Abstract
Recent studies revealed that posttranslational modifications (e.g., phosphorylation and methylation) of the small hepatitis delta antigen (SHDAg) are required for hepatitis delta virus (HDV) replication from antigenomic to genomic RNA. The phosphorylation of SHDAg at serine 177 (Ser177) is involved in this step, and this residue is crucial for interaction with RNA polymerase II (RNAP II), the enzyme assumed to be responsible for antigenomic RNA replication. This study demonstrated that SHDAg dephosphorylated at Ser177 interacted preferentially with hypophosphorylated RNAP II (RNAP IIA), which generally binds at the transcription initiation sites. In contrast, the Ser177-phosphorylated counterpart (pSer177-SHDAg) exhibited preferential binding to hyperphosphorylated RNAP II (RNAP IIO). In addition, RNAP IIO associated with pSer177-SHDAg was hyperphosphorylated at both the Ser2 and Ser5 residues of its carboxyl-terminal domain (CTD), which is a hallmark of the transcription elongation isoform. Moreover, the RNAP II CTD kinase inhibitor 5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole (DRB) not only blocked the interaction between pSer177-SHDAg and RNAP IIO but also inhibited HDV antigenomic replication. Our results suggest that the phosphorylation of SHDAg at Ser177 shifted its affinity toward the RNA RNAP IIO isoform and thus is a switch for HDV antigenomic RNA replication from the initiation to the elongation stage.
The hepatitis delta virus (HDV) is a negative-stranded RNA virus with a 1.7-kb circular, single-stranded RNA genome. The single-stranded RNA genome is folded into an unbranched rod-like structure because of a high degree of intramolecular self-complementarity (10, 32, 44, 63). HDV is a defective virus (58) that requires surface antigen (HBsAg) of helper hepatitis B virus (HBV) for virion assembly and infectivity (3, 58, 60). In addition to the HDV RNA genome and the HBV envelope, the HDV particle also contains hepatitis delta antigen (HDAg), which is the sole known protein encoded by the open reading frame of the HDV RNA (26, 48). There are two forms of HDAg, the small HDAg of 195 amino acids (SHDAg; 24 kDa) and the large HDAg of 214 amino acids (LHDAg; 27 kDa) (64). The amino acid sequences of these two forms of HDAg are identical, with the exception that LHDAg is 19 amino acids longer than SHDAg at its C terminus, which results from an RNA-editing event during viral replication (40). Although these two isoforms of HDAg share similar amino acid sequences, they play different functions in the life cycle of HDV. SHDAg is an essential activator of HDV RNA replication (9, 33), while LHDAg promotes virion assembly (6).
The replication of HDV RNA is independent from its helper HBV (33) and does not involve any DNA intermediates (10). It occurs in the nucleus, probably via a double-rolling-circle mechanism (42, 62). As neither HDV nor mammalian cells encode an RNA-dependent RNA polymerase (RdRp), the replication of HDV RNA may rely on a redirection of a host DNA-dependent RNA polymerase to become the RdRp of HDV (34, 62). The host RNA polymerase II (RNAP II) is assumed to be responsible for HDV RNA replication, as HDV replication is sensitive to low-dose alpha-amanitin treatment (8, 21, 22, 25, 27, 41, 49, 65) and to anti-RNAP II antibody (38). Recently, a crystallography study revealed that RNAP II recognizes RNA scaffolds and possesses RdRp activity that leads to RNA elongation (37). In addition, RNAP II interacts directly with both the C terminus of SHDAg (65) and with the terminal stem-loop regions of the HDV RNA genome in vitro (25). An in vitro transcription assay was used to show that SHDAg stimulates HDV antigenomic RNA-templated elongation (65). Therefore, it generally is believed that HDV RNA-dependent RNA replication is regulated positively by SHDAg and is carried out by cellular RNAP II; however, the mechanism underlying the involvement of SHDAg in this process remains unclear.
Interestingly, both SHDAg and RNAP II are nuclear phosphoproteins. In both prokaryotic and eukaryotic organisms, the reversible phosphorylation of proteins is an important regulatory mechanism that is used to switch protein function on or off (1, 14). Reversible phosphorylation often results in a conformational change in the structure of many proteins, leading to their activation or inactivation. RNAP II is a 12-subunit enzyme that is responsible for the synthesis of all eukaryotic mRNAs (15, 16, 24). A unique feature of RNAP II (compared to the features of the other two RNA polymerases) is that the carboxyl-terminal domain of the largest RNAP II subunit (Rpb1) contains multiple tandem heptapeptide repeats of the consensus sequence YSPTSPS and is rich in potential phosphorylation sites (18). The two forms of RNAP II, hypophosphorylated RNAP IIA and hyperphosphorylated RNAP IIO, can be distinguished from each other by different levels of phosphorylation at their CTDs. The switch from RNAP IIA to RNAP IIO is a hallmark of RNAP II during the transition from the initiation to the elongation stage of cellular transcription (5).
Similarly to RNAP II, SHDAg is also a phosphoprotein with multiple phosphorylation sites (most are serine and threonine residues). Among them, serine 177 (Ser177) is the predominant phosphorylation site used in vivo (52, 54). Our previous studies revealed that a Ser177 mutant of SHDAg lost the ability to interact with RNAP II and to support HDV replication from antigenomic to genomic RNA. Moreover, we found that extracellular signal-regulated kinase 1/2 (ERK1/2) is an SHDAg-associated kinase, and that its activation enhances the phosphorylation levels of Ser177 of SHDAg and the antigenomic RNA replication of HDV in vivo (11, 52). However, the role of SHDAg phosphorylation in regulating HDV antigenomic RNA replication remains unclear. In this study, we found that the phosphorylation of the Ser177 residue of SHDAg led to a distinct localization of the protein and changed its preferential interaction with different isoforms of RNAP II. These findings suggest that the phosphorylation of the Ser177 residue of SHDAg regulates viral antigenomic RNA replication in specific cellular compartments via interacting with cellular factors, especially with its RdRp, RNAP II.
MATERIALS AND METHODS
Cell culture and DNA transfection.
Human embryonic kidney 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine in 5% CO2 at 37°C. Transfections were performed using Lipofectamine 2000 (Invitrogen) reagent according to the manufacturer's protocol. Actinomycin D (ActD) and 5,6-dichloro-1-β-d-ribofuranosyl-benzimidazole (DRB) were purchased from Sigma. Media containing ActD or DRB were prepared from an aqueous 1-mg/ml or 100 mM stock solution in dimethyl sulfoxide (DMSO), respectively. Plasmids used in this study were described previously (52). Briefly, pCD2AG (2AGwt) can produce a dimer form of HDV antigenomic RNA and HDAg; pCD2AGm (2AGm) has a two-base deletion in the HDAg open reading frame (ORF); pCD2AG177 (2AG177) has a serine-to-alanine mutation at Ser177 encoded in the HDAg ORF; pCDAg-S, pCDAg-S177A, and pCDAg-S177D can express wild-type and mutant SHDAg, respectively.
Coimmunoprecipitation.
Cell lysates of each 100-mm dish were prepared in 1 ml NP-40 lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.2% NP-40, 1 mM EDTA, 100 mM sodium butyrate, 100 mM sodium fluoride, 2 mM sodium orthovanadate, 20 mM sodium pyrophosphate, 20 mM β-glycerophosphate) supplemented with 2× protease inhibitor cocktail (Roche Applied Science) by gentle pipetting 30 times. The cell lysate then was incubated on ice for 30 min and centrifuged at 13,000 rpm for 10 min at 4°C. The supernatant was collected and precleared with rabbit preimmune serum/protein A-agarose for 30 min at 4°C with agitation. The precleared aliquot then was immunoprecipitated with rabbit anti-HDAg- or anti-pSer177-HDAg-conjugated protein A-agarose for 2 h at 4°C with agitation. Beads were washed three times with lysis buffer, and pellets were boiled for 5 min in 10 μl 2× SDS-PAGE sample buffer for the analysis and identification described below.
Western blot analysis.
For the detection of SHDAg and RNAP II, proteins were subjected to electrophoresis in SDS-6 to 12% PAGE. After electrotransfer, blots were blocked with 5% nonfat dry milk-1× TBST (20 mM Tris base, 140 mM NaCl, 0.1% Tween-20, pH 7.2) for 1 h at room temperature with shaking and then were incubated with the primary antibodies monoclonal anti-RNAP II N terminus (N-ter) (ab57300; Abcam) (1:1,000), H5 and H14 (Covance) (1:500), and anti-HDAg or anti-pSer177-HDAg (1:3,000). Finally, blots were probed with secondary antibodies (1:5,000) for 1 h at room temperature, and proteins were visualized by chemiluminescence detection.
RNA preparation and Northern blotting.
Total cellular RNAs of 293T cells were extracted using TRIzol reagent (Invitrogen) according to the instructions of the user manual. For Northern blot analysis, 5 μg of total cellular RNA was analyzed by electrophoresis on a 6% formaldehyde-1% agarose RNA gel and then transferred to a nylon membrane. The membranes then were hybridized with digoxigenin (DIG)-labeled strand-specific HDV RNA probes at 68°C overnight. Signals were detected using the DIG detection kit (Roche) according to the manufacturer's protocol.
Glycerol gradient sedimentation.
Cell lysates (0.4 ml) from HDV-transfected 293T cells were layered on 3.6 ml of linear 15 to 30% glycerol density gradients prepared with NP-40 lysis buffer. Centrifugation was performed on a Beckman SW60 rotor at 40,000 rpm for 3.5 h at 4°C. Fractions were collected from the top of the tube. Aliquots were analyzed for refractive index. To identify viral RNA distribution, even-numbered fractions were extracted and analyzed by Northern blotting. To identify the cellular factors associated with viral RNPs, odd-numbered fractions were subjected to immunoprecipitation with rabbit anti-HDAg antibody and analyzed by Western blotting as described above.
Lambda protein phosphatase reactions.
Lambda protein phosphatase (Biolabs) was used to dephosphorylate SHDAg purified from immunoprecipitation with rabbit anti-HDAg antibody. Dephosphorylation was performed by incubation with 400 U of lambda protein phosphatase in phosphatase reaction buffer (50 mM HEPES, pH 7.5, 0.1 mM Na2EDTA, 5 mM dithiothreitol, 0.01% Brij 35, and 2 mM MnCl2) for 30 min at 30°C. The reaction was stopped by three washes with 1% NP-40 lysis buffer containing phosphatase inhibitor (100 mM sodium fluoride, 2 mM sodium orthovanadate, 20 mM sodium pyrophosphate, 20 mM β-glycerophosphate).
Immunofluorescence assay.
Immunofluorescence staining was performed as described previously (69), with minor modifications. Briefly, cells grown on chamber slides (Nunc) were washed twice with cold phosphate-buffered saline (PBS), permeabilized for 1 to 2 min with 0.5% Triton X-100 in CSK buffer [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 7, 1 mM EGTA, 3 mM MgCl2, 20% sucrose] on ice, and fixed with 2.5% paraformaldehyde in the same buffer at room temperature for 30 min, and the reaction was stopped with three washes with 100 mM glycine in PBS. For dual-immunofluorescence staining, slides were blocked and incubated with properly diluted antibodies with 1% bovine serum albumin in PBS at 37°C for 30 min. In this study, the following primary antibodies were used for immunofluorescence: monoclonal antibody anti-HDAg (1:400) and rabbit anti-pSer177-HDAg (1:400) (LTK Laboratories, Taiwan). After being washed with PBS three times, cells were incubated with diluted secondary antibodies (1:400) conjugated with fluorescein isothiocyanate (FITC) or Texas red (Jackson Immuno-Research) at 37°C for 30 min and then washed and counterstained with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) in PBS for 5 min. Slides finally were mounted with Vectashield mounting medium (Vector Laboratories). Images were obtained by fluorescent microscopy (Leica DMR).
In vitro transcription and RNA transfection.
SHDAg-encoding mRNA and dimer forms of HDV antigenomic RNA were transcribed from pGEM3Zf(+)/HDV934-1651 with mMESSAGE mMACHINE (Ambion) and from pGEM3Zf(+)/HDV2G with SP6 MEGAscript (Ambion), respectively, after linearization by BamHI digestion. HDV RNA transfection experiments were performed by the use of the DMRIE-C transfection reagent (Invitrogen) according to the manufacturer's directions.
RESULTS
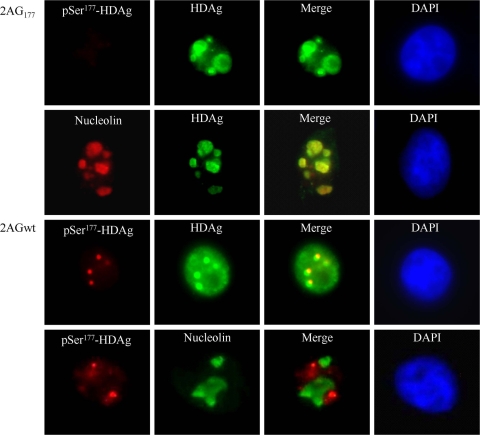
Ser177-phosphorylated HDAg was localized predominantly to nuclear bodies.
Our previous studies revealed that SHDAg is a phosphorylated protein, and that Ser177 is the residue that is phosphorylated predominantly (52, 54). In addition, the S177A SHDAg mutant (replacement of Ser177 with Ala177) exhibits the disruption of the synthesis of HDV genomic RNA with the preservation of the synthesis of antigenomic RNA (11, 52), which suggests that the phosphorylation of Ser177 plays a key role in regulating the synthesis of HDV genomic RNA. As the phosphorylation of SHDAg at Ser177 may be accompanied by the active transcription of the viral antigenome, it would be possible to locate the subcellular compartments where HDV genome synthesis occurs by staining HDV-transfected cells with an antibody that recognizes Ser177-phosphorylated HDAg (pSer177-HDAg) specifically. 293T cells were transfected with HDV-expressing plasmids that were either antigenomic RNA replication competent (2AGwt) or replication deficient (2AG177). We performed dual-immunofluorescence staining at 3 days posttransfection. As shown in Fig. 1, wild-type HDAg was localized throughout the nucleoplasm and nuclear speckles, which is consistent with previous studies reported by others (2, 38). Interestingly, pSer177-HDAg was localized predominantly to distinct nuclear structures, which was not observed for the S177A mutant of HDAg. In contrast, this mutant, which exhibits single-direction replication from genomic to antigenomic RNA, accumulated in nucleoli. Taken together, these results suggest that pSer177-HDAg regulates HDV genomic RNA synthesis within a distinct nuclear structure.
FIG. 1.
Localization of Ser177-phosphorylated HDAg was restricted to nuclear bodies, while its unphosphorylated counterpart accumulated predominantly in nucleoli. 293T cells were transfected either with an HDV replication-competent construct, 2AGwt, or with an HDV antigenomic RNA replication-deficient construct, 2AG177. At 2 days posttransfection, cells were dually immunostained with antibodies against HDAg, Ser177-phosphorylated HDAg (pSer177-HDAg), or nucleolin and were counterstained with 4′,6-diamidino-2-phenylindol (DAPI). pSer177-HDAg is indicated in red, HDAg in green, and DAPI counterstaining in blue.
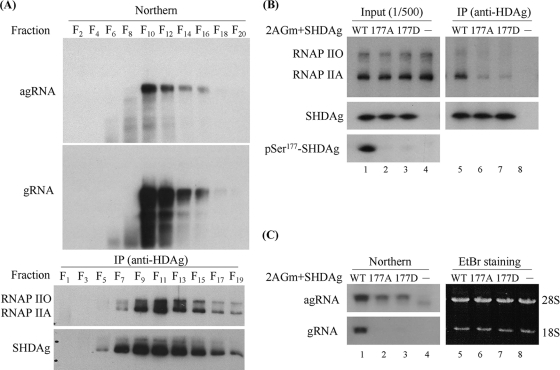
The Ser177 residue of SHDAg was important for its interaction with RNAP IIA and RNAP IIO.
The specific localization of pSer177-HDAg implied that the phosphorylated isoform participates in different replication complexes (RNAP IIA and IIO) during viral replication. To test this hypothesis, we first investigated the interaction between host RNAP II and SHDAg. 293T cells were transfected with an HDV replication-competent plasmid (2AGwt) and harvested after 3 days, and cell lysates were subjected to 15 to 30% glycerol-gradient ultracentrifugation. Even-numbered fractions were extracted for viral RNA analysis, and odd-numbered fractions were immunoprecipitated using an anti-HDAg antibody to verify its association with RNAP II. Although the majority of RNAP II had a lower density distribution (data not shown), two forms of RNAP II cosedimented with HDV ribonucleoproteins (RNPs), as assessed using coimmunoprecipitation after glycerol-gradient ultracentrifugation (Fig. 2A). This result was consistent with previous in vitro (65) and in vivo (7) studies. To further investigate the importance of SHDAg Ser177 phosphorylation in the binding to RNAP II, plasmids designed to express wild-type SHDAg or the Ser177 mutants of SHDAg (pCDAg-S, pCDAg-S177A, and pCDAg-S177D) were cotransfected with nonreplicative 2AGm into 293T cells. The lysates then were subjected to immunoprecipitation using a rabbit anti-HDAg antibody after 3 days of transfection. We found that the substitution of Ala or Asp for Ser177 in SHDAg dramatically reduced its interaction with RNAP II and impaired HDV antigenomic RNA replication (Fig. 2B, lanes 6 and 7, and C, lanes 2 and 3). Our result is consistent with a previous observation that the evolutionarily conserved C terminus of SHDAg is critical for RNAP II binding (66); however, we cannot exclude the possibility that these two mutants at residue 177 of SHDAg induce a conformational change of SHDAg and then lose the ability to interact with RNAP II.
FIG. 2.
Ser177 residue of SHDAg was an essential factor for its interaction with both RNAP IIA and RNAP IIO. (A) Interaction between SHDAg and RNAP II assessed by glycerol gradient sedimentation. 2AGwt-transfected 293T cells were harvested at 3 days posttransfection, and cell lysates were subjected to 15 to 30% glycerol gradient ultracentrifugation. Fractions were collected to detect either viral RNA by Northern blotting or viral antigen and RNAP II isoforms by immunoprecipitation using an anti-HDAg antibody followed by Western blotting (IP-Western). Fractions from the top (F1) to bottom (F20) are indicated. (B and C) Effects of SHDAg Ser177 variants on the interaction with RNAP II and on HDV antigenomic RNA replication. The manipulated 2AGm construct, which can produce HDV antigenomic RNA but not viral antigen, was cotransfected into 293T cells with a plasmid expressing either wild-type (WT) or mutant types (177A and 177D) of SHDAg. After 3 days of transfection, RNAP II isoforms coimmunoprecipitated with an anti-HDAg antibody were detected using Western blotting (B), and HDV RNA was detected by Northern blotting (C), using strand-specific probes.
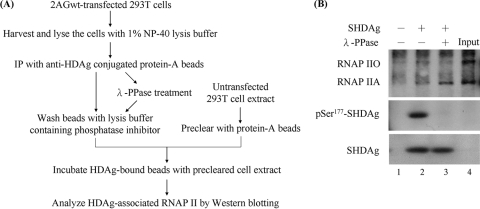
Phosphorylation of SHDAg regulated its binding to different RNAP II complexes in vitro.
To exclude potential conformational changes of mutant proteins and dissect further the possible role of SHDAg Ser177 phosphorylation in the binding to RNAP II and viral replication, we performed an in vitro binding assay of an in vivo protein (Fig. 3A). Using 2AGwt-transfected 293T cells, SHDAg was immunoprecipitated, dephosphorylated by lambda protein phosphatase, and then subjected to an in vitro binding assay of whole-cell extracts. As shown in Fig. 3B, dephosphorylated SHDAg interacted preferentially with RNAP IIA (lane 3), while the phosphorylated counterpart exhibited a preference for RNAP IIO (lane 2). Although we could not rule out the existence of other phosphorylated residues in SHDAg, we provided a new perspective suggesting that SHDAg plays different roles in viral antigenomic RNA replication via the control of its phosphorylation status.
FIG. 3.
Dephosphorylated and phosphorylated isoforms of SHDAg had different binding affinities to RNAP IIA and RNAP IIO in vitro. (A) Flowchart of the in vitro binding assay. The lysate of 2AGwt-transfected 293T cells was immunopurified using an anti-HDAg antibody and/or dephosphorylated using lambda protein phosphatase (λ-PPase). The dephosphorylation reaction was stopped by three washes with 1% NP-40 lysis buffer containing a phosphatase inhibitor. The HDAg-bound protein A beads then were incubated with a whole-cell extract to pull down the HDAg-binding proteins. (B) The bound isoforms of RNAP II in the anti-HDAg immunoprecipitates were detected using immunoblotting. The phosphatase activity on Ser177-phosphorylated SHDAg (pSer177-SHDAg) also was evaluated using a specific anti-pSer177-HDAg antibody.
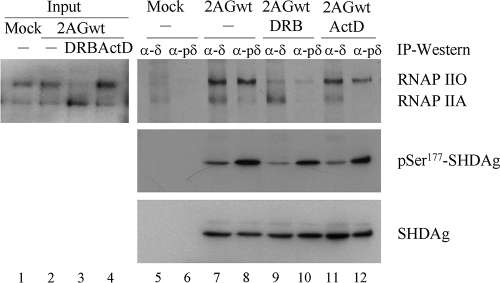
SHDAg phosphorylated at Ser177 interacted preferentially with RNAP IIO complexes in vivo.
It has been proposed that RNAP IIA interacts with promoters to form a stable preinitiation complex, whereas RNAP IIO is generated after the onset of the initiation of transcription and remains phosphorylated during elongation (13, 19, 56, 68). Previous studies also found that RNAP IIO accumulates in the presence of ActD, and RNAP IIA is increased in cells treated with CTD-kinase inhibitors, such as the nucleoside analog DRB (20). Therefore, we treated 2AGwt-transfected 293T cells with 0.1 μg/ml of ActD or 200 μM DRB for 2 h to transiently change the phosphorylation status of RNAP II in vivo. Subsequently, we used the pSer177-HDAg-specific antibody to verify the phosphorylation-dependent interaction between SHDAg and RNAP II. Two forms of RNAP II were coimmunoprecipitated with SHDAg (Fig. 4, lane 7), and a preferential interaction of pSer177-SHDAg with RNAP IIO was observed in untreated HDV-transfected 293T cells (Fig. 4, lane 8). Consistent results were found after ActD and DRB treatment (Fig. 4, lanes 9 to 12). Thus, these results suggest that the phosphorylation of the Ser177 residue of SHDAg modulates its binding affinity to RNAP IIO and IIA.
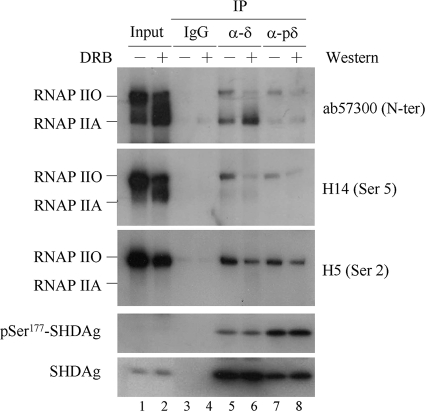
FIG. 4.
pSer177-SHDAg interacted preferentially with RNAP IIO in vivo. Three days after transfection with 2AGwt, 293T cells were treated with 200 μM DRB (lanes 3, 9, and 10) or 0.1 μg/ml of ActD (lanes 4, 11, and 12) for 2 h. Total cellular proteins were extracted and then immunoprecipitated with rabbit anti-HDAg (α-δ; lanes 5, 7, 9, and 11) or anti-pSer177-HDAg (α-pδ; lanes 6, 8, 10, and 12) antibody. Bound RNAP II and SHDAg were detected with anti-RNAP II, anti-HDAg, or anti-pSer177-HDAg antibody. Inputs from transfected or mock-transfected cells are indicated.
SHDAg phosphorylated at Ser177 associated with the processive RNAP II complexes, which were hyperphosphorylated at both the Ser5 and Ser2 residues of their CTD.
During transcription, RNAP II initially carries a largely unphosphorylated CTD at preinitiation. Early in the transition from preinitiation to elongation, the CTD is phosphorylated at the Ser5 residue by cyclin-dependent kinase 7 (CDK7) in the general transcription factor TFIIH to create the second CTD phosphorylation state. After initiation, CDK9, which is a catalytic subunit of positive transcription elongation factor (P-TEFb), phosphorylates Ser2 residues mainly to generate elongation-proficient RNAP II. Finally, it is widely believed that, near the 3′ end of the gene, CTD phosphorylation is dominated by phospho-Ser2 residues (31, 36, 45, 50). To examine a similar scenario for HDV RNA transcription, RNAP II coimmunoprecipitated with pSer177-SHDAg from 2AGwt-transfected 293T cells was analyzed using Ser5- or Ser2-specific antibody, respectively. As shown in Fig. 5, RNAP II associated with pSer177-SHDAg was hyperphosphorylated at both Ser5 and Ser2 residues of its CTD (Fig. 5, lane 7). This suggests that SHDAg Ser177 phosphorylation regulates viral antigenomic RNA replication via interaction with the processive RNAP IIO.
FIG. 5.
pSer177-SHDAg associated with the transcription elongation complexes of RNAP IIO. Three days after transfection with 2AGwt, 293T cells were treated with 100 μM DRB or with the same volume of DMSO for 2 h. Immunoprecipitation was performed as described in the legend to Fig. 4, and bound proteins were detected with specific antibodies against the N terminus, the phospho-Ser5 (H14) CTD, or the phospho-Ser2 (H5) CTD of RNAP II. Immunoprecipitation with nonspecific immunoglobulin G (IgG), which was used as a negative control, is indicated.
HDV antigenomic RNA replication was inhibited by DRB treatment and was reversed by DRB clearance in vivo.
The effect of DRB on viral antigenomic RNA replication was examined to test the role of RNAP II CTD phosphorylation in this process. 293T cells were cotransfected with a dimer form of antigenomic RNA and SHDAg mRNA. At 4 h posttransfection, cells were treated with different concentrations of DRB, and culture medium containing DRB was refreshed every 8 h. Three days later, total RNA was extracted and analyzed by Northern blotting to verify viral antigenomic RNA replication. As shown in Fig. 6, HDV replication from antigenome to genome was inhibited by DRB treatment in a dose-dependent manner (lanes 5 to 8). The subsequent removal of DRB led to the reversal of this effect (lanes 10 to 12). In addition, a high dose (>100 μM) of DRB inhibited the phosphorylation of both SHDAg and RNAP II at the C terminus (lanes 7 and 8). Therefore, these findings imply that the phosphorylation of both SHDAg and RNAP II at the C terminus was sensitive to DRB treatment and was essential for viral antigenomic RNA replication.
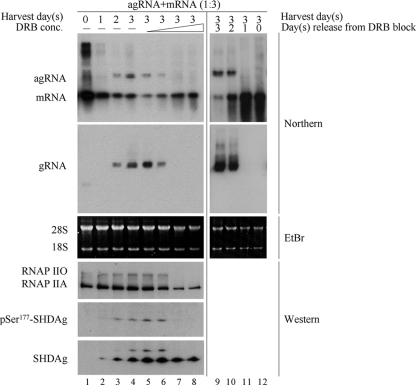
FIG. 6.
HDV antigenomic RNA replication was arrested by DRB treatment and resumed after DRB withdrawal. 293T cells were transfected with both SHDAg mRNA and a dimer form of HDV antigenomic RNA at a ratio of 3:1. At 4 h posttransfection, DRB was added to cells at a concentration of 25, 50, 100, or 200 μM (lanes 5 to 8, respectively) and was refreshed every 8 h. Total RNA and protein were extracted from untreated cells at the indicated days after transfection (lanes 1 to 4) and from DRB-treated cells at 3 days posttransfection. To analyze the effect of DRB withdrawal, the drug (200 μM) was removed from transfected cells at days 0, 1, 2, and 3, and total RNA was extracted at 3 days posttransfection (lanes 9 to 12). Five micrograms of total RNA was analyzed by Northern blotting. Strand-specific HDV RNAs labeled with DIG were used as probes to detect HDV genomic RNA (gRNA), antigenomic RNA (agRNA), and mRNA. 28S/18S RNA was used as a loading control. Twenty-five micrograms of total protein was detected by Western blotting using specific antibody against RNAP II, pSer177-HDAg, or HDAg.
DISCUSSION
A growing body of evidence suggests a role for RNAP II in the RNA-dependent genomic RNA synthesis of HDV. In this report, we showed that the phosphorylation of the Ser177 residue of SHDAg changed its binding preference to different phosphorylated forms of RNAP II complexes. The inhibition of the phosphorylation of SHDAg and of the CTD of RNAP II by DRB treatment led to the arrest of HDV genomic RNA synthesis. These results indicate that DRB blocks the transition of HDV genomic RNA synthesis from the hypo- to the hyperphosphorylated replication complexes. In addition, we observed that pSer177-SHDAg accumulated in unidentified nuclear bodies, which may act as factories of HDV genomic RNA synthesis. Therefore, we propose a hypothetical model of the SHDAg-mediated regulation of viral genomic RNA synthesis via its phosphorylation, based on the analogy to cellular RNA transcription by RNAP II (Fig. 7). Briefly, unphosphorylated SHDAg and RNAP II bind to HDV antigenomic RNA independently until they become associated with each other at the replication initiation site(s) to form a preinitiation complex. Specific cellular kinase(s) then phosphorylate SHDAg and/or RNAP II complexes, thereby forcing hyperphosphorylated transcription complexes to depart from the initiation site and engaging productive elongation. If this model is correct, it will be possible to separately identify the components involved in the unphospho-enriched initiation complex and in the phospho-enriched elongation complex in future studies.
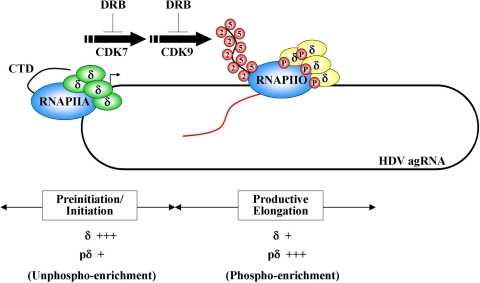
FIG. 7.
Proposed model of HDV antigenomic RNA replication by RNAP II. Briefly, unphosphorylated SHDAg binds viral RNA through its RNA-binding domain and interacts with RNAP IIA at the initiation site to promote antigenomic RNA replication. At the preinitiation/initiation stage, the HDV antigenomic RNA replication complex is unphospho-enriched. The phosphorylation of SHDAg by a certain cellular kinase(s) stimulates the transition of viral antigenomic RNA replication from the initiation into the elongation complex, which is phospho-enriched.
Although it was suggested that SHDAg interacts with RNAP II during both the initiation and elongation phases of transcription (66), the possible interaction of different forms of RNAP II with different isoforms of SHDAg during transcription has not been examined. Interestingly, RNAP II undergoes conformational changes via physical interaction with general transcription factors and mediators or via CTD phosphorylation (29). However, a previous study reported that the C terminus of SHDAg interacts with the clamp, but not with the CTD, of RNAP II (66). This implies that phosphorylation at the C terminus of SHDAg is a checkpoint for its interacting preference for RNAP II. On the basis of the crystal structure and function of RNAP II complexes, it is assumed that SHDAg, negative elongation factor (NELF), and the general transcription factor TFIIF interact with RNAP II in the overlapping regions of clamp submodules of the RNAP II active center (12, 55, 66). However, NELF and TFIIF play opposite roles during transcription. NELF often acts together with DRB sensitivity-inducing factor (DSIF) to cause the transcriptional pausing of RNAP II complexes (67), which can be reversed by the P-TEFb-dependent phosphorylation of RNAP II (57) and NELF (23). In contrast, the phosphorylation of TFIIF regulates its affinity to RNAP II, thereby stimulating transcription initiation and elongation (30, 61). The transcription factor-mediated increase or decrease of gene transcription may depend on their interacting partners. Thus, our findings suggest that SHDAg, by acting as a viral TFIIF counterpart, is regulated by phosphorylation and modulates its binding affinity to RNAP II as well as viral genome synthesis. Of course, our results cannot rule out a role for other posttranslational modifications of SHDAg (e.g., methylation and acetylation) in regulating HDV antigenomic RNA replication cooperatively. Previous studies have revealed that the methylation and acetylation of SHDAg affected its subcellular localization (39, 53). However, whether the methylation or acetylation of SHDAg has any effect on its binding affinity to RNAP II needs further study.
During viral replication, HDV RNAs exist in three forms: genome, antigenome, and mRNA. The synthesis of these RNA species is independent and probably is mediated by different machineries (43). Although SHDAg may regulate viral genome synthesis via phosphorylation, it is not stringent enough to differentiate among these three machineries. Currently, it has become clear that the nucleus is highly compartmentalized, and many nuclear factors are localized in distinct structures (47, 59). It also has been indicated that nuclear speckles correspond to active sites of RNAP II transcription and RNA processing (4, 46, 51), as a portion of the Ser2-phosphorylated RNAP II was observed in these structures (35). Several reports revealed that, during HDV replication, HDV RNA and SHDAg are colocalized in the nucleoplasm and in nuclear speckles. A previous study speculated that these speckles, which colocalize with SC-35, are the sites at which HDV RNA is transcribed by RNA polymerase II and/or represent sites of HDV RNA processing (2). Another study demonstrated that HDV antigenomic RNA is recruited into the promyelocytic leukemia (PML) speckles, whereas genomic RNA is localized to nucleoli (38). However, one previous report showed that the speckles are unlikely to be the major sites for HDV viral transcription or replication, as 5-bromouridine triphosphate (BrUTP) was not incorporated into viral RNA in nuclear speckles (17). In addition, a recent study concluded that the majority of HDAg in the nucleoplasm colocalizes with cellular RNAP II and is an indication of HDV RNP accumulation rather than of ongoing viral transcription (28). Although several lines of evidence indicate that the replication of HDV RNA is carried out by different cellular machineries, there still is much controversy about whether HDV genomic and antigenomic RNA replication take place in the same cellular compartment. Our study showed that the localization of pSer177-HDAg (as a surrogate marker for a productive elongation of HDV antigenomic RNA replication) was restricted to nuclear bodies, rather than to the PML or SC35-positive speckles (data not shown). In addition, the Ser177 mutation of SHDAg accumulated in nucleoli could support single-direction replication from genomic to antigenomic RNA. These suggest that the fate-determining posttranslational modifications can create uniquely modified SHDAg isoforms and direct them to specific locations and to exert specific functions. Whether the sequential accumulation of SHDAg is correlated with the active transcription of viral RNA needs further investigation. Nevertheless, our findings shed light on the relationship between the phosphorylation of SHDAg and the subnuclear localization of the HDV RNP. The identification of the unique nuclear compartment will provide more information to decipher the exact cellular machineries for viral replication.
HDV, which is the simplest RNA virus, employs a unique strategy for viral replication and is the only known pathogen capable of using a DNA-dependent polymerase as an RdRp in eukaryotic cells. Although RdRp has not been found in animal cells, it is possible that RNAP II functions as an RdRp to amplify small RNAs for posttranscriptional gene silencing during early development and differentiation. Consequently, the identification and characterization of this RdRp using HDV RNA as a model system may shed light on these fundamental questions.
Acknowledgments
This work was supported by a grant from the National Science Council, Taiwan (NSC 97-2311-B-002-008-MY3).
Footnotes
Published ahead of print on 18 November 2009.
REFERENCES
- 1.Barford, D., A. K. Das, and M. P. Egloff. 1998. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. 27:133-164. [DOI] [PubMed] [Google Scholar]
- 2.Bichko, V. V., and J. M. Taylor. 1996. Redistribution of the delta antigens in cells replicating the genome of hepatitis delta virus. J. Virol. 70:8064-8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonino, F., B. Hoyer, J. W. Shih, M. Rizzetto, R. H. Purcell, and J. L. Gerin. 1984. Delta hepatitis agent: structural and antigenic properties of the delta-associated particle. Infect. Immun. 43:1000-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bregman, D. B., L. Du, S. van der Zee, and S. L. Warren. 1995. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 129:287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buratowski, S. 2003. The CTD code. Nat. Struct. Biol. 10:679-680. [DOI] [PubMed] [Google Scholar]
- 6.Chang, F. L., P. J. Chen, S. J. Tu, C. J. Wang, and D. S. Chen. 1991. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 88:8490-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, J., X. Nie, H. E. Chang, Z. Han, and J. Taylor. 2008. Transcription of hepatitis delta virus RNA by RNA polymerase II. J. Virol. 82:1118-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, J., and J. Taylor. 2002. In vivo RNA-directed transcription, with template switching, by a mammalian RNA polymerase. EMBO J. 21:157-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao, Y. C., M. F. Chang, I. Gust, and M. M. Lai. 1990. Sequence conservation and divergence of hepatitis delta virus RNA. Virology 178:384-392. [DOI] [PubMed] [Google Scholar]
- 10.Chen, P. J., G. Kalpana, J. Goldberg, W. Mason, B. Werner, J. Gerin, and J. Taylor. 1986. Structure and replication of the genome of the hepatitis delta virus. Proc. Natl. Acad. Sci. USA 83:8774-8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, Y. S., W. H. Huang, S. Y. Hong, Y. G. Tsay, and P. J. Chen. 2008. ERK1/2-mediated phosphorylation of small hepatitis delta antigen at serine 177 enhances hepatitis delta virus antigenomic RNA replication. J. Virol. 82:9345-9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung, W. H., J. L. Craighead, W. H. Chang, C. Ezeokonkwo, A. Bareket-Samish, R. D. Kornberg, and F. J. Asturias. 2003. RNA polymerase II/TFIIF structure and conserved organization of the initiation complex. Mol. Cell 12:1003-1013. [DOI] [PubMed] [Google Scholar]
- 13.Corden, J. L. 1993. RNA polymerase II transcription cycles. Curr. Opin. Genet. Dev. 3:213-218. [DOI] [PubMed] [Google Scholar]
- 14.Cozzone, A. J. 1988. Protein phosphorylation in prokaryotes. Annu. Rev. Microbiol. 42:97-125. [DOI] [PubMed] [Google Scholar]
- 15.Cramer, P., D. A. Bushnell, J. Fu, A. L. Gnatt, B. Maier-Davis, N. E. Thompson, R. R. Burgess, A. M. Edwards, P. R. David, and R. D. Kornberg. 2000. Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288:640-649. [DOI] [PubMed] [Google Scholar]
- 16.Cramer, P., D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292:1863-1876. [DOI] [PubMed] [Google Scholar]
- 17.Cunha, C., J. Monjardino, D. Cheng, S. Krause, and M. Carmo-Fonseca. 1998. Localization of hepatitis delta virus RNA in the nucleus of human cells. RNA 4:680-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dahmus, M. E. 1995. Phosphorylation of the C-terminal domain of RNA polymerase II. Biochim. Biophys. Acta 1261:171-182. [DOI] [PubMed] [Google Scholar]
- 19.Dahmus, M. E. 1994. The role of multisite phosphorylation in the regulation of RNA polymerase II activity. Prog. Nucleic Acid Res. Mol. Biol. 48:143-179. [DOI] [PubMed] [Google Scholar]
- 20.Dubois, M. F., V. T. Nguyen, S. Bellier, and O. Bensaude. 1994. Inhibitors of transcription such as 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole and isoquinoline sulfonamide derivatives (H-8 and H-7) promote dephosphorylation of the carboxyl-terminal domain of RNA polymerase II largest subunit. J. Biol. Chem. 269:13331-13336. [PubMed] [Google Scholar]
- 21.Filipovska, J., and M. M. Konarska. 2000. Specific HDV RNA-templated transcription by pol II in vitro. RNA 6:41-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu, T. B., and J. Taylor. 1993. The RNAs of hepatitis delta virus are copied by RNA polymerase II in nuclear homogenates. J. Virol. 67:6965-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujinaga, K., D. Irwin, Y. Huang, R. Taube, T. Kurosu, and B. M. Peterlin. 2004. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 24:787-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gnatt, A. L., P. Cramer, J. Fu, D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science 292:1876-1882. [DOI] [PubMed] [Google Scholar]
- 25.Greco-Stewart, V. S., P. Miron, A. Abrahem, and M. Pelchat. 2007. The human RNA polymerase II interacts with the terminal stem-loop regions of the hepatitis delta virus RNA genome. Virology 357:68-78. [DOI] [PubMed] [Google Scholar]
- 26.Gudima, S., S. Y. Wu, C. M. Chiang, G. Moraleda, and J. Taylor. 2000. Origin of hepatitis delta virus mRNA. J. Virol. 74:7204-7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gudima, S. O., J. Chang, and J. M. Taylor. 2005. Reconstitution in cultured cells of replicating HDV RNA from pairs of less than full-length RNAs. RNA 11:90-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han, Z., C. Alves, S. Gudima, and J. Taylor. 2009. Intracellular localization of hepatitis delta virus proteins in the presence and absence of viral RNA accumulation. J. Virol. 83:6457-6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirose, Y., and Y. Ohkuma. 2007. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J. Biochem. 141:601-608. [DOI] [PubMed] [Google Scholar]
- 30.Kitajima, S., T. Chibazakura, M. Yonaha, and Y. Yasukochi. 1994. Regulation of the human general transcription initiation factor TFIIF by phosphorylation. J. Biol. Chem. 269:29970-29977. [PubMed] [Google Scholar]
- 31.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kos, A., R. Dijkema, A. C. Arnberg, P. H. van der Meide, and H. Schellekens. 1986. The hepatitis delta (delta) virus possesses a circular RNA. Nature 323:558-560. [DOI] [PubMed] [Google Scholar]
- 33.Kuo, M. Y., M. Chao, and J. Taylor. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 63:1945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai, M. M. 2005. RNA replication without RNA-dependent RNA polymerase: surprises from hepatitis delta virus. J. Virol. 79:7951-7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamond, A. I., and D. L. Spector. 2003. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4:605-612. [DOI] [PubMed] [Google Scholar]
- 36.Lee, J. M., and A. L. Greenleaf. 1997. Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J. Biol. Chem. 272:10990-10993. [DOI] [PubMed] [Google Scholar]
- 37.Lehmann, E., F. Brueckner, and P. Cramer. 2007. Molecular basis of RNA-dependent RNA polymerase II activity. Nature 450:445-449. [DOI] [PubMed] [Google Scholar]
- 38.Li, Y. J., T. Macnaughton, L. Gao, and M. M. Lai. 2006. RNA-templated replication of hepatitis delta virus: genomic and antigenomic RNAs associate with different nuclear bodies. J. Virol. 80:6478-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, Y. J., M. R. Stallcup, and M. M. Lai. 2004. Hepatitis delta virus antigen is methylated at arginine residues, and methylation regulates subcellular localization and RNA replication. J. Virol. 78:13325-13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo, G. X., M. Chao, S. Y. Hsieh, C. Sureau, K. Nishikura, and J. Taylor. 1990. A specific base transition occurs on replicating hepatitis delta virus RNA. J. Virol. 64:1021-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacNaughton, T. B., E. J. Gowans, S. P. McNamara, and C. J. Burrell. 1991. Hepatitis delta antigen is necessary for access of hepatitis delta virus RNA to the cell transcriptional machinery but is not part of the transcriptional complex. Virology 184:387-390. [DOI] [PubMed] [Google Scholar]
- 42.MacNaughton, T. B., and M. M. Lai. 2002. Genomic but not antigenomic hepatitis delta virus RNA is preferentially exported from the nucleus immediately after synthesis and processing. J. Virol. 76:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacNaughton, T. B., S. T. Shi, L. E. Modahl, and M. M. Lai. 2002. Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J. Virol. 76:3920-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Makino, S., M. F. Chang, C. K. Shieh, T. Kamahora, D. M. Vannier, S. Govindarajan, and M. M. Lai. 1987. Molecular cloning and sequencing of a human hepatitis delta (delta) virus RNA. Nature 329:343-346. [DOI] [PubMed] [Google Scholar]
- 45.Marshall, N. F., J. Peng, Z. Xie, and D. H. Price. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271:27176-27183. [DOI] [PubMed] [Google Scholar]
- 46.Mintz, P. J., and D. L. Spector. 2000. Compartmentalization of RNA processing factors within nuclear speckles. J. Struct. Biol. 129:241-251. [DOI] [PubMed] [Google Scholar]
- 47.Misteli, T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science 291:843-847. [DOI] [PubMed] [Google Scholar]
- 48.Modahl, L. E., and M. M. Lai. 1998. Transcription of hepatitis delta antigen mRNA continues throughout hepatitis delta virus (HDV) replication: a new model of HDV RNA transcription and replication. J. Virol. 72:5449-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moraleda, G., and J. Taylor. 2001. Host RNA polymerase requirements for transcription of the human hepatitis delta virus genome. J. Virol. 75:10161-10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morris, D. P., G. A. Michelotti, and D. A. Schwinn. 2005. Evidence that phosphorylation of the RNA polymerase II carboxyl-terminal repeats is similar in yeast and humans. J. Biol. Chem. 280:31368-31377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mortillaro, M. J., B. J. Blencowe, X. Wei, H. Nakayasu, L. Du, S. L. Warren, P. A. Sharp, and R. Berezney. 1996. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc. Natl. Acad. Sci. USA 93:8253-8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mu, J. J., D. S. Chen, and P. J. Chen. 2001. The conserved serine 177 in the delta antigen of hepatitis delta virus is one putative phosphorylation site and is required for efficient viral RNA replication. J. Virol. 75:9087-9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mu, J. J., Y. G. Tsay, L. J. Juan, T. F. Fu, W. H. Huang, D. S. Chen, and P. J. Chen. 2004. The small delta antigen of hepatitis delta virus is an acetylated protein and acetylation of lysine 72 may influence its cellular localization and viral RNA synthesis. Virology 319:60-70. [DOI] [PubMed] [Google Scholar]
- 54.Mu, J. J., H. L. Wu, B. L. Chiang, R. P. Chang, D. S. Chen, and P. J. Chen. 1999. Characterization of the phosphorylated forms and the phosphorylated residues of hepatitis delta virus delta antigens. J. Virol. 73:10540-10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Palangat, M., D. B. Renner, D. H. Price, and R. Landick. 2005. A negative elongation factor for human RNA polymerase II inhibits the anti-arrest transcript-cleavage factor TFIIS. Proc. Natl. Acad. Sci. USA 102:15036-15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Payne, J. M., P. J. Laybourn, and M. E. Dahmus. 1989. The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIa. J. Biol. Chem. 264:19621-19629. [PubMed] [Google Scholar]
- 57.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rizzetto, M., B. Hoyer, M. G. Canese, J. W. Shih, R. H. Purcell, and J. L. Gerin. 1980. Delta agent: association of delta antigen with hepatitis B surface antigen and RNA in serum of delta-infected chimpanzees. Proc. Natl. Acad. Sci. USA 77:6124-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spector, D. L. 2001. Nuclear domains. J. Cell Sci. 114:2891-2893. [DOI] [PubMed] [Google Scholar]
- 60.Sureau, C., B. Guerra, and R. E. Lanford. 1993. Role of the large hepatitis B virus envelope protein in infectivity of the hepatitis delta virion. J. Virol. 67:366-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan, S., R. C. Conaway, and J. W. Conaway. 1995. Dissection of transcription factor TFIIF functional domains required for initiation and elongation. Proc. Natl. Acad. Sci. USA 92:6042-6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taylor, J. M. 2006. Structure and replication of hepatitis delta virus RNA. Curr. Top. Microbiol. Immunol. 307:1-23. [DOI] [PubMed] [Google Scholar]
- 63.Wang, K. S., Q. L. Choo, A. J. Weiner, J. H. Ou, R. C. Najarian, R. M. Thayer, G. T. Mullenbach, K. J. Denniston, J. L. Gerin, and M. Houghton. 1986. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature 323:508-514. [DOI] [PubMed] [Google Scholar]
- 64.Weiner, A. J., Q. L. Choo, K. S. Wang, S. Govindarajan, A. G. Redeker, J. L. Gerin, and M. Houghton. 1988. A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24 delta and p27 delta. J. Virol. 62:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamaguchi, Y., J. Filipovska, K. Yano, A. Furuya, N. Inukai, T. Narita, T. Wada, S. Sugimoto, M. M. Konarska, and H. Handa. 2001. Stimulation of RNA polymerase II elongation by hepatitis delta antigen. Science 293:124-127. [DOI] [PubMed] [Google Scholar]
- 66.Yamaguchi, Y., T. Mura, S. Chanarat, S. Okamoto, and H. Handa. 2007. Hepatitis delta antigen binds to the clamp of RNA polymerase II and affects transcriptional fidelity. Genes Cells 12:863-875. [DOI] [PubMed] [Google Scholar]
- 67.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 68.Zawel, L., and D. Reinberg. 1992. Advances in RNA polymerase II transcription. Curr. Opin. Cell Biol. 4:488-495. [DOI] [PubMed] [Google Scholar]
- 69.Zeng, C., E. Kim, S. L. Warren, and S. M. Berget. 1997. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 16:1401-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]