Abstract
Hepatitis delta virus (HDV) RNA forms an unbranched rod structure that is associated with hepatitis delta antigen (HDAg) in cells replicating HDV. Previous in vitro binding experiments using bacterially expressed HDAg showed that the formation of a minimal ribonucleoprotein complex requires an HDV unbranched rod RNA of at least about 300 nucleotides (nt) and suggested that HDAg binds the RNA as a multimer of fixed size. The present study specifically examines the role of HDAg multimerization in the formation of the HDV ribonucleoprotein complex (RNP). Disruption of HDAg multimerization by site-directed mutagenesis was found to profoundly alter the nature of RNP formation. Mutant HDAg proteins defective for multimerization exhibited neither the 300-nt RNA size requirement for binding nor specificity for the unbranched rod structure. The results unambiguously demonstrate that HDAg binds HDV RNA as a multimer and that the HDAg multimer is formed prior to binding the RNA. RNP formation was found to be temperature dependent, which is consistent with conformational changes occurring on binding. Finally, analysis of RNPs constructed with unbranched rod RNAs successively longer than the minimum length indicated that multimeric binding is not limited to the first HDAg bound and that a minimum RNA length of between 604 and 714 nt is required for binding of a second multimer. The results confirm the previous proposal that HDAg binds as a large multimer and demonstrate that the multimer is a critical determinant of the structure of the HDV RNP.
Human hepatitis delta virus (HDV) is an unusual subviral agent that increases the severity of acute and chronic liver disease in those infected with its helper, hepatitis B virus (23). The HDV genome is a 1,680-nucleotide (nt) single-stranded circular RNA that is replicated by a double-rolling-circle mechanism (reviewed in references 15 and 28). Both the genome and antigenome RNAs form a characteristic unbranched rod structure due to 70% sequence complementarity between the noncoding and coding regions of the RNA (10, 11, 31). HDV encodes just one protein, hepatitis delta antigen (HDAg), which forms ribonucleoprotein (RNP) complexes with both the genome and the antigenome in cells replicating HDV (3, 5, 30). These complexes play fundamental roles in viral RNA replication and packaging and their characterization is essential for understanding these processes, which are not well characterized.
HDAg has been shown to form dimers and higher order multimers, even in the absence of HDV RNA (25, 30, 32). The multimerization activity has been localized to the amino-terminal third of the 195-amino-acid (aa) protein (12, 24, 30, 32). X-ray crystallographic analysis of a peptide comprised of aa 12 to 60 indicated that antiparallel dimers are stabilized by a coiled coil (aa 16 to 48), as well as a hydrophobic core region (aa 50 to 60) that also stabilizes interactions between dimers such that an octameric structure may form (35). Zuccola et al. found that bacterially expressed HDAg could be cross-linked in an octameric structure, and Cornillez-Ty et al. obtained evidence supporting such a structure in cells replicating HDV (7, 35). Site-directed mutations of HDAg amino acids critical for dimerization and/or multimerization abolish the ability of HDAg to support RNA replication (18, 32), indicating that the formation of HDAg multimers is essential for this process.
We recently showed that bacterially expressed, C-terminally truncated HDAg forms stable RNP complexes in vitro with segments of HDV RNA that form unbranched rod structures (8). No particular sequences or structures in the RNA, other than the HDV unbranched rod, were essential for complex formation, but, remarkably, binding required that the RNA have a minimum length of at least about 300 nt. Overall, the results were consistent with the formation of a large RNP containing multiple copies of the 19-kDa protein that bound to the RNA either in a highly cooperative manner or as a preformed multimer. On the other hand, based on indirect measures of the RNA-binding activity of site-directed HDAg mutations in cells, others have found that HDAg multimerization might not be required for RNA-binding activity (18).
Here, we directly analyze the role of HDAg multimerization in the formation of the HDV RNP complex. We find that HDAg binds to HDV unbranched rod RNA as a preformed multimer. Site-directed mutations that disrupted protein multimerization did not abolish binding but profoundly altered the nature of the RNA-protein complex. In particular, we found that multimerization is associated with RNA-binding specificity, including the RNA length requirement for binding. For the wild-type protein, RNP formation was found to be strongly temperature dependent, suggesting that conformational changes occur on binding, and providing a plausible explanation of the RNA length requirement for binding. Furthermore, we show that the protein binds as multiple multimeric units on longer RNAs, provided the length of the RNA is sufficient. We conclude that the HDAg multimer plays a critical role in the formation of properly structured HDV RNPs.
MATERIALS AND METHODS
Bacterial expression and purification of HDAg.
For the expression of wild-type HDAg-160 and HDAg-145, the HDAg open reading frames for amino acids 2 to 160 and amino acids 2 to 145, respectively, were amplified by PCR from the plasmid template pCMV-AgS (21) with additional sequences encoding for six histidines at the N terminus added to the 5′ end of the forward primer (8). These sequences were cloned between the NdeI and HindIII sites of pET30c (Novagen) to generate the bacterial expression plasmids pET-H6HDAg-160 and pET-H6HDAg-145. Mutations 37/44G and W50A were created in the pET-H6HDAg-160 plasmid by site-directed mutagenesis using PCR primers containing the desired mutations and the unique PstI site in the HDAg cDNA. For the clone used (31), the 37/44G double mutation included substitution of G for T37 and of G for I44. All mutations created by site-directed mutagenesis using the PCR were verified by sequencing. Expression and purification of His6-tagged proteins were as previously described (8). Briefly, expression was carried out in Rosetta(DE3)BL21/pLysS cells induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubated overnight at 25°C. Cells were lysed by sonication, and proteins were purified on ProBond Ni-NTA columns (Invitrogen, Carlsbad, CA), according to the manufacturer's specifications. Proteins were analyzed by SDS-PAGE and quantified by SYPRO (Invitrogen) staining intensity in comparison to bovine serum albumin (BSA) concentration standards.
Rate zonal ultracentrifugation.
Protein samples were diluted in phosphate-buffered saline (200 μl) and layered on top of 5 to 30% (wt/vol) linear sucrose gradients at 4°C. HDAg-160 (wild-type and mutant) samples were treated with RNase A (10 μg/ml) for 15 m at 37°C prior to placing on the gradient. Protein standards were 20 μg of chicken lysozyme (Lys), BSA, and yeast alcohol dehydrogenase (ADH) (Sigma-Aldrich, St. Louis, MO). Samples were centrifuged at 32,000 rpm in an SW41 rotor for 20 h at 4°C. Fractions (500 μl) were recovered from the top of the gradient. HDAg was quantified by immunoblotting with the anti-HD monoclonal antibody T1/39 (19) and the horseradish peroxidase-conjugated anti-human IgG (KPL, Gaithersburg, MD) and ECL-Plus chemifluorescent reagent (Amersham); chemifluorescent signals were detected with a Molecular Dynamics Storm 840 PhosphorImager and quantified by using ImageJ (W. S. Rasband, 1997-2009, ImageJ [http://rsb.info.nih.gov/ij/ed; National Institutes of Health, Bethesda, MD]). Protein standards were analyzed by SDS-PAGE, followed by staining with PageBlue (Fermentas, Glen Burnie, MD).
RNA preparation.
Transcription reaction conditions and RNA purification procedures were as previously described (8). RNAs were transcribed from PCR products into which the T7 promoter sequence was incorporated via the forward primer; transcription templates were amplified from the plasmid pCMV-DC1×1.2-AgS(-) (4). All RNAs used were antigenomic sense and were named according to the length in nucleotides. RNAs 395L, 311L, 253L, and 207L were described previously (8). The RNA 497L includes the 497 nt from nt 203 (5′) to nt 1389 (3′); 604L includes nt 257 (5′) to nt 1336 (3′); and 714L includes nt 314 (5′) to nt 1283 (3′) (see the figures for schematic representations). Prior to synthesis, RNA sequences were analyzed by using the folding algorithm mfold (16, 36) to confirm that the unbranched rod structure was the most thermodynamically favored structure; in all cases, unbranched rod structures were favored over those with short branches (2 to 6 bp) by more than 4.7 kcal. Transcripts were purified from native polyacrylamide gels to minimize contributions of undesired alternative secondary structures formed during transcription; in all cases the major species (at least ∼90%) in the gel corresponded to the unbranched rod conformation. Heating RNAs to 90°C, followed by either quench cooling on ice or slow cooling, had no effect on the mobility of RNAs in the gel, binding by HDAg, or the mobility of RNPs formed. RNA dimers were ruled out based on the lack of an effect on gel mobility of heating, followed by quench cooling, and on the observation that the mobility of the 395L RNA was the same as that of the annealed 390NL duplex (8), which could not form the equivalent of a dimer. The double-stranded RNA ds395 was prepared by annealing two cRNA transcripts. Templates for transcription of the genomic and antigenomic RNAs from nt 1441 to 1637 were amplified by PCR. RNA transcripts were purified by native polyacrylamide gel electrophoresis. Equimolar amounts were annealed by overnight incubation at 45°C in 40 mM PIPES (pH 6.7,) 400 mM NaCl, 1 mM dithiothreitol (DTT) after denaturation at 75°C for 5 min. Annealed dsRNAs were gel purified again.
Electrophoretic mobility shift assays.
Electrophoretic mobility shift assays were performed as previously described (8). Briefly, binding reactions included 10 mM Tris-HCl (pH 7.5), 25 mM KCl, 10 mM NaCl, 0.1 μg of bovine serum albumin/μl, 5% glycerol, 0.5 mM DTT, 0.2 U of RNase inhibitor (Applied Biosystems)/μl, and 1 mM phenylmethylsulfonyl fluoride solution. Radiolabeled RNA concentrations were 5 pM, unless indicated otherwise; protein concentrations were as indicated in figure legends. Reactions were assembled on ice, incubated at 37°C for 1 h, and electrophoresed on 6% native polyacrylamide gels in 0.5× Tris-borate-EDTA at 250 V for 2 h at room temperature unless otherwise noted. Lower electrophoresis temperatures did not alter the results. The results were visualized by using a Molecular Dynamics Storm 840 PhosphorImager.
RESULTS
HDAg forms large multimeric complexes in vitro.
We previously detected a stable, discrete, RNP complex of HDAg with HDV unbranched rod RNA in an electrophoretic mobility shift assay using purified recombinant HDAg, expressed in bacteria as a 19-kDa form truncated by 35 aa from the C terminus; we refer to this protein as HDAg-160 because it terminates at HDAg aa 160 (8). Several characteristics of the RNP complex and its formation, including the minimum length RNA required for binding, the electrophoretic mobility of the RNP, and the stability of the complex to micrococcal nuclease digestion, led to the suggestion that the RNP contains an HDAg multimer (8). The formation of such multimers would be consistent with observations that HDAg forms multimeric complexes, even in the absence of HDV RNA (5, 7, 24, 35).
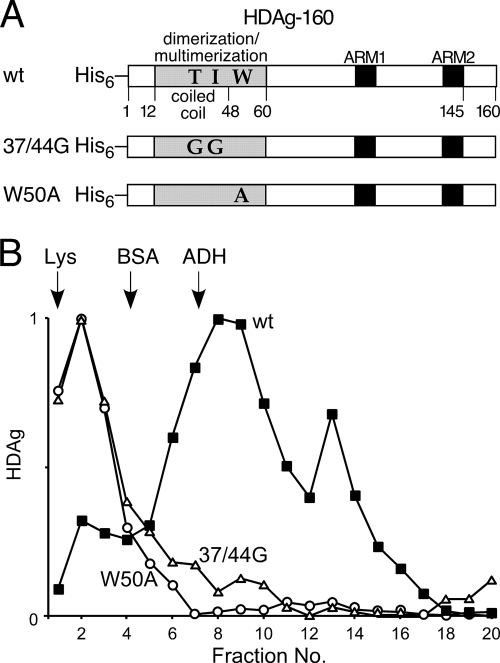
To more directly analyze whether HDAg binds HDV RNA as a multimer and to determine the role of HDAg multimerization in RNA binding, we created two mutant forms of HDAg-160, in which site-directed mutations were introduced in the region responsible for the formation of HDAg multimers (Fig. 1A). The mutation 37/44G is based on a double mutation created by Xia et al. (32), who introduced glycine substitutions at positions 37 and 44, which are present in the coiled coil dimerization region. As measured by glutaraldehyde cross-linking in vitro, this double mutation was observed to disrupt dimer formation (32). The crystal structure of the multimerization domain (aa 12 to 60) obtained by Zuccola et al. indicated that aa W50 plays a critical role in a hydrophobic core region that stabilizes both antiparallel dimers and octamers formed by interactions between pairs of dimers (35). Using cell-based packaging and coimmunoprecipitation assays, Moraleda et al. showed that mutation of W50 to A substantially reduced the ability of the protein to form complexes (18).
FIG. 1.
(A) Schematic of bacterially expressed HDAg proteins. All proteins were expressed with N-terminal His6 tags. The protein multimerization region (32, 35) is shaded gray; the two arginine-rich motifs (ARM1 and ARM2) involved in RNA binding are shaded black. The amino acid identities of positions 37, 44, and 50 in the wild-type (wt) isolate used in the present study are indicated, as are the identities of these positions in the 37/44G and W50A mutants. (B) Rate zonal sucrose gradient centrifugation of bacterially expressed HDAg-160. Samples were prepared and centrifuged as described in Materials and Methods. Fraction numbers increase from the top to the bottom of the gradient. Arrows indicate the positions of peak fractions of the protein standards (Lys, lysozyme, 14.3 kDa; BSA, bovine serum albumin, 66 kDa; ADH, yeast alcohol dehydrogenase, 150 kDa).
We analyzed the effects of the W50A and 37/44G mutations on the ability of bacterially expressed HDAg-160 to form multimers in vitro in the absence of HDV RNA using rate zonal centrifugation on 5 to 30% sucrose gradients. Representative results are shown in Fig. 1B. The peak fractions for wild-type HDAg-160 were just below those of alcohol dehydrogenase (molecular weight, 150,000), indicating that the 19-kDa protein forms a large multimeric complex. This result is consistent with the observed glutaraldehyde cross-linking of an octameric complex formed by full-length HDAg expressed and purified from bacteria (35), as well as sucrose gradient sedimentation of HDAg complexes isolated from infected liver and transfected cells (5, 14, 25, 30). The protein complex did not require RNA, because samples were treated with RNase prior to centrifugation. The second, smaller HDAg peak in fraction 13 was not observed consistently and may have been due to incomplete RNase digestion.
The 37/44G and W50A mutant proteins produced nearly identical gradient profiles that differed markedly from that of the wild type (Fig. 1B): the peak fractions for both mutant proteins were at the top of the gradient. Based on the migration of the molecular weight standards, the peak position of both mutants is consistent with the expected behavior of an HDAg-160 monomer, indicating that disruption of either the coiled-coil (37/44G) or the hydrophobic core region (W50A) interferes with the formation of HDAg multimers, including dimers. These results agree with previous observations of others for these mutations (18, 32). Although Moraleda observed heterodimers formed by wild-type and W50A mutant HDAg, the ability of the W50A mutant protein to form homodimers in cells was severely reduced (18).
Disruption of HDAg multimerization alters the binding of HDAg to HDV RNA.
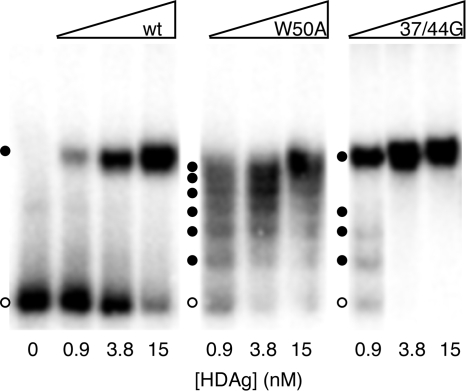
To examine the role of HDAg multimerization in forming complexes with HDV RNA, we used an electrophoretic mobility shift assay to analyze binding of the wt and mutant HDAg-160 proteins to 395L RNA. As shown previously (8), this RNA, which forms an unbranched rod structure, forms a discrete, stable complex with HDAg-160 in vitro that is readily observed by native gel electrophoresis (Fig. 2, left). The W50A and 37/44G mutations exhibited two marked effects on the binding of HDAg to 395L RNA (Fig. 2). First, comparison of the amounts of unbound RNA in the presence of low protein concentrations indicates that both mutant proteins exhibited higher affinity binding for the RNA. For example, in the presence of 0.9 nM wild-type protein, ca. 90% of the RNA remained unbound, whereas for both mutants the amount of unbound RNA at this protein concentration was less than 20%. Second, while wild-type HDAg-160 formed just one complex with about half the electrophoretic mobility of unbound RNA in the native 6% acrylamide gel, both mutant proteins produced a series of complexes with intermediate mobilities. These complexes were particularly prominent for the W50A mutation (Fig. 2, middle panel). The observation that the mutant proteins exist as monomers in the absence of RNA (Fig. 1) could suggest that the complexes differ by the successive addition of monomers; however, we cannot rule out the possibility that, despite the mutations, monomers dimerize in the process of binding the RNA.
FIG. 2.
Electrophoretic mobility shift of an HDV RNA segment with multimerization-defective HDAg mutants. 395L RNA (10 pM) was incubated with indicated concentrations of wild type (wt), W50A, or 37/44G HDAg-160 for 1 h at 37°C prior to electrophoresis in a 6% native acrylamide gel for 2 h. Open circles denote the locations of unbound RNA; solid circles indicate the locations of RNP complexes.
As the protein concentration was increased, the mobilities of the complexes formed by 395L RNA and the W50A mutant decreased, which is consistent with the interpretation that more protein was bound to the RNA in the slower-migrating complexes. The pattern of mobilities of these complexes suggested that they differ by the addition of a regular number of HDAg units. At least six complexes with mobilities faster than that of the RNP formed by the wild-type protein were clearly visible for the W50A mutant; possibly, more higher-molecular-weight complexes are present at higher protein concentrations but are not resolved in the gel. The 37/44G mutant also yielded complexes of intermediate mobility (Fig. 2, right panel), although these were not as numerous as those observed for W50A. Overall, these results clearly indicate that wild-type HDAg binds the 395L RNA as a large multimer.
HDAg binds unbranched RNA as a preformed multimer.
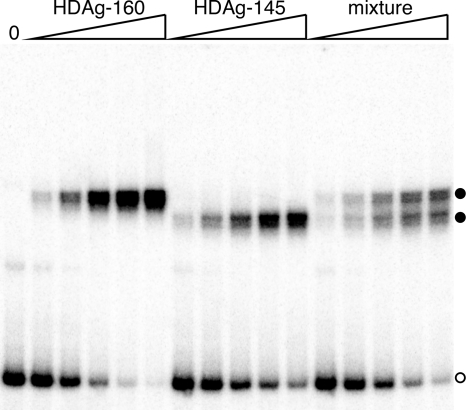
Given that HDAg exists primarily as a multimer in the absence of RNA and that no complexes of intermediate mobility were observed for the wild-type HDAg-160 protein, it seems most likely that HDAg binds the RNA as a preformed multimer. However, it is also possible that binding to the RNA is preceded by multimer dissociation and that the protein binds to the RNA with high cooperativity, such that no intermediate complexes are observed. To determine whether HDAg binds the RNA as a preformed multimer or whether binding is preceded by multimer dissociation, we compared RNP complexes formed by HDAg-160, HDAg-145, an additional truncated form of the protein, and a mixture of these two truncated forms of HDAg. Like HDAg-160, HDAg-145 is a C-terminally truncated protein that retains all sequences known to be involved in RNA binding (Fig. 1A). Analysis of RNP formation in an electrophoretic mobility shift assay indicated that HDAg-145 exhibits binding activity with 395L RNA similar to that of HDAg-160 (Fig. 3). Consistent with the lower molecular weight of the protein, RNA-protein complexes formed by HDAg-145 migrate faster in the gel than those formed by HDAg-160. With the extended electrophoresis times used in Fig. 3 (3.5 h), the mobilities of the RNP complexes formed by 395L RNA with these two proteins are readily distinguishable. We also observed with this extended electrophoresis that complexes formed by both proteins resolve into two species with similarly spaced mobilities, indicating that there might be some degree of heterogeneity in the RNP complexes formed. The nature of this heterogeneity is currently uncertain but could potentially be related to heterogeneity in the protein itself (including posttranslational modifications such as phosphorylation [33]), or even in the number of HDAg monomers present.
FIG. 3.
HDAg binds an HDV RNA segment as a preformed multimer. 395L RNA was incubated with increasing concentrations of either HDAg-160 (left lanes), HDAg-145 (middle lanes), or a mixture of equal amounts of the two proteins. The total protein concentrations were 0.5, 1.7, 6.1, 23, and 84 nM. RNP complexes formed were electrophoresed on a 6% native polyacrylamide gel for 3.5 h. The open circle denotes the location of unbound RNA; the solid circles indicate the locations of RNP complexes formed by the two proteins.
We reasoned that if HDAg multimers dissociate prior to binding the RNA, then we would expect that incubation of the RNA with a mixture of the HDAg-145 and HDAg-160 proteins would produce heterogeneous RNP complexes composed of various ratios of HDAg-145 and HDAg-160, with mobilities intermediate between those formed by HDAg-145 and HDAg-160. Also, these mixed complexes would be more abundant than those formed by HDAg-145 or HDAg-160 alone, and an equimolar mixture—with mobility intermediate between those produced by HDAg-145 and HDAg-160 alone—would be the most abundant. If, on the other hand, HDAg binds the RNA as a preformed multimer that does not readily dissociate, then we would expect the mixture of the HDAg-145 and HDAg-160 proteins to yield a mixture of two RNA-protein complexes with mobilities identical to those formed by the individual proteins with the RNA. As shown in Fig. 3, we observed that the mixture of HDAg-145 and HDAg-160 produced RNPs with the latter pattern of mobilities. Thus, we conclude that HDAg binds this RNA as a single preformed multimer and does not dissociate before assembling on the RNA.
The HDAg multimer is an important determinant of RNA binding specificity, including the RNA length requirement for binding.
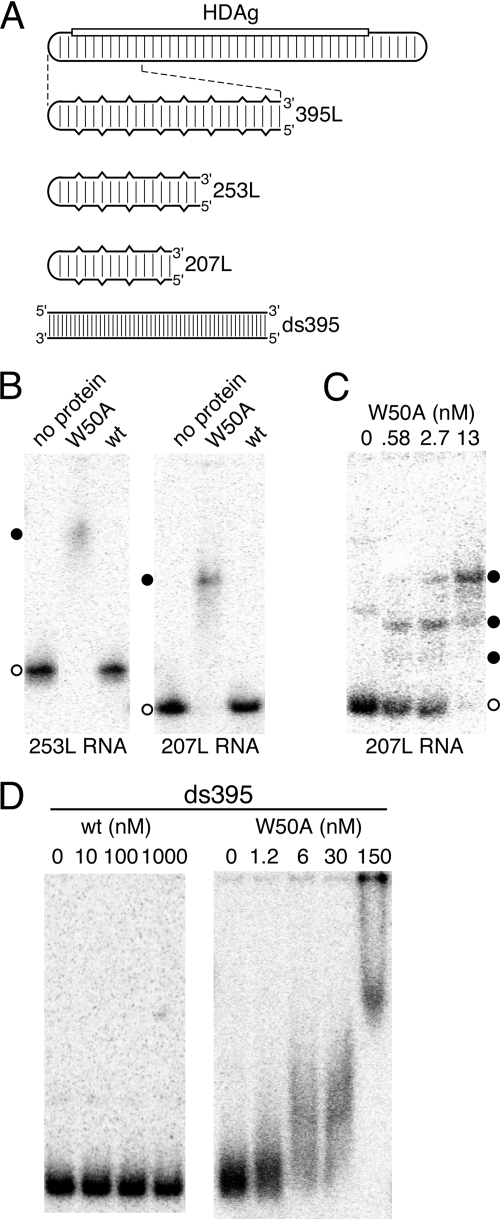
We previously showed that binding of HDAg to HDV RNA requires a minimum length unbranched rod RNA of about 300 nt (8). Possibly, this remarkable length requirement is related to the multimeric nature of the protein prior to binding. The W50A mutant protein, which is defective for multimer formation in the absence of RNA (Fig. 1), produces RNA-protein complexes with mobilities consistent with binding of smaller HDAg units, perhaps as small as a monomer. This result suggests the possibility that the multimerization-defective mutant protein might bind shorter RNAs.
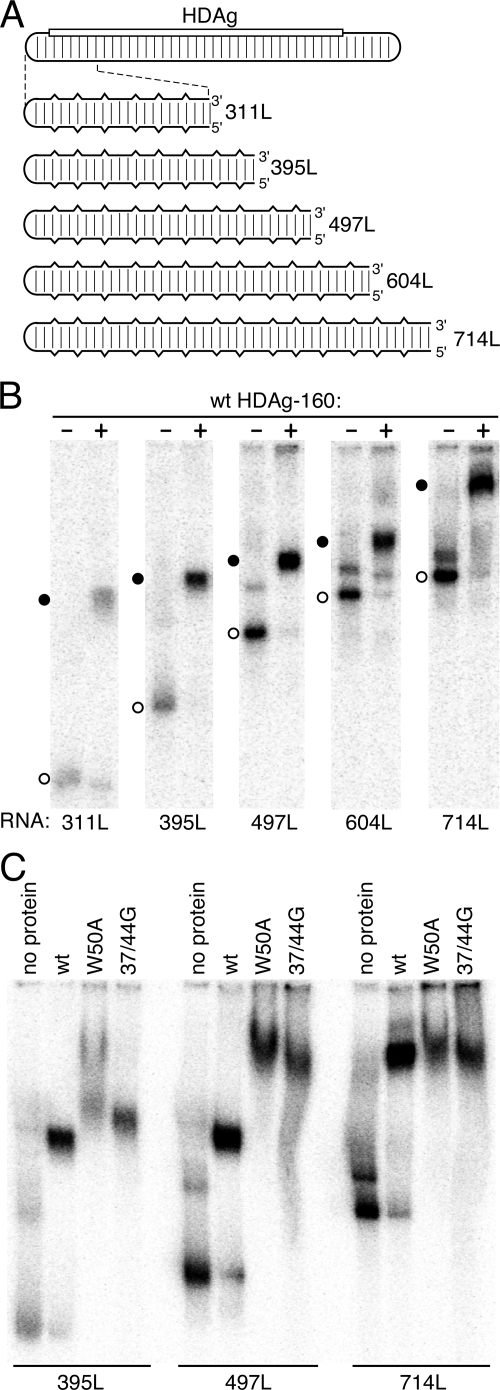
To examine whether HDAg multimerization affects the RNA length requirement for binding, we examined binding of the W50A mutant protein to two HDV RNA unbranched rod segments, 253L and 207L, which are shorter than the minimum length required for binding wild-type HDAg-160. As shown previously (8), neither of these RNAs forms detectable complexes with HDAg-160, even at a protein concentration of 60 nM (Fig. 4B), which is well above the ∼2 nM kDa for binding RNAs of ≥311 nt (8). However, both RNAs were bound by the W50A protein, as indicated by the disappearance of unbound RNAs in the gel and the appearance of diffuse, more slowly migrating species (Fig. 4B and C). The loss of signal intensity and the diffuse appearance of the more slowly migrating bands in the gel likely indicate that the RNP complexes formed are either heterogeneous or not completely stable during the 2-h electrophoresis. We observed similar binding of the 37/44G mutant protein to 207L RNA (not shown). The W50A mutant protein also bound to the short 207 nt RNA at lower protein concentrations and increasingly larger RNP complexes were formed as the protein concentration was increased (Fig. 4C), similar to the pattern observed for this protein with the longer 395L RNA (Fig. 2). These results indicate that the RNA length requirement for binding to wild-type HDAg is related to the multimeric nature of the protein, which binds the RNA as a preformed multimer (Fig. 3).
FIG. 4.
HDAg RNA binding specificity is related to protein multimerization. (A) Schematic of the 1,679-nt HDV RNA genome and RNAs analyzed for binding. The rounded rectangular lines represent HDV RNA. The location of the HDAg gene is indicated by the open rectangle. Regularly spaced vertical lines indicate 70% base-pairing between the coding and noncoding portions of the genome. In the expanded diagrams below, the partial base-pairing is indicated schematically by vertical breaks in the horizontal lines. The bottom schematic, with more closely spaced vertical lines, indicates that the RNA ds395, derived from the coding portion of the HDV RNA present in the RNA 395L, is completely double stranded. RNAs are named according to nucleotide length (8). (B to D) Native polyacrylamide gel electrophoresis after incubation of indicated 32P-labeled RNAs with the indicated concentrations of either wild-type (wt) HDAg-160 or the W50A mutant protein. Incubations were 1 h at 37°C. Protein concentrations in panel B were 30 nM. In panels B and C, open and filled circles indicate free RNA and RNPs, respectively.
We previously reported that HDAg-160 binding is specific for the unbranched rod structure, in which ca. 70% of bases are paired. The maximum number of contiguous canonical base pairs (including G-U pairs) predicted for 395L RNA is 9, and the median number is 5. Binding was not observed for a non-HDV RNA, or for completely double-stranded RNA, even when derived from HDV sequences (8). To determine whether the multimeric HDAg structure is also an important determinant of specificity for the unbranched rod structure, we examined binding of the W50A mutant protein to ds395, a completely double-stranded RNA derived from HDV sequences (Fig. 4). Although no binding was observed for the wild type at concentrations as high as 1 μM, the W50A mutant did bind ds395 (Fig. 4D). Thus, the HDAg multimer contributes to both the secondary structure specificity of binding and the length requirement for binding.
Binding of HDAg to HDV RNA is strongly temperature dependent.
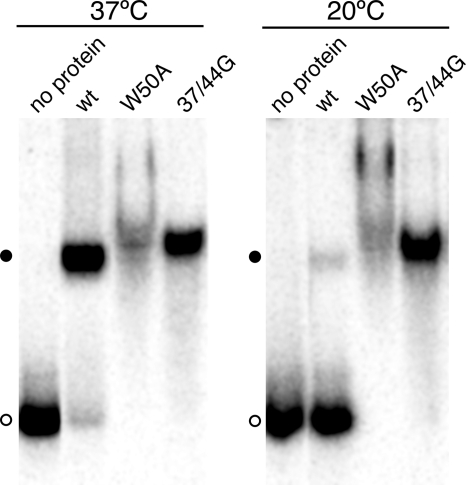
The ability of the W50A mutant protein to bind RNAs shorter than the minimum length required for binding by wild-type HDAg raises the question of why the wild-type protein is unable to bind such RNAs, particularly because the wild-type protein binds as a multimer that contains multiple RNA-binding sites. A possible explanation of this apparent paradox is that binding of the multimer involves a conformational change. Because conformational changes are often temperature dependent, we examined whether the binding of HDAg-160 to HDV unbranched rod RNA was affected by the incubation temperature. When incubated for 1 h in the presence of 30 nM HDAg-160, >95% of 395L RNA was bound at 37°C; however, <10% of the RNA was bound at 20°C (Fig. 5). Thus, the binding of wild-type HDAg to HDV RNA is strongly temperature dependent. Neither the W50A nor the 37/44G mutant proteins exhibited temperature-dependent RNA binding; both of these mutant proteins bound more than 95% of 395L RNA at both temperatures (Fig. 5). Thus, the observed temperature dependence of RNA binding for wild-type HDAg is related to the multimeric nature of the protein.
FIG. 5.
Temperature dependence of binding wild-type (wt) HDAg to HDV RNA. 32P-labeled 395L RNA was incubated with 30 nM HDAg-160 (wild type, W50A, or 37/44G mutant, as indicated) for 1 h at either 20°C or 37°C, as indicated. Samples were electrophoresed for 2 h at room temperature.
HDAg binds HDV unbranched rod RNA in discrete multimeric units depending on the RNA length.
Based on the size of RNP complexes protected against nuclease digestion, we previously suggested that HDAg binds HDV RNA as multimers of fixed size. Thus, the amount of HDAg bound to the minimum length RNA 311L appeared to be the same as that bound to the longer 395 nt RNA 395L. This conclusion is consistent with the observations that HDAg binds the RNA as a preformed multimer (Fig. 3) and that a minimum RNA size is required for binding (8) but further suggests that binding of additional HDAg to RNAs significantly longer than the minimum length also occurs as multimers. To test this concept and to determine the approximate length at which the RNA can bind additional HDAg multimers, we analyzed binding to a series of unbranched rod RNAs of increasing length (Fig. 6).
FIG. 6.
HDAg binds HDV unbranched rod RNA in discrete multimeric units depending on the RNA length. (A) Schematic of the HDV RNA genome and the series of RNAs analyzed for binding, labeled as in Fig. 4. (B and C) Native polyacrylamide gel electrophoresis after incubation of the indicated 32P-labeled RNAs (20 pM) with either no protein or 30 nM HDAg-160 (wild type, W50A mutant, or 37/44G mutant, as indicated). Circles indicate unbound RNA (open) or RNP complexes (filled). In panel B, the time of electrophoresis was 3 h. In panel C, the electrophoresis times were varied: 395L, 2.65 h; 497L, 3.1 h; and 714L, 4 h.
For RNPs formed by wt HDAg-160 with RNAs from 311 to 604 nt, we observed a consistent pattern of electrophoretic mobilities that was not maintained for the 714 nt RNA (Fig. 6B). The electrophoretic mobility of the RNP formed by RNAs from 311 to 604 nt with wild-type HDAg-160 decreased in regular increments of ca. 15% for each increase in the size of the RNA of ∼100 nt. Concomitantly, the extent of the mobility shift due to protein binding (i.e., the difference in the mobility of the RNP versus that of the free RNA) decreased progressively as the RNA length was increased from 311 to 604 nt. This pattern indicates a progressive decrease in the contribution of the molecular weight of the protein to the electrophoretic mobility of the RNP as the RNA size is increased from 311 to 604 nt and is consistent with the interpretation that all of these RNAs bind the same amount of HDAg. However, when the length of the RNA was increased from 604 nt to 714 nt, the mobility of the RNP decreased by more than 60% relative to that formed by the 604L RNA. Moreover, the extent of the mobility shift of the RNA due to HDAg binding increased markedly for 714L compared to 604L.
In contrast to the discontinuity in the behavior of the RNP as the RNA length was increased beyond 604 nt, the mobility of the unbound RNAs exhibited a regular pattern from 311 to 714 nt that was consistent with the progressive increase in RNA length, thus indicating that the 714RNA most likely formed an unbranched rod structure that was similar to that of the other RNAs, as predicted by the mfold algorithm (16, 36). Overall, the pattern of mobility shifts observed in Fig. 6B is consistent with the interpretation that RNAs from 311 to 604 nt bind an HDAg protein complex of identical size and that the longer 714L RNA binds at least one additional HDAg complex. Thus, the minimum length RNA required for binding more than one HDAg multimer is between 604 and 714 nt—approximately twice the length required for binding of one multimer. It is interesting that for the 604-nt RNA, but not the shorter RNAs, a minor fraction of the RNA bound to protein was present in a band with significantly reduced mobility. This result could indicate that a fraction of the 604-nt RNAs binds two HDAg complexes, perhaps because this RNA is close to the minimum size required for binding two HDAg multimers.
We further explored the possibility that HDAg binds unbranched rod RNA as a multimer regardless of RNA length—as long as the RNA is greater than the minimum length requirement of ∼300 nt—by comparing the mobilities of complexes formed by wild-type HDAg-160 and the multimerization-defective HDAg-160 mutants with the 395L, 497L, and 714L RNAs (Fig. 6C). Complexes formed by 395L RNA with the W50A and 37/44G mutant proteins primarily migrated slightly slower than that formed by the wild-type HDAg-160. For W50A, some of the RNA also formed complexes that migrated even more slowly; these complexes appeared only at high protein concentrations. With the longer 497L RNA, the differences between the migration of complexes formed by the wild-type and mutant proteins were much greater; the majority of the RNPs formed with the mutant proteins migrated with less than half the mobility of complexes formed with the wild-type protein. This result likely indicates that the longer 497L RNA binds more of the mutant proteins than wild type. Complexes formed by the wild-type and mutant proteins on the 714L RNA exhibited nearly identical migration in the gel, which was more similar to the situation observed with 395L RNA than with 497L. These results indicate that the 714L RNA binds similar or identical amounts of the wild-type and mutant HDAg-160 in the presence of 60 nM protein. Overall, the results presented in Fig. 6B and C strongly suggest that the 714-nt RNA 714L binds two HDAg multimers, while the RNAs from 311 to 604 nt bind just one multimer.
DISCUSSION
We previously showed that bacterially expressed, C-terminally truncated HDAg forms a stable, discrete complex with HDV RNA in vitro (8). Binding was specific in that it required an unbranched rod structure and a minimum RNA length of ∼300 nt. HDAg is known to form large multimers, even in the absence of HDV RNA, and we previously suggested that HDAg binds to HDV RNA as a multimer. Here, we show that binding indeed occurs as a multimer and that this multimer is formed prior to RNA binding. Moreover, the multimeric nature of the protein is a critical determinant of binding specificity, including both the requisite unbranched rod structure and the remarkable RNA size requirement for binding.
Although the results presented here were obtained using a C-terminally truncated form of HDAg (8), we expect that these findings will be relevant for the full-length protein in cells replicating HDV. For reasons that are not yet clear, the full-length bacterially expressed HDAg formed aggregates and did not exhibit specific RNA binding (8); thus, this form of the protein was unsuitable for the current studies. On the other hand, the in vitro binding activity of the C-terminally truncated forms of HDAg, HDAg-145, and HDAg-160 is critically dependent on the length of the unbranched rod RNA, just like that of the full-length protein expressed in cultured cells (8). Moreover, the C-terminal sequences removed in these expressed proteins have not been implicated in either RNA binding or HDAg multimerization by any of the numerous studies of HDAg function (6, 12, 13, 20, 29, 30, 32).
The methods used here (native gel electrophoresis and rate zonal centrifugation) are not suitable for accurate determination of molecular weight, but our results are certainly consistent with the octameric structure proposed by Zuccola et al. for HDAg (35). In the absence of HDV RNA, the 19.2-kDa protein was observed to sediment just below ADH (150 kDa, Fig. 1B). Because HDAg binds the RNA as a preassembled multimer, it seems likely that the size of the HDAg multimer is the same regardless of whether it is bound to the RNA. Further studies using additional experimental approaches will be required to fully address the size of the HDAg multimer in the RNP.
The mechanism by which the multimer contributes to the specificity of binding is not clear, but the observed temperature dependence (Fig. 5) suggests the possibility that a conformational change is associated with RNP formation. Conformation-dependent assembly has been observed in other RNA-protein complexes (2, 22). In the case of the HDV RNP, conformational changes are likely to occur in the protein because the multimerization-defective mutants did not exhibit temperature-dependent binding (Fig. 5). According to this model, RNA binding by an HDAg subunit is accompanied by a conformational change that, perhaps due to structural constraints in the multimer, induces conformational changes in other subunits, regardless of whether those other subunits are bound to the RNA. The energetic cost of the conformational change in the multimer would require engagement of a minimum number of RNA binding sites in order for RNP formation to be energetically favored, necessitating binding to an RNA long enough (>300 nt) to contact a sufficient number of HDAg subunits in the multimer. The flexibility of the unbranched rod structure could play an important role in facilitating contacts between multiple binding sites, but this model does not rule out the possibility that conformational changes occur in the RNA also. The nature of the binding sites in the RNA remains to be determined. In earlier work, we found that no single specific structure in the RNA was required for binding, but it is certainly possible that the protein recognizes multiple repeated motifs in the unbranched rod structure, such as occurs in the interaction between the B. subtilis, trp RNA-binding attenuation protein and trp leader RNA (1).
The data presented here and in our previous work (8) do not appear to support a model proposed by Zuccola et al. (35), who suggested the possibility that HDV RNA could bind in a central hole formed by the arrangement of HDAg subunits in an octamer, such that the plane of the octamer is perpendicular to the axis of the unbranched rod. Binding in this manner would likely involve more than one multimer per minimum 300-nt RNA segment. However, the results presented in Fig. 3 indicate that just one multimer binds the 395-nt RNA 395L. An alternative model is that the RNA is wrapped around the HDAg multimer in such a way that the RNA is protected against micrococcal nuclease digestion (8). These models may be evaluated more explicitly in future studies using biophysical and imaging approaches.
Others have shown that mutations that disrupt HDAg multimer formation abolish HDV RNA replication (18, 32), but the specific role of HDAg multimerization in HDV RNA replication has not been clear. Indirect measures of RNA binding have indicated that multimerization-defective HDAg proteins still bind HDV RNA (18); this result might imply that multimerization-defective mutations do not affect RNA binding. However, based on the effects of the W50A and 37/44G mutations on RNP formation found here, we suggest that the effect of multimerization mutations on replication is due, at least in part, to defects in the RNP formed.
The observation that HDAg continues to bind as multimeric units as the RNA length is extended (Fig. 6) indicates that the HDAg multimer structure is likely to be an essential determinant of the overall structure of the HDV RNP. We previously identified a minimum RNA length of ∼300 nt for binding. Here, we report that this initial binding involves a single multimer and that the minimum length for binding an additional multimeric unit is between 604 and 714 nt (Fig. 6), which is approximately twice the length necessary for a single multimer to bind. Attempts to use the mixture of HDAg-145 and HDAg-160 to confirm the conclusion that 714L RNA binds two multimers were unsuccessful due to poor resolution of bands in the gel on extended electrophoresis. Ultimately, further experiments using different methods, such as light scattering or atomic force microscopy, will be required to definitively resolve the number of complexes bound to progressively longer RNAs. At present, it is not clear whether binding of subsequent multimers requires a linker RNA sequence, such as is found in nucleosomes.
If the same length requirements apply for binding of additional HDAg multimers as the RNA is lengthened, we expect four or five such multimers to be bound per full-length HDV RNA. The total number of multimers bound in the full-length RNP will depend to a large extent on whether protein binding is highly ordered on the RNA. Also, although the sequences truncated from HDAg-160 are not known to be involved in RNA binding, it is possible that the larger size of the full-length protein could affect the spacing between HDAg multimers. Nevertheless, we can estimate an upper limit for the number of HDAg subunits bound per full-length RNA. If the HDAg multimer consists of 8 HDAg subunits, as suggested by Zuccola et al. (35), then we would expect between 32 and 40 HDAg per full-length RNA. This ratio is lower than, but more similar to earlier estimates (25), than more recent ones (9).
Our analysis of the HDV RNP suggests that it adds yet another dimension to the array of RNPs formed by RNA viruses. Although several viral nucleocapsid proteins bind RNA as monomers (see, for example, reference 34), others, such as HDV, bind as multimers. Rotavirus NSP2, and the related NS2 of bluetongue virus, have both been shown to bind viral RNA as preformed multimers of fixed size, most likely hexamers (26, 27). However, in these cases, it is not clear whether the multimeric nature of the protein plays a role in specificity for the viral RNA. The trimeric hantavirus nucleoprotein has been shown to specifically recognize the panhandle structure formed by the viral RNA (17); this interaction appears to differ from that of HDAg in that binding occurs to a single specific structure in the RNA and may serve to initiate RNP formation. It seems likely that the structure of the HDV viral RNP, assembled from either four or five HDAg multimers, will be distinct from that of other viral RNPs. We expect that further analysis of this structure will provide the necessary framework for understanding the mechanisms of viral RNA replication and packaging of this unique virus.
Acknowledgments
B.C.L. was supported by a grant to Georgetown University from the Undergraduate Program at the Howard Hughes Medical Institute.
We thank Laura Matthews for creating the site-directed mutations, Leah Louisville for excellent technical assistance, and York Tomita for helpful discussions.
Footnotes
Published ahead of print on 18 November 2009.
REFERENCES
- 1.Antson, A. A., E. J. Dodson, G. Dodson, R. B. Greaves, X. Chen, and P. Gollnick. 1999. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature 401:235-242. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, C., J. Otridge, and P. Gollnick. 1996. Kinetic and thermodynamic analysis of the interaction between TRAP (trp RNA-binding attenuation protein) of Bacillus subtilis and trp leader RNA. J. Biol. Chem. 271:12269-12274. [DOI] [PubMed] [Google Scholar]
- 3.Bichko, V. V., S. M. Lemon, J. G. Wang, S. Hwang, M. M. Lai, and J. M. Taylor. 1996. Epitopes exposed on hepatitis delta virus ribonucleoproteins. J. Virol. 70:5807-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casey, J. L., and J. L. Gerin. 1998. Genotype-specific complementation of hepatitis delta virus RNA replication by hepatitis delta antigen. J. Virol. 72:2806-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, J., X. Nie, H. E. Chang, Z. Han, and J. Taylor. 2008. Transcription of hepatitis delta virus RNA by RNA polymerase II. J. Virol. 82:1118-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, M. F., C. H. Chen, S. L. Lin, C. J. Chen, and S. C. Chang. 1995. Functional domains of delta antigens and viral RNA required for RNA packaging of hepatitis delta virus. J. Virol. 69:2508-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornillez-Ty, C. T., and D. W. Lazinski. 2003. Determination of the multimerization state of the hepatitis delta virus antigens in vivo. J. Virol. 77:10314-10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Defenbaugh, D. A., M. Johnson, R. Chen, Y. Y. Zheng, and J. L. Casey. 2009. Hepatitis delta antigen requires a minimum length of the hepatitis delta virus unbranched rod RNA structure for binding. J. Virol. 83:4548-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudima, S., J. Chang, G. Moraleda, A. Azvolinsky, and J. Taylor. 2002. Parameters of human hepatitis delta virus genome replication: the quantity, quality, and intracellular distribution of viral proteins and RNA. J. Virol. 76:3709-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kos, A., R. Dijkema, A. C. Arnberg, P. H. van der Meide, and H. Schellekens. 1986. The hepatitis delta (delta) virus possesses a circular RNA. Nature 323:558-560. [DOI] [PubMed] [Google Scholar]
- 11.Kuo, M. Y., J. Goldberg, L. Coates, W. Mason, J. Gerin, and J. Taylor. 1988. Molecular cloning of hepatitis delta virus RNA from an infected woodchuck liver: sequence, structure, and applications. J. Virol. 62:1855-1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lazinski, D. W., and J. M. Taylor. 1993. Relating structure to function in the hepatitis delta virus antigen. J. Virol. 67:2672-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee, C. Z., J. H. Lin, M. Chao, K. McKnight, and M. M. Lai. 1993. RNA-binding activity of hepatitis delta antigen involves two arginine-rich motifs and is required for hepatitis delta virus RNA replication. J. Virol. 67:2221-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macnaughton, T. B., E. J. Gowans, B. Reinboth, A. R. Jilbert, and C. J. Burrell. 1990. Stable expression of hepatitis delta virus antigen in a eukaryotic cell line. J. Gen. Virol. 71:1339-1345. [DOI] [PubMed] [Google Scholar]
- 15.Macnaughton, T. B., and M. M. Lai. 2006. HDV RNA replication: ancient relic or primer? Curr. Top. Microbiol. Immunol. 307:25-45. [DOI] [PubMed] [Google Scholar]
- 16.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 17.Mir, M. A., and A. T. Panganiban. 2004. Trimeric hantavirus nucleocapsid protein binds specifically to the viral RNA panhandle. J. Virol. 78:8281-8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moraleda, G., K. Dingle, P. Biswas, J. Chang, H. Zuccola, J. Hogle, and J. Taylor. 2000. Interactions between hepatitis delta virus proteins. J. Virol. 74:5509-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pohl, C., B. M. Baroudy, K. F. Bergmann, P. J. Cote, R. H. Purcell, J. Hoofnagle, and J. L. Gerin. 1987. A human monoclonal antibody that recognizes viral polypeptides and in vitro translation products of the genome of the hepatitis D virus. J. Infect. Dis. 156:622-629. [DOI] [PubMed] [Google Scholar]
- 20.Poisson, F., P. Roingeard, and A. Goudeau. 1995. Direct Investig. of protein RNA-binding domains using digoxigenin-labeled RNAs and synthetic peptides: application to the hepatitis delta antigen. J. Virol. Methods 55:381-389. [DOI] [PubMed] [Google Scholar]
- 21.Polson, A. G., H. L. Ley III, B. L. Bass, and J. L. Casey. 1998. Hepatitis delta virus RNA editing is highly specific for the amber/W site and is suppressed by hepatitis delta antigen. Mol. Cell. Biol. 18:1919-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Powers, T., and H. F. Noller. 1995. A temperature-dependent conformational rearrangement in the ribosomal protein S4.16 S rRNA complex. J. Biol. Chem. 270:1238-1242. [DOI] [PubMed] [Google Scholar]
- 23.Rizzetto, M. 1983. The delta agent. Hepatology 3:729-737. [DOI] [PubMed] [Google Scholar]
- 24.Rozzelle, J. E., Jr., J. G. Wang, D. S. Wagner, B. W. Erickson, and S. M. Lemon. 1995. Self-association of a synthetic peptide from the N terminus of the hepatitis delta virus protein into an immunoreactive alpha-helical multimer. Proc. Natl. Acad. Sci. U. S. A. 92:382-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryu, W. S., H. J. Netter, M. Bayer, and J. Taylor. 1993. Ribonucleoprotein complexes of hepatitis delta virus. J. Virol. 67:3281-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taraporewala, Z., D. Chen, and J. T. Patton. 1999. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J. Virol. 73:9934-9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taraporewala, Z. F., D. Chen, and J. T. Patton. 2001. Multimers of the bluetongue virus nonstructural protein, NS2, possess nucleotidyl phosphatase activity: similarities between NS2 and rotavirus NSP2. Virology 280:221-231. [DOI] [PubMed] [Google Scholar]
- 28.Taylor, J. M. 2006. Structure and replication of hepatitis delta virus RNA. Curr. Top. Microbiol. Immunol. 307:1-23. [DOI] [PubMed] [Google Scholar]
- 29.Wang, H. W., P. J. Chen, C. Z. Lee, H. L. Wu, and D. S. Chen. 1994. Packaging of hepatitis delta virus RNA via the RNA-binding domain of hepatitis delta antigens: different roles for the small and large delta antigens. J. Virol. 68:6363-6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, J. G., and S. M. Lemon. 1993. Hepatitis delta virus antigen forms dimers and multimeric complexes in vivo. J. Virol. 67:446-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, K. S., Q. L. Choo, A. J. Weiner, J. H. Ou, R. C. Najarian, R. M. Thayer, G. T. Mullenbach, K. J. Denniston, J. L. Gerin, and M. Houghton. 1986. Structure, sequence, and expression of the hepatitis delta (delta) viral genome. Nature 323:508-514. [DOI] [PubMed] [Google Scholar]
- 32.Xia, Y. P., and M. M. Lai. 1992. Oligomerization of hepatitis delta antigen is required for both the trans-activating and trans-dominant inhibitory activities of the delta antigen. J. Virol. 66:6641-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, L., and Z. R. Liu. 2004. Bacterially expressed recombinant p68 RNA helicase is phosphorylated on serine, threonine, and tyrosine residues. Protein Expr. Purif. 35:327-333. [DOI] [PubMed] [Google Scholar]
- 34.Ye, Q., R. M. Krug, and Y. J. Tao. 2006. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 444:1078-1082. [DOI] [PubMed] [Google Scholar]
- 35.Zuccola, H. J., J. E. Rozzelle, S. M. Lemon, B. W. Erickson, and J. M. Hogle. 1998. Structural basis of the oligomerization of hepatitis delta antigen. Structure 6:821-830. [DOI] [PubMed] [Google Scholar]
- 36.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]