows indicate the amino acid residues required for E6/FADD binding, * indicates homologous amino acids, and : indicates similar amino acids. (B) Sequence alignment of the DEDs of procaspase 8 and FADD; two peptide inhibitors designed to inhibit E6/FADD binding, peptides A and B (54); and a negative control peptide, peptide D. (C) Peptide A inhibits both the E6/FADD and E6/caspase 8 DED interactions but does not affect the E6/E6AP interaction. A total of 1 × 10−3 μM GST-tagged E6large was incubated with 0.5 μM His-tagged caspase 8 DED, 0.4 μM His-tagged FADD, or 0.3 μM His-tagged E6AP along with various concentrations of peptide A (0, 10, and 25 μM) at room temperature for 1 h. GSH-coated donor beads and nickel-coated acceptor beads were then added, and plates were read on an EnVision multilabel plate reader after an overnight incubation period in the absence of light. Measurements for each point were made in triplicate, and the reduction of binding in the presence of peptide A was calculated as a percentage of the observed binding in the absence of peptide.
Previously, we identified a novel E6 binding site on the FADD DED through the creation and use of site-directed and deletion mutants as well as by employing blocking peptides in biological assays. We found that the E6 binding domain localized to the N-terminal 23 amino acids of the FADD DED and incorporated serine 16, serine 18, and leucine 20 of the protein and that a peptide mimicking the E6 binding motif, peptide A, significantly impaired the ability of E6large to efficiently bind to the FADD DED (54). Due to the similarities in the DED sequences of FADD and procaspase 8, it seemed plausible that the blocking peptide used to obstruct the E6/FADD interaction might also hinder E6/procaspase 8 binding.
Figure 4B shows the aligned sequences of FADD and procaspase 8 along with the amino acid sequences of two peptides used to test for the obstruction of E6/FADD binding in vitro (peptides A and B) together with that of peptide D, which contains 2 mutations that disrupt E6large/FADD binding. To test the possibility that peptide A could block E6large/procaspase 8 binding, 0, 10, or 25 μM peptide A was incubated with the indicated mixtures of E6/FADD, E6/caspase 8, or E6/E6AP. E6AP is a known cellular binding partner of HPV16 E6, and together, these two proteins form the ubiquitin ligase moiety responsible for the ubiquitination and subsequent degradation of the tumor suppressor p53 (11, 38, 51). E6AP therefore served as a positive control for determining the effectiveness of the assay for detecting protein-protein interactions and also allowed us to assess the specificity of peptide A. Figure 4C demonstrates that in the absence of peptide A, all proteins tested bound maximally to E6. However, when peptide A was added to the mixtures containing either purified E6/FADD or E6/procaspase 8, there was a significant decrease in the level of E6 binding. In contrast, the interaction between E6 and E6AP remained unchanged at both of the tested concentrations of peptide A, indicating the specificity of this peptide for obstructing E6 binding to FADD and procaspase 8. These results support our earlier findings regarding the identification of a novel E6 binding site that differs from the E6/E6AP binding motif and provide evidence that the residues on procaspase 8 that localize to the E6large binding site are somewhat homologous to those that govern the E6/FADD interaction.
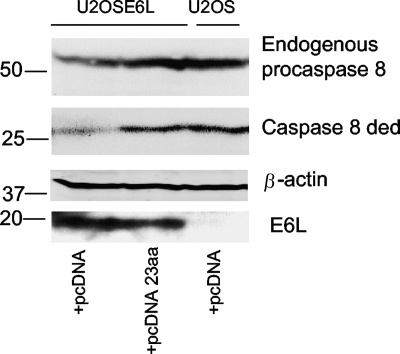
Figure 4 collectively demonstrates not only that the binding of E6large to both the FADD DED and procaspase 8 DED is mediated by a similar set of amino acids but also that a peptide composed of residues within this novel binding domain can block E6 binding in vitro. We have previously shown that the cellular overexpression of the region of FADD involved in E6 binding can obstruct E6/FADD binding, prevent the subsequent degradation of FADD, and rescue FADD in E6-expressing cells (54). Since peptide A is able to block E6/procaspase 8 binding (Fig. 4C), it seemed possible that the overexpression of this same 23-aa sequence in vivo could lead to the restoration of procaspase 8 DED levels in a similar manner. To test this hypothesis, E6large-expressing U2OSE6L cells were transfected with either control plasmid (pcDNA) or pcDNA 23 aa, a plasmid encoding the E6large binding domain. U2OS cells transfected with pcDNA served as a control. An immunoblot of the collected lysates (Fig. 5) demonstrates that in U2OSE6L cells transfected with vector alone (pcDNA), lowered levels of both the endogenous procaspase 8 and the exogenously expressed caspase 8 DED can be detected due to E6-mediated protein degradation. The overexpression of the 23 aa involved in E6large binding, however, restores the expression of both endogenous procaspase 8 and the plasmid-encoded caspase 8 DED, consistent with the peptide effectively blocking E6/procaspase 8 binding and the ensuing protein degradation.
FIG. 5.
Overexpression of a 23-amino-acid peptide homologous to the E6large binding domain of FADD restores cellular levels of both endogenous procaspase 8 and the exogenously expressed caspase 8 DED in E6-expressing cells. U2OS and E6-expressing U2OSE6L cells were transfected with the empty pcDNA vector or with pcDNA 23 aa, a plasmid encoding 23 amino acids from the FADD E6 binding domain, along with pcDNA/cas 8 ded. After 48 h of incubation at 37°C, lysates were prepared and subjected to SDS-PAGE analysis. The resulting immunoblot was probed with antibodies to procaspase 8 and β-actin. The levels of endogenous procaspase 8 (top) and the exogenous caspase 8 DED (middle) were detected by using antibodies directed against caspase 8. The membrane was stripped and reblotted by using antibodies directed against β-actin to demonstrate equal loading.
Both E6large and E6* bind to the caspase 8 DED.
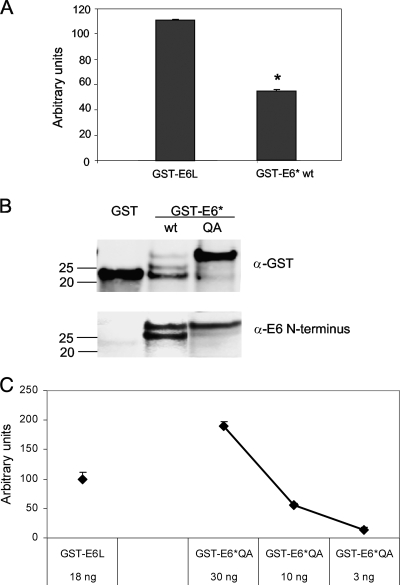
We have previously reported that both isoforms of HPV16 E6 can interact with procaspase 8 and influence its stability and activity (20). To verify binding between the E6 and procaspase 8 proteins, Perkin-Elmer's AlphaScreen technology was used to assess these protein-protein interactions in vitro. Bacterially expressed and purified GST, GST-E6large, GST-E6*, and His-caspase 8 DED proteins were used to demonstrate that both isoforms of E6 bind to procaspase 8, as determined by the emission of a detectable signal following the incubation of the bead-bound proteins with each other (Fig. 6A). These data also suggested that the level of binding between GST-E6* and the His-caspase 8 DED was lower than that of the binding of GST-E6large to the His-caspase 8 DED, in contrast to our previous findings using coimmunoprecipitation and in vitro pulldown assays where we found that the binding between E6* and the caspase 8 DED was at least as robust as that of the larger isoform (20). Interestingly, we also found that although a binding signal was consistently obtained for each of the two E6 isoforms, the intensity of the E6*/caspase 8 signal varied from one experiment to another.
FIG. 6.
Both splice isoforms of E6 bind to the caspase 8 DED. (A) E6large and E6* each bind to procaspase 8, as assessed by the AlphaScreen bead-based assay. A total of 1 × 10−3 μM GST, GST-tagged E6large, or GST-tagged E6* was incubated for 1 h with 0.5 μM His-tagged caspase 8 DED at room temperature. GSH-coated donor beads and nickel-coated acceptor beads were then added, and plates were read on an EnVision multilabel plate reader after an overnight incubation period in the absence of light. Counts recorded in the presence of GST and His-caspase 8 were considered to represent nonspecific background and were subtracted from the raw experimental data. Assays were performed in triplicate, and error bars represent the standard deviations. (B) GST-E6*QA is more resistant to proteolytic degradation than is GST-E6*wt. GST, GST-E6*wt, and GST-E6*QA were expressed in E. coli cells and purified. Approximately 1 μg of each sample was separated by SDS-PAGE, and detection was performed by using antibodies directed against GST (top) or the N terminus of E6 (bottom). (C) GST-E6*QA binds to the His-caspase 8 DED with an affinity comparable to that of GST-E6large. A total of 30 (0.05 × 10−3 μM), 10 (0.02 × 10−3 μM), or 3 ng (0.005 × 10−3 μM) of GST-E6*QA or 18 ng (0.02 × 10−3 μM) of GST-E6large was combined with 6 ng (0.01 × 10−3 μM) of the caspase 8 DED and assessed for binding by using the bead-based assay. GST-E6*QA in the absence of the His-caspase 8 DED was used as the background, and the signal from this incubation mixture was subtracted from the raw data. Assays were performed in triplicate, and error bars represent the standard deviations.
One possible explanation for this decreased and variable signal was that GST-E6* was particularly subject to proteolytic cleavage during purification and/or performance of the bead-based binding assay. Figure 6B shows that the degradation of GST-E6* does indeed occur, resulting in the presence of GST and GST-truncated E6* within the reaction mixture. These species are likely to compete with the intact GST-E6* for binding to the donor beads. To reduce this degradation and to increase the yield of our full-length GST-E6*, mutagenesis of H23 and L24, the amino acids located at the site predicted for the major breakpoint, was carried out. H23 was mutated to Q, and L24 was mutated to A; these changes significantly reduced the degree of GST-E6* degradation, as determined by immunoblotting using antibodies directed against either GST or the N terminus of E6 (Fig. 6B). To ensure that this mutated form of E6* retained the ability to bind to the caspase 8 DED, various amounts of GST-E6*QA were incubated with the His-caspase 8 DED, and binding was measured by using the bead-based binding assay (Fig. 6C). These results show that GST-E6*QA binds approximately as well to the His-caspase 8 DED as does GST-E6L, and subsequent experiments using the bead-based binding assay were therefore carried out using GST-E6*QA.
E6large and E6* binding interactions localize to different regions of procaspase 8.
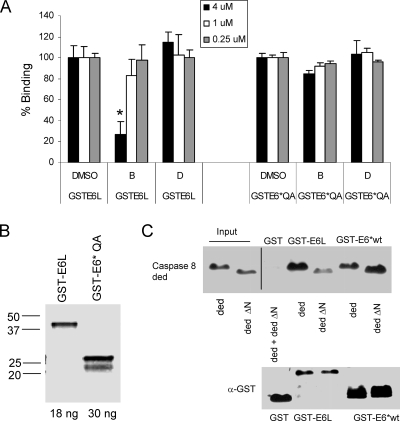
Figure 4C shows that the binding between E6large and the caspase 8 DED can be inhibited by a peptide homologous to the FADD binding motif, demonstrating that the regions on FADD and procaspase 8 to which E6large binds are similar. The next question was whether the large and small isoforms of E6 bound to the same region or to different regions of procaspase 8. To address this question, we first asked whether a peptide capable of blocking the E6large/caspase 8 interaction would also inhibit the E6*/caspase 8 interaction. Reaction mixtures containing GST-E6large or GST-E6*QA together with the His-caspase 8 DED were incubated in the presence or absence of either peptide B, a heptameric peptide that matches the FADD binding motif, or peptide D, a similar heptameric peptide that incorporates two amino acid changes (Fig. 4B). These two amino acid changes correspond to two of the three mutations that disable the ability of FADD to bind to E6large and were therefore expected to prevent this peptide from blocking binding. Reaction mixtures were then analyzed by using the AlphaScreen bead-based binding assay. The results from this experiment are shown in Fig. 7A and demonstrate that, as expected, peptide B but not peptide D can block the binding between E6large and the caspase 8 DED. Interestingly, however, neither peptide was able to block the binding between E6*QA and the caspase 8 DED, suggesting that the two E6 isoforms employ different binding sites. The amounts of the two E6 isoforms used in this experiment are shown in Fig. 7B.
FIG. 7.
The binding sites of caspase 8 to which E6large and E6* bind are not identical. (A) Peptide B impedes the binding of E6large, but not of E6*QA, to the caspase 8 DED. Eighteen nanograms (0.02 × 10−3 μM) of GST-E6large or 30 ng (0.05 × 10−3 μM) GST-E6*QA was incubated in the absence or presence of 6 ng (0.01 × 10−3 μM) of the His-caspase 8 DED and assessed for binding by using the bead-based assay. Where indicated, peptide B or D at the indicated concentrations was added to the reaction mixture. GST-E6*QA or GST-E6large in the absence of the His-caspase 8 DED was used as a background, and the signal from this incubation was subtracted from the raw data. Dimethyl sulfoxide (DMSO), used to dissolve peptides B and D, was used as a negative control for peptide inhibition. Binding between GST-E6 and the His-caspase 8 DED in the presence of DMSO was set at 100%, and the ability of the two peptides to inhibit this binding was calculated as a percentage of this initial value. Assays were performed in triplicate, and the error bars represent the standard deviations. (B) Immunoblot detection of GST-E6L and GST-E6*QA. Eighteen nanograms (0.02 × 10−3 μM) of GST-E6large and 30 ng (0.05 × 10−3 μM) of GST-E6*QA were separated by SDS-PAGE, and immunoblotting was performed by using anti-GST antibodies. (C) Deletion of the 16 N-terminal amino acids of the caspase 8 DED significantly impairs binding to E6large but not to E6*. U2OS cells were transfected with plasmids coding for either the wt caspase 8 DED (pTriEx-cas8 ded) or a mutant form from which the 16 N-terminal amino acids had been removed (pTriEx-cas8 ded ΔN). Glutathione bead-bound GST, GST-E6large, and GST-E6* were then used to pull down wild-type (wt) or mutant caspase 8. Lanes 1 and 2 show that equivalent amounts of procaspase 8 proteins were used. (Top) Immunoblot analysis using antibodies directed against caspase 8. (Bottom) The same membrane was stripped and reprobed with antibodies directed against GST.
We then asked whether amino acids in the N terminus of the caspase 8 DED were required for binding to either the full-length protein or the shorter isoform of the protein by using an in vitro pulldown assay. Because the region of the caspase 8 DED that corresponds to the E6 binding region on FADD resides within the N terminus of this protein, we expected that a deletion of this region would reduce or eliminate the ability of E6large to bind to the caspase 8 DED. However, if the binding sites for the two isoforms are different, as suggested by the experiment described above, it seemed likely that the same deletion might affect the binding of E6* either to a lesser extent or not at all. To test this prediction, a deletion of the caspase 8 DED in which the N-terminal 16 aa were removed (ΔN) was constructed, and this mutant was then tested for its ability to bind to both E6 isoforms in an in vitro pulldown assay. It should be noted that in this experiment, we used the wt version of E6* rather than E6*QA. This was possible because GST and GST-truncated E6* breakdown products do not compete for binding to the caspase 8 DED in an in vitro pulldown assay, while these breakdown products do compete for binding to the donor beads in the bead-based binding assay. The results from this experiment are shown in Fig. 7C and demonstrate that the binding of E6large to the caspase 8 DED is indeed affected to a much greater degree than is the binding of E6* to the caspase 8 DED, again indicating that the two isoforms bind to different sites on caspase 8.
Specific amino acids within the N terminus of the caspase 8 DED contribute to the E6large/caspase 8 interaction.
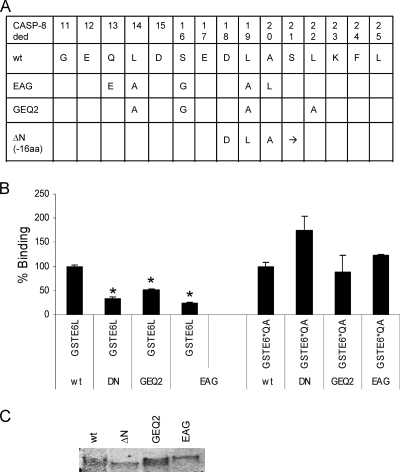
To further define the region within the N terminus of the caspase 8 DED that is involved in the binding to E6large, we created a series of point mutations. Target amino acids were chosen on the basis of their alignment with or close proximity to the E6 binding domain of the FADD DED (Fig. 8A), and the GEQ2 and EAG constructs consist of either four or five point mutations, respectively, within this region. These mutant constructs were then used in an AlphaScreen assay along with purified GST-E6large or GST-E6*QA to assess the abilities of the individual E6 isoforms to recognize and bind to the various mutant proteins. Figure 8B demonstrates that the binding between each of the three mutant proteins and E6large is hampered, while binding between the same caspase 8 mutants and GST-E6*QA is not. Figure 8C shows the input levels of the various His-caspase 8 DED proteins used in this experiment as estimated by immunoblotting. These results suggest that some combination of the amino acids within the N terminus of the procaspase 8 DED plays a role in mediating the binding of E6large, but not the binding of E6*, to procaspase 8.
FIG. 8.
Amino acids in the N terminus of the procaspase 8 DED facilitate HPV16 E6large but not E6* binding to the caspase 8 DED. (A) The protein sequences of the various deletion and mutant constructs created to help localize the amino acids involved in E6 binding to the procaspase 8 DED are shown. (B) The EAG, GEQ2, and ΔN mutations impair the binding of E6large but not E6*QA to the caspase 8 DED. Eighteen nanograms (0.02 × 10−3 μM) of GST-E6large or 30 ng (0.05 × 10−3 μM) of GST-E6*QA was combined with ∼6 ng (0.01 × 10−3 μM) of the His-caspase 8 DED (Cas8 wt), His-caspase 8 DED ΔN (DN), His-caspase 8 DED GEQ2 (GEQ2), or His-caspase 8 DED EAG (EAG). Incubation mixtures containing only GST-E6*QA or GST-E6large in the absence of the His-caspase 8 DED were considered to represent the background signal, and this signal was subtracted from the raw experimental data. Binding between GST-E6 and the His-caspase 8 DED wt was set at 100%, and the binding between GST-E6 and other His-caspase 8 proteins was expressed as a percentage of that binding. Assays were performed in triplicate, and error bars represent the standard deviations. (C) Expression of the caspase 8 proteins. Equivalent amounts of the proteins used in the experiment shown in B were separated by SDS-PAGE, and the His-caspase 8 proteins were detected by using antibodies directed against caspase 8.
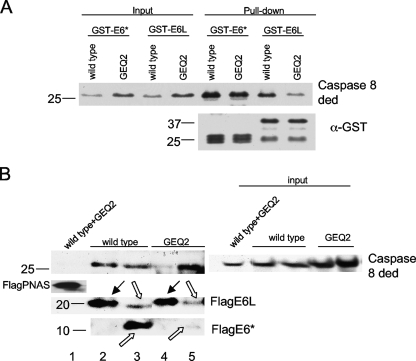
To further evaluate the effect of the GEQ2 set of mutations on the binding of the two isoforms of E6, this construct was tested in in vitro pulldown assays with bead-bound GST-E6large and GST-E6*. Figure 9A demonstrates that although the protein encoded by the GEQ2 mutant construct binds to both isoforms of E6, binding to E6large was significantly impaired, while binding to E6* was unchanged. To confirm these findings, a coimmunoprecipitation was also performed. U2OS cells stably expressing either Flag-tagged E6large or E6* were transfected with a plasmid encoding either the GEQ2 mutant or the wild-type protein. Forty-eight hours posttransfection, the ability of both isoforms to bind to GEQ2 and to the wt caspase 8 DED was assessed. Immunoprecipitation was performed with anti-Flag-agarose, which recognizes and binds to Flag-E6. Following SDS-PAGE, the resultant immunoblot was probed with antibodies directed against caspase 8 and against E6. Figure 9B verifies our previously reported findings and shows that despite its ability to strongly bind to the wild-type procaspase 8 DED, E6large binds to GEQ2 at a much lower efficiency, as shown by the decrease in the procaspase 8 pulldown band (top). In contrast to E6large, the mutations in GEQ2 did not impair the binding of E6* since both proteins are efficiently bound by the shorter isoform (Fig. 9B, top). Immunoblotting with an antibody that recognizes the N terminus of E6, and, thus, both isoforms of the protein, confirms that the immunoprecipitation of Flag-tagged E6 was successful (Fig. 9B, middle and bottom). Taken together, these results confirm that E6* and E6large utilize different amino acids to mediate their binding and that some of the amino acids in the binding pocket of E6large are those located within the N-terminal 16 amino acids of the procaspase 8 DED.
FIG. 9.
E6* and E6large bind to different regions on the procaspase 8 DED. (A) The GEQ2 set of mutations impairs binding to E6large but not to E6*, as assessed by a pulldown assay. The His-caspase 8 wt and GEQ2 mutant proteins were overexpressed in U2OS cells, and lysates were then prepared and incubated with GST-E6large or GST-E6* bound to glutathione beads. The detection of the bound caspase 8 DED wt and GEQ2 mutants was performed by immunoblotting using antibodies directed against caspase 8. The membrane was then stripped and reblotted using antibodies directed against GST (bottom). (B) The GEQ2 set of mutations impairs binding to E6large but not to E6*, as assessed by coimmunoprecipitation. U2OS cells transiently expressing either Flag-tagged E6large or E6* were transfected with plasmids encoding the GEQ2 mutant (pTriEx-cas8 ded GEQ2) or the wild-type caspase 8 DED (pTriExcas8 ded wt). Forty-eight hours posttransfection, immunoprecipitation was performed by using anti-Flag agarose, which recognizes and binds to Flag-E6. Following SDS-PAGE, the resultant immunoblot was probed with antibodies directed against caspase 8 and E6. Flag-PNAS was included as a negative control. Black arrows indicate Flag-E6large, and uncolored arrows indicate Flag-E6*. E6* can migrate as both monomers and dimers in SDS-PAGE gels.
DISCUSSION
Viruses have evolved numerous ways by which to escape recognition and subsequent elimination by the host immune response following infection; this facilitates their own survival and persistence. Many viruses produce molecules that target various steps in the apoptotic cascade, such as caspase inhibitors or homologues of cytokine or chemokine receptors (1, 5, 12, 49). In some cases, viruses can expand their existing collection of proteins via the use of polycistronic transcripts (30, 57), alternative splicing (10, 14, 37), or posttranslational processing (13, 33). For example, the splicing of the HPV16 early gene product E6 into an ∼0.5-kb and an approximately ∼0.3-kb transcript was reported previously by several laboratories, including our own (20, 43, 52). Combined with data from our previous report (20), the results reported here provide strong evidence that both isoforms of E6 have significant and separable effects on cellular signaling pathways, supporting the idea that splicing variability is an important mechanism for expanding the number of actions that this virus can carry out.
We have demonstrated that the expression of E6large confers cells with protection from cell death-inducing stimuli (Fig. 3A), causes decreased levels of procaspase 8 (Fig. 2), and binds to procaspase 8 (Fig. 6 to 8). We therefore sought to localize the binding site of the large isoform of E6, E6large, to procaspase 8 in the hope that inhibitors of this interaction could be generated, used to obstruct E6 binding, and sensitize cells to apoptosis. We recently identified a novel E6 binding site on the FADD DED that mediates oncoprotein binding (54). Since FADD and procaspase 8 both contain a DED, the possibility that similar residues facilitate E6/procaspase 8 binding was considered. An initial sequence alignment of the two proteins revealed regions of conserved amino acids and thus provided a basis for the creation of deletion and mutant constructs to help localize the E6 binding site. The level of binding to E6large was significantly reduced either when the N-terminal 16 amino acids were removed or when two separate sets of point mutations were created within this 16-amino-acid sequence, as assessed by three independent in vitro binding assays (Fig. 8 and 9). However, this binding was not completely eliminated. Therefore, although some of the targeted amino acids in the N terminus of the procaspase 8 DED appear to be involved in binding to E6large, other residues or regions are also likely to contribute. However, the overall results from this set of mutation experiments support the idea that the regions of the FADD DED and caspase 8 DED that bind to E6large are similar.
The E6 binding domain on FADD is composed of the serine residues at positions 16 and 18 and the leucine residue at position 20 of the protein sequence and localizes to the outer surface of the molecule. It was previously reported that the amino acids at positions 25, 33, 34, and 35 of the FADD DED are important for procaspase 8 binding (29). These residues are in close proximity to those in the E6 binding site, which implies that both E6 and procaspase 8 bind to the same surface of FADD. Additionally, F24 and L25 of the procaspase 8 DED are those that were previously suggested to interact with the FADD DED (9). According to our sequence alignment, these residues are in the vicinity of those that may play a role in E6 binding.
The similarity between the FADD and caspase 8 binding sites suggested that inhibitors of the E6/FADD interaction might also hinder the E6/caspase 8 interaction. To test this prediction, we utilized the AlphaScreen assay to detect protein-protein interactions in the absence and presence of a peptide inhibitor. Figure 4C demonstrates that peptide A, which specifically impairs E6 binding to FADD, also impairs the binding of E6 to procaspase 8 while leaving E6/E6AP binding undisturbed. This confirms the results that we obtained from our mutational analysis, which indicated that the E6 binding domain on FADD is similar to that of procaspase 8. Furthermore, this experiment also shows that this domain differs from the region of E6AP required for E6large binding.
One of the most interesting findings from this work was that the two E6 splice isoforms bind to different regions of the caspase 8 DED. Two separate lines of evidence led us to this conclusion. First, a peptide that blocks E6large/caspase 8 binding does not inhibit E6*/caspase binding (Fig. 7A). Next, three separate mutations, ΔN, EAG, and GEQ2, affected the binding of E6large to caspase 8 much more than the binding of E6* to caspase 8, as assessed in three ways: the bead-binding assay, an in vitro pulldown assay, and coimmunoprecipitation (Fig. 7 to 9). This is somewhat remarkable in view of the fact that E6* is approximately equivalent to the N-terminal half of E6large. The actual binding site on caspase 8 with which E6* interacts has not yet been identified; when it is, it may be possible to explore how it is that a protein that nearly matches the N terminus of another protein can utilize what appears to be a separate binding site.
Our results from the bead-based binding assay demonstrate the effectiveness of peptide inhibitors in obstructing protein-protein binding and suggest that peptides could be used to block oncoprotein binding to FADD and procaspase 8 in HPV-infected cells and thus to resensitize these cells to apoptosis-inducing agents. However, several laboratories reported a low efficiency of peptide entry into cells. Therefore, different routes have been explored to increase uptake. These include the addition of the HIV-1 Tat peptide or the herpesvirus VP22 peptide to the amino or carboxy terminus of the peptide inhibitor. Also, the incorporation of a liposome moiety into the peptide prior to administration into cells was previously reported to facilitate peptide trafficking across the cell membrane (7, 16, 47). Nonetheless, problems with peptide stability and solubility as well as target specificity remain with regard to peptide entry into cells.
Overall, our results demonstrate that the HPV16 E6large and E6* isoforms bind differentially to procaspase 8, supporting and providing a molecular explanation for our previously reported findings regarding the variable effect that each entity has on procaspase 8 stability. Furthermore, the ability of our peptide inhibitor to impede E6large binding to both FADD and procaspase 8 contributes to the current literature regarding the design of potential therapeutic agents to treat HPV16 infection.
Acknowledgments
This study was supported by NIH grant R01 CA-095461 (P.J.D.-H.).
Footnotes
Published ahead of print on 11 November 2009.
REFERENCES
- 1.Alcami, A. 2003. Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. Immunol. 3:36-50. [DOI] [PubMed] [Google Scholar]
- 2.Be, X., Y. Hong, J. Wei, E. J. Androphy, J. J. Chen, and J. D. Baleja. 2001. Solution structure determination and mutational analysis of the papillomavirus E6 interacting peptide of E6AP. Biochemistry 40:1293-1299. [DOI] [PubMed] [Google Scholar]
- 3.Bedell, M. A., K. H. Jones, and L. A. Laimins. 1987. The E6-E7 region of human papillomavirus type 18 is sufficient for transformation of NIH 3T3 and rat-1 cells. J. Virol. 61:3635-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belanger, C., A. Gravel, A. Tomoiu, M. E. Janelle, J. Gosselin, M. J. Tremblay, and L. Flamand. 2001. Human herpesvirus 8 viral FLICE-inhibitory protein inhibits Fas-mediated apoptosis through binding and prevention of procaspase-8 maturation. J. Hum. Virol. 4:62-73. [PubMed] [Google Scholar]
- 5.Bertin, J., R. C. Armstrong, S. Ottilie, D. A. Martin, Y. Wang, S. Banks, G. H. Wang, T. G. Senkevich, E. S. Alnemri, B. Moss, M. J. Lenardo, K. J. Tomaselli, and J. I. Cohen. 1997. Death effector domain-containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR1-induced apoptosis. Proc. Natl. Acad. Sci. U. S. A. 94:1172-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boccardo, E., and L. L. Villa. 2007. Viral origins of human cancer. Curr. Med. Chem. 14:2526-2539. [DOI] [PubMed] [Google Scholar]
- 7.Brooks, H., B. Lebleu, and E. Vives. 2005. Tat peptide-mediated cellular delivery: back to basics. Adv. Drug Deliv. Rev. 57:559-577. [DOI] [PubMed] [Google Scholar]
- 8.Butel, J. S. 2000. Viral carcinogenesis: revelation of molecular mechanisms and etiology of human disease. Carcinogenesis 21:405-426. [DOI] [PubMed] [Google Scholar]
- 9.Carrington, P. E., C. Sandu, Y. Wei, J. M. Hill, G. Morisawa, T. Huang, E. Gavathiotis, Y. Wei, and M. H. Werner. 2006. The structure of FADD and its mode of interaction with procaspase-8. Mol. Cell 22:599-610. [DOI] [PubMed] [Google Scholar]
- 10.Cereseto, A., Z. Berneman, I. Koralnik, J. Vaughn, G. Franchini, and M. E. Klotman. 1997. Differential expression of alternatively spliced pX mRNAs in HTLV-I-infected cell lines. Leukemia 11:866-870. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, B., S. Schneider, J. Bohl, Y. Jiang, A. Beaudet, and S. Vande Pol. 2003. Requirement of E6AP and the features of human papillomavirus E6 necessary to support degradation of p53. Virology 306:87-99. [DOI] [PubMed] [Google Scholar]
- 12.Cunnion, K. M. 1999. Tumor necrosis factor receptors encoded by poxviruses. Mol. Genet. Metab. 67:278-282. [DOI] [PubMed] [Google Scholar]
- 13.de Turenne-Tessier, M., and T. Ooka. 2007. Post-translational modifications of Epstein Barr virus BARF1 oncogene-encoded polypeptide. J. Gen. Virol. 88:2656-2661. [DOI] [PubMed] [Google Scholar]
- 14.Dowling, D., S. Nasr-Esfahani, C. H. Tan, K. O'Brien, J. L. Howard, D. A. Jans, D. F. Purcell, C. M. Stoltzfus, and S. Sonza. 2008. HIV-1 infection induces changes in expression of cellular splicing factors that regulate alternative viral splicing and virus production in macrophages. Retrovirology 5:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elston, R. C., S. Napthine, and J. Doorbar. 1998. The identification of a conserved binding motif within human papillomavirus type 16 E6 binding peptides, E6AP and E6BP. J. Gen. Virol. 79(Pt. 2):371-374. [DOI] [PubMed] [Google Scholar]
- 16.Fawell, S., J. Seery, Y. Daikh, C. Moore, L. L. Chen, B. Pepinsky, and J. Barsoum. 1994. Tat-mediated delivery of heterologous proteins into cells. Proc. Natl. Acad. Sci. U. S. A. 91:664-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsani, A., A. M. Mileo, and M. G. Paggi. 2006. Retinoblastoma family proteins as key targets of the small DNA virus oncoproteins. Oncogene 25:5277-5285. [DOI] [PubMed] [Google Scholar]
- 18.Filippova, M., T. A. Brown-Bryan, C. A. Casiano, and P. J. Duerksen-Hughes. 2005. The human papillomavirus 16 E6 protein can either protect or further sensitize cells to TNF: effect of dose. Cell Death Differ. 12:1622-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filippova, M., V. A. Filippov, M. Kagoda, T. Garnett, N. Fodor, and P. J. Duerksen-Hughes. 2009. Complexes of human papillomavirus type 16 E6 proteins form pseudo-death-inducing signaling complex structures during tumor necrosis factor-mediated apoptosis. J. Virol. 83:210-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Filippova, M., M. M. Johnson, M. Bautista, V. Filippov, N. Fodor, S. S. Tungteakkhun, K. Williams, and P. J. Duerksen-Hughes. 2007. The large and small isoforms of human papillomavirus type 16 E6 bind to and differentially affect procaspase 8 stability and activity. J. Virol. 81:4116-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filippova, M., L. Parkhurst, and P. J. Duerksen-Hughes. 2004. The human papillomavirus 16 E6 protein binds to Fas-associated death domain and protects cells from Fas-triggered apoptosis. J. Biol. Chem. 279:25729-25744. [DOI] [PubMed] [Google Scholar]
- 22.Filippova, M., H. Song, J. L. Connolly, T. S. Dermody, and P. J. Duerksen-Hughes. 2002. The human papillomavirus 16 E6 protein binds to tumor necrosis factor (TNF) R1 and protects cells from TNF-induced apoptosis. J. Biol. Chem. 277:21730-21739. [DOI] [PubMed] [Google Scholar]
- 23.Gardiol, D., S. Galizzi, and L. Banks. 2002. Mutational analysis of the discs large tumour suppressor identifies domains responsible for human papillomavirus type 18 E6-mediated degradation. J. Gen. Virol. 83:283-289. [DOI] [PubMed] [Google Scholar]
- 24.Garland, S. M. 2002. Human papillomavirus update with a particular focus on cervical disease. Pathology 34:213-224. [DOI] [PubMed] [Google Scholar]
- 25.Garvey, T., J. Bertin, R. Siegel, M. Lenardo, and J. Cohen. 2002. The death effector domains (DEDs) of the molluscum contagiosum virus MC159 v-FLIP protein are not functionally interchangeable with each other or with the DEDs of caspase-8. Virology 300:217-225. [DOI] [PubMed] [Google Scholar]
- 26.Gillison, M. L., W. M. Koch, R. B. Capone, M. Spafford, W. H. Westra, L. Wu, M. L. Zahurak, R. W. Daniel, M. Viglione, D. E. Symer, K. V. Shah, and D. Sidransky. 2000. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 92:709-720. [DOI] [PubMed] [Google Scholar]
- 27.Hafner, N., C. Driesch, M. Gajda, L. Jansen, R. Kirchmayr, I. B. Runnebaum, and M. Durst. 2008. Integration of the HPV16 genome does not invariably result in high levels of viral oncogene transcripts. Oncogene 27:1610-1617. [DOI] [PubMed] [Google Scholar]
- 28.Hoppe-Seyler, F., and K. Butz. 1995. Molecular mechanisms of virus-induced carcinogenesis: the interaction of viral factors with cellular tumor suppressor proteins. J. Mol. Med. 73:529-538. [DOI] [PubMed] [Google Scholar]
- 29.Kaufmann, M., D. Bozic, C. Briand, J. L. Bodmer, O. Zerbe, A. Kohl, J. Tschopp, and M. G. Grutter. 2002. Identification of a basic surface area of the FADD death effector domain critical for apoptotic signaling. FEBS Lett. 527:250-254. [DOI] [PubMed] [Google Scholar]
- 30.Lamb, R. A., and C. M. Horvath. 1991. Diversity of coding strategies in influenza viruses. Trends Genet. 7:261-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langelier, Y., S. Bergeron, S. Chabaud, J. Lippens, C. Guilbault, A. M. Sasseville, S. Denis, D. D. Mosser, and B. Massie. 2002. The R1 subunit of herpes simplex virus ribonucleotide reductase protects cells against apoptosis at, or upstream of, caspase-8 activation. J. Gen. Virol. 83:2779-2789. [DOI] [PubMed] [Google Scholar]
- 32.Lee, C., and L. A. Laimins. 2004. Role of the PDZ domain-binding motif of the oncoprotein E6 in the pathogenesis of human papillomavirus type 31. J. Virol. 78:12366-12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, L., N. Bastien, and Y. Li. 2007. Intracellular processing, glycosylation, and cell surface expression of human metapneumovirus attachment glycoprotein. J. Virol. 81:13435-13443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, Y., and J. D. Baleja. 2008. Structure and function of the papillomavirus E6 protein and its interacting proteins. Front. Biosci. 13:121-134. [DOI] [PubMed] [Google Scholar]
- 35.Liu, Y., Z. Liu, E. Androphy, J. Chen, and J. D. Baleja. 2004. Design and characterization of helical peptides that inhibit the E6 protein of papillomavirus. Biochemistry 43:7421-7431. [DOI] [PubMed] [Google Scholar]
- 36.Lopes, V., L. S. Young, and P. G. Murray. 2003. Epstein-Barr virus-associated cancers: aetiology and treatment. Herpes 10:78-82. [PubMed] [Google Scholar]
- 37.Lutzelberger, M., L. S. Reinert, A. T. Das, B. Berkhout, and J. Kjems. 2006. A novel splice donor site in the gag-pol gene is required for HIV-1 RNA stability. J. Biol. Chem. 281:18644-18651. [DOI] [PubMed] [Google Scholar]
- 38.Matsumoto, Y., S. Nakagawa, T. Yano, S. Takizawa, K. Nagasaka, K. Nakagawa, T. Minaguchi, O. Wada, H. Ooishi, K. Matsumoto, T. Yasugi, T. Kanda, J. M. Huibregtse, and Y. Taketani. 2006. Involvement of a cellular ubiquitin-protein ligase E6AP in the ubiquitin-mediated degradation of extensive substrates of high-risk human papillomavirus E6. J. Med. Virol. 78:501-507. [DOI] [PubMed] [Google Scholar]
- 39.Melton, J. L., and J. E. Rasmussen. 1991. Clinical manifestations of human papillomavirus infection in nongenital sites. Dermatol. Clin. 9:219-233. [PubMed] [Google Scholar]
- 40.Morris, J. D., A. L. Eddleston, and T. Crook. 1995. Viral infection and cancer. Lancet 346:754-758. [DOI] [PubMed] [Google Scholar]
- 41.Motoyama, S., C. A. Ladines-Llave, S. L. Villanueva, and T. Maruo. 2004. The role of human papilloma virus in the molecular biology of cervical carcinogenesis. Kobe J. Med. Sci. 50:9-19. [PubMed] [Google Scholar]
- 42.Nomine, Y., M. Masson, S. Charbonnier, K. Zanier, T. Ristriani, F. Deryckere, A. P. Sibler, D. Desplancq, R. A. Atkinson, E. Weiss, G. Orfanoudakis, B. Kieffer, and G. Trave. 2006. Structural and functional analysis of E6 oncoprotein: insights in the molecular pathways of human papillomavirus-mediated pathogenesis. Mol. Cell 21:665-678. [DOI] [PubMed] [Google Scholar]
- 43.Rohlfs, M., S. Winkenbach, S. Meyer, T. Rupp, and M. Durst. 1991. Viral transcription in human keratinocyte cell lines immortalized by human papillomavirus type-16. Virology 183:331-342. [DOI] [PubMed] [Google Scholar]
- 44.Scheurer, M. E., G. Tortolero-Luna, and K. Adler-Storthz. 2005. Human papillomavirus infection: biology, epidemiology, and prevention. Int. J. Gynecol. Cancer 15:727-746. [DOI] [PubMed] [Google Scholar]
- 45.Schiffman, M., P. E. Castle, J. Jeronimo, A. C. Rodriguez, and S. Wacholder. 2007. Human papillomavirus and cervical cancer. Lancet 370:890-907. [DOI] [PubMed] [Google Scholar]
- 46.Schultz, D. R., and W. J. Harrington, Jr. 2003. Apoptosis: programmed cell death at a molecular level. Semin. Arthritis Rheum. 32:345-369. [DOI] [PubMed] [Google Scholar]
- 47.Schwartz, J. J., and S. Zhang. 2000. Peptide-mediated cellular delivery. Curr. Opin. Mol. Ther. 2:162-167. [PubMed] [Google Scholar]
- 48.Shiratori, Y., S. Shiina, M. Imamura, N. Kato, F. Kanai, T. Okudaira, T. Teratani, G. Tohgo, N. Toda, M. Ohashi, et al. 1995. Characteristic difference of hepatocellular carcinoma between hepatitis B- and C-viral infection in Japan. Hepatology 22:1027-1033. [DOI] [PubMed] [Google Scholar]
- 49.Shisler, J. L., and B. Moss. 2001. Molluscum contagiosum virus inhibitors of apoptosis: the MC159 v-FLIP protein blocks Fas-induced activation of procaspases and degradation of the related MC160 protein. Virology 282:14-25. [DOI] [PubMed] [Google Scholar]
- 50.Skaletskaya, A., L. M. Bartle, T. Chittenden, A. L. McCormick, E. S. Mocarski, and V. S. Goldmacher. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. U. S. A. 98:7829-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talis, A. L., J. M. Huibregtse, and P. M. Howley. 1998. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J. Biol. Chem. 273:6439-6445. [DOI] [PubMed] [Google Scholar]
- 52.Tang, S., M. Tao, J. P. McCoy, Jr., and Z. M. Zheng. 2006. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J. Virol. 80:4249-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tungteakkhun, S. S., and P. J. Duerksen-Hughes. 2008. Cellular binding partners of the human papillomavirus E6 protein. Arch. Virol. 153:397-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tungteakkhun, S. S., M. Filippova, J. W. Neidigh, N. Fodor, and P. J. Duerksen-Hughes. 2008. The interaction between human papillomavirus type 16 and FADD is mediated by a novel E6 binding domain. J. Virol. 82:9600-9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wajant, H. 2002. The Fas signaling pathway: more than a paradigm. Science 296:1635-1636. [DOI] [PubMed] [Google Scholar]
- 56.Zhou, Q., S. Snipas, K. Orth, M. Muzio, V. M. Dixit, and G. S. Salvesen. 1997. Target protease specificity of the viral serpin CrmA. Analysis of five caspases. J. Biol. Chem. 272:7797-7800. [DOI] [PubMed] [Google Scholar]
- 57.Ziff, E. B. 1985. Splicing in adenovirus and other animal viruses. Int. Rev. Cytol. 93:327-358. [DOI] [PubMed] [Google Scholar]
- 58.Zimmermann, K. C., C. Bonzon, and D. R. Green. 2001. The machinery of programmed cell death. Pharmacol. Ther. 92:57-70. [DOI] [PubMed] [Google Scholar]
- 59.Zimmermann, K. C., and D. R. Green. 2001. How cells die: apoptosis pathways. J. Allergy Clin. Immunol. 108:S99-S103. [DOI] [PubMed] [Google Scholar]
- 60.zur Hausen, H. 1991. Viruses in human cancers. Science 254:1167-1173. [DOI] [PubMed] [Google Scholar]