Abstract
The optimization of immune responses (IR) induced by HIV DNA vaccines in humans is one of the great challenges in the development of an effective vaccine against AIDS. Ideally, this vaccine should be delivered in a single dose to immunize humans. We recently demonstrated that the immunization of mice with a single dose of a DNA vaccine derived from pathogenic SHIVKU2 (Δ4SHIVKU2) induced long-lasting, potent, and polyfunctional HIV-specific CD8+ T-cell responses (G. Arrode, R. Hegde, A. Mani, Y. Jin, Y. Chebloune, and O. Narayan, J. Immunol. 178:2318-2327, 2007). In the present work, we expanded the characterization of the IR induced by this DNA immunization protocol to rhesus macaques. Animals immunized with a single high dose of Δ4SHIVKU2 DNA vaccine were monitored longitudinally for vaccine-induced IR using multiparametric flow cytometry-based assays. Interestingly, all five immunized macaques developed broad and polyfunctional HIV-specific T-cell IR that persisted for months, with an unusual reemergence in the blood following an initial decline but in the absence of antibody responses. The majority of vaccine-specific CD4+ and CD8+ T cells lacked gamma interferon production but showed high antigen-specific proliferation capacities. Proliferative CD8+ T cells expressed the lytic molecule granzyme B. No integrated viral vector could be detected in mononuclear cells from immunized animals, and this high dose of DNA did not induce any detectable autoimmune responses against DNA. Taken together, our comprehensive analysis demonstrated for the first time the capacity of a single high dose of HIV DNA vaccine alone to induce long-lasting and polyfunctional T-cell responses in the nonhuman primate model, bringing new insights for the design of future HIV vaccines.
The development of a vaccine that substantially decreases the viral load set points and reduces the transmission of HIV-1 appears to be the long-term solution to control the persistently growing epidemic of this virus in the world (10). In the past, vaccines against challenging infectious diseases, including smallpox, polio, measles, and yellow fever, have been the most effective strategies for fighting these human pandemics. However, and unlike these traditional vaccines that mostly rely on the production of neutralizing antibodies (Abs) for protection from pathogenic infections, the control of HIV infection strongly depends on the development of high-frequency, broadly targeted, polyfunctional T-cell responses specific to the virus (11, 28, 45). Live-attenuated simian immunodeficiency virus (SIV)/HIV vaccines so far have been the best inducers of potent T-cell responses that correlate with protection against AIDS following challenge with pathogenic strains in nonhuman primate (NHP) models (24, 39, 47, 61), although the exact correlates of such protection remain to be fully delineated. However, the persistence, integration, and possible reversion to pathogenic forms of these replication-competent vaccines comprise a risk that will not be acceptable for their use in humans.
Instead, the use of DNA-based vaccines as a strategy to induce protective responses to control infectious diseases, including HIV-1/AIDS, is very attractive, based on its safety, the absence of infection even in immunocompromised recipients, and its capacity to induce both humoral and T-cell immune responses. For many years, numerous plasmid DNAs encoding HIV proteins have been developed and tested in animal models, and some of them have been tested in humans (14, 18, 42, 49). However, unlike that in rodents, the immune responses induced in humans and NHPs by these DNA vaccines were dramatically weak despite successive immunizations with multiple doses of DNA (30). To circumvent this limitation, new strategies currently are used to improve the immunogenicity of DNA vaccines, including the incorporation of signal-to-target dendritic cells (43), the codon optimization of HIV antigens (Ag) (14), the coexpression of adjuvant (15), and new tools that optimize the delivery of DNA in target cells in the muscle (34).
We have developed a noninfectious DNA vaccine derived from the highly pathogenic SHIVKU2 expressing seven proteins of HIV under the control of the SIV 5′ long terminal repeat (LTR) promoter (35). This design mimics the natural expression of the viral proteins and leads to the formation of numerous viral-like particles that are extruded out of expressing cells (4). Repeated low-dose immunizations with this vaccine without heterologous boost protected macaques from progression to AIDS following challenge with pathogenic SHIV. However, enzyme-linked immunospot (ELISPOT) assay responses to HIV antigens before challenge were sporadic and weak (35, 54). In contrast, T-cell responses specific to HIV antigens induced by our construct in immunized mice were substantially higher (21). Using the mouse model, we developed a more sensitive immunity-monitoring assay that measures proliferative capacity, cytotoxic potential, and other immune functions (gamma interferon [IFN-γ] and interleukin-2 [IL-2] secretion) and provides more robust indications regarding the immunogenicity induced by the vaccine. We reported that the intramuscular immunization of mice with a single dose of this HIV DNA vaccine induced long-lasting and polyfunctional CD8+ T-cell responses directed against all HIV antigens expressed by the construct. Interestingly, in the absence of any additional immunization, we observed a primary peak of immune responses (IR) within 2 to 4 weeks postinfection (p.i.), followed by a contraction phase and then the late reemergence of responses after 14 to 20 weeks p.i. and lasting until the end of the experiment (more than 63 weeks p.i.). This is a typical pattern of vaccine-specific T-cell responses induced by nonpersistent vectors that progressively elicit secondary lymphoid tissue-based memory T cells as the expressed antigen becomes rare (9, 38, 58). Importantly, the major proportion of these HIV-specific CD8+ T cells was not producing IFN-γ but proliferated vigorously following antigen stimulation and produced the lytic molecule granzyme B (5). The contribution of this type of antigen-specific T cell to viral control remains to be fully elucidated.
The surprising lack of efficacy of the human STEP trial conducted by Merck using the Ad-5 vectors expressing HIV antigens that elicit sustained effector T-cell responses has been disappointing for strategies designed to induce T-cell responses to prevent HIV-1 infections (29, 48, 53, 59). However, we learned from this failure that the characteristics of the T-cell responses induced by candidate vaccines could be critical for immediate as well as long-term protection (20). To address this issue, we developed a multiparametric flow-cytometric assay in the NHP model that was similar to that used in the mouse model. Using this assay, we performed a longitudinal characterization of HIV-specific T-cell IR induced in rhesus macaques immunized with a single high dose of Δ4SHIVKU2 DNA vaccine given intramuscularly (i.m.). We also assessed the antibody responses against HIV antigens. We also addressed potential safety concerns, since this is the first report using one high dose of DNA in NHP, and we tested the animals for the integration of the vaccine genome into that of the circulating mononuclear cells and assessed whether anti-DNA antibodies were produced in all immunized monkeys.
MATERIALS AND METHODS
HIV peptides.
Overlapping 15-mer peptides, with 11-amino-acid (aa) overlaps, spanning the entire molecules of HIV Gag, Env, Tat, Rev, and Nef proteins were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (catalog nos. 8117, 6451, 5138, 6445, and 5189, respectively). These peptides are based on consensus sequences from clade B HIV genomes. Our HIV DNA vaccine encodes Gag and Nef from the SF2 HIV strain and Tat, Rev, and Env from the HXB2 HIV strain (5, 35). These two strains both are clade B viruses.
Animals.
Five 3- to 5-year-old Indian rhesus macaques were purchased and housed in the Laboratory Animal Resources of the University of Kansas Medical Center. All macaques were used in accordance with National Institutes of Health and the University of Kansas Medical Center Institutional Animal Care and Use Committee guidelines.
Vaccine Δ4SHIVKU2 and IL-15 plasmid DNAs.
The construction procedures of SHIVKU2 plasmid DNA have been described earlier (36). The inserted sequences were derived from SHIVKU2 (GenBank accession no. AY751799) and from HIV-1 SF2. The strategy of the construction of Δ4SHIVKU2 plasmid DNA has been described previously (21, 35, 54). This construct carries vpx and vpr genes of SIVmac239 and gag, pro, vpu, tat, rev, env, and nef genes of HIV-1 under the transcriptional control of the SIV 5′ LTR promoter and the poly(A) sequences of simian virus 40 (SV40). The reverse transcriptase integrase and vif genes and the 3′LTR were removed from the SHIVKU2 genome.
The rmamuIL-15 plasmid was kindly provided by François Villinger (Resource for NHP Immune Reagents, Emory University, Atlanta, GA). The detailed construction of this plasmid designed for expression in rhesus macaque IL-15 has been reported previously (15, 57). DNA of this plasmid was prepared under conditions similar to those used for the isolation of Δ4SHIVKU2 DNA. At week 45 p.i. of macaque AL49 with Δ4SHIVKU2, this animal was injected with a 5-mg single dose of IL-15 expression plasmid delivered by the i.m. route.
Inoculation of macaques.
Macaques were inoculated i.m. with a single dose of 30 mg of DNA vaccine at a 6-mg/ml concentration. Endotoxin-free vaccine DNA was produced using a BIOFLO 110 modular Fermentor (New Brunswick Scientific) that routinely produced high yields of DNA following plasmid DNA extraction using the standard methods with the Qiagen Giga kit. All DNAs used to inject the macaques contained at least 90% of the supercoiled (ccc) form of plasmid. DNA solution was prepared in 5 ml of phosphate-buffered saline (PBS) (0.1 M [pH 7.4]) and injected intramuscularly at 10 different sites of the rear legs using a 21-gauge needle.
Processing of blood for plasma and PBMC isolations.
Peripheral blood collected by venipuncture in EDTA or sodium heparin-coated tubes was centrifuged to separate plasma and blood cells. Plasma was frozen and used for the detection of antibodies against HIV proteins and autoantibodies against DNA. Peripheral blood mononuclear cells (PBMCs) were isolated from Buffy coats by centrifugation through Ficoll-Hypaque density gradients. Cells then were divided in three fractions and used to extract DNA for the detection of integrated DNA to perform ELISPOT and multiparametric flow-cytometry assays.
Detection of IFN-γ-producing cells by ELISPOT assay.
We used the quantitative ELISPOT assay, as we previously described in Yankee et al. (64), to evaluate IFN-γ-producing PBMCs in response to groups of overlapping peptides. Millipore multiscreen Immobilon-P opaque hydrophobic high-protein-binding 96-well plates (0.45 μm) were coated with anti-monkey IFN-γ (Mabtech). Cells then were incubated for 18 h at 37°C with medium alone (used as negative control), medium containing peptides, or medium containing concanavalin A (used as a positive control). Cells were washed away, and the membranes were rinsed three times with PBS prior to 3 h of incubation at room temperature with 50 μl of biotinylated anti-IFN-γ antibodies (2 μg/ml) (Mabtech). After six successive rinses, the membranes were incubated for 1 h at room temperature with 50 μl of Vectastain AB (Vector Laboratories). Color development was performed by the addition of 100 μl/well of Nova-Red, incubation for 4 min at room temperature, and then a final rinse with running tap water. Spot-forming IFN-γ-excreting cells (SFC) were counted with a stereomicroscope and reported as the number of spots/106 PBMC. The cutoff for positivity in this assay was >12 SFC/million PBMCs. This corresponds to the average of spots obtained in cultured medium controls ± three standard deviations (SD) (i.e., three times the value of the SD).
Flow-cytometry assays for HIV-specific immune T cells.
Polychromatic (six- to seven-color) flow-cytometry analysis was performed on a three-laser BD LSRII instrument with standard setup. Data files were collected and analyzed using the FACSDiva software program (version 4.1.2; BD Biosciences). To monitor the expansion and proliferation of HIV-specific T cells, carboxy fluoroscein succinimidyl ester (CFSE) (107 cells/ml in 1 μM CFSE for 10 min at 37°C; Molecular Probes)-labeled PBMCs were seeded in deep-well tissue culture plates (96 wells; Nunc) at a density of 2 × 106 cells/well in 1 ml of serum-free medium (AIM V; Invitrogen) alone or loaded with 2 μg/ml of HIV peptides and incubated for 5 days at 37°C. After 5 days of incubation, cells were restimulated for 6 h with medium only or by adding relevant HIV peptides in the presence of 0.5 μg/ml of costimulatory CD28 (clone 37.51) and CD49 (clone 9F10) monoclonal antibodies (MAbs), as well as brefeldin A (Sigma-Aldrich). Cells then were washed and stained with Alexa Fluor 405-conjugated anti-CD3 (clone SP34-2), allophycocyanin-Cy7-conjugated anti-CD8 (clone SK1), and phycoerythrin (PE)-conjugated anti-CD4 (clone L200) MAbs for 20 min at 4°C. Additionally, ethidium monoazide (EMA; Molecular Probes) was added at 0.5 μg/ml during the surface-labeling step to allow the exclusion of dead cells in samples that have been cultured for 5 days and restimulated for 6 h. In such a case, all samples were exposed to light for 15 min at room temperature to allow EMA to covalently link to the DNA in dead cells prior to permeabilization. The cells then were fixed/permeabilized (Cytofix/Cytoperm Plus; BD Biosciences) and stained with PE-Cy7-conjugated anti-IFN-γ (clone B27) and/or Alexa 647-conjugated anti-granzyme B (clone GB11) MAbs for 30 min at room temperature. Cells were washed again (Perm/Wash; BD Biosciences), fixed in 1% paraformaldehyde in PBS, and stored at 4°C until flow-cytometry analysis. All Abs were purchased from BD Biosciences. For each experiment, unstained and all single-color controls were processed to allow proper compensation as well as all-fluorescence-minus-one controls to determine proper population gates. Each analysis was gated on low forward and side scatter lymphocytes (FSC/SSC), EMA−, and CD3+ and a high CD8+ or CD4+ population to allow the collection of 25,000 to 50,000 CD8+ events (>106 total events). Data were displayed as two-color or density dot plots to measure the proportion of the single-positive or double-positive cells in the highly CD3+ CD8+ population (orange) or CD3+ CD4+ population (blue). Bioexponential display also was used to show each population in its entirety.
Assays to detect anti-HIV and anti-DNA antibodies.
To examine whether immunized animals have developed specific antibodies to HIV, we used a commercial enzyme-linked immunosorbent assay (ELISA) kit for the detection of HIV-1 and HIV-2 Abs (BioChain Institute). To examine whether DNA immunization has induced autoimmune anti-DNA responses, we also used a commercial kit that detects anti-double-stranded DNA (dsDNA) Abs (Diagnostic Automation). Plasma samples from macaques were diluted between 1/50 and 1/400 by serial 2-fold dilutions and were used in duplicate to load 96-well polystyrene plates precoated with either HIV antigens for HIV Ab detection or purified dsDNA for anti-DNA Ab detection. Mixtures were incubated for 30 min at 37°C, and then unbound material was washed away. A secondary Ab, horseradish peroxidase (HRP)-conjugated goat anti-human immunoglobulin G (IgG), was added and incubated for 30 min (as recommended by the supplier) and detected with the addition of peroxidase substrate.
Sera from patients diagnosed with HIV or systemic lupus erythematosus (SLE) or from healthy donors were provided by each manufacturer and used as positive and negative controls, respectively. As additional controls we also used stored samples from SIV-infected rhesus macaques. For the anti-HIV Ab kit, the cutoff value calculation (COV) was equal to the average OD (optical density) of negative controls + 0.1, and the OD at 450 nm (OD450) of the sample is positive when the value is greater than the COV. For the detection of anti-DNA Abs, the results were calculated as (IU of calibrator/OD of calibrator) × OD of test sample. A positive response is indicated by >25 IU.
Both assays were read at 450 nm using an Dynatech MR4000 ELISA reader instrument.
Quantitative PCR for detection of plasmid DNA integration.
DNA was extracted from PBMCs (10 × 106) of immunized macaques throughout the 49 weeks after immunization using Qiagen DNA isolation reagents. DNA copy numbers were determined by real-time PCR using specific gag primers (sense, 5′-GCA GAG GAG GAA ATT ACC CAG TAC-3′; antisense, 5′-CAA TTT TAC CCA GGC ATT TAA TGT T-3′) and TaqMan probe (6-carboxyfluorescein-TGT CCA CCT GCC ATT AAG CCC GA-carboxytetramethylrhodamine) and universal PCR Mastermix (Applied Biosystems) in duplicate 25-μl reaction mixtures in the ABI PRISM 7700 Sequence Detection System. Proviral copy numbers were normalized to the quantity of total cellular DNA used in the reaction. DNA real-time conditions were the same as those described by Smith et al. (55). Serial 10-fold dilutions of cloned HIV gag plasmid of more than six orders of magnitude were used as standards. The minimum detectable level of proviral DNA was 30 copies.
RESULTS
IFN-γ ELISPOT assay responses induced by a single dose of Δ4SHIVKU2 DNA vaccine.
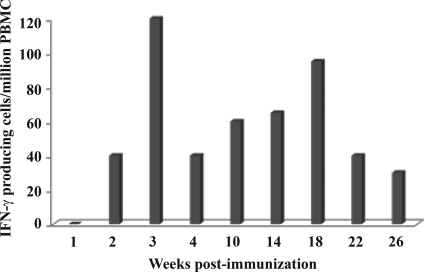
Our DNA vaccine derived from the pathogenic SHIVKU2 is noninfectious, is replication defective, and expresses seven HIV proteins under the control of the SIV 5′ LTR promoter (21). A single dose of intramuscular immunization with this vaccine induced sustained and polyfunctional T-cell responses against all expressed HIV antigens in mice (5). In this study, we evaluated whether similar responses could be induced in a nonhuman primate model using a single dose of HIV DNA vaccine. Since the optimal dose used in mice was 1 mg of DNA per 100 g of body weight, we estimated that the equivalent dose for immunizing macaques would be 30 to 50 mg of DNA. We first tested this single high dose to immunize one rhesus macaque (identified as AL49) with 30 mg of Δ4SHIVKU2 DNA injected intramuscularly. We then used the classical IFN-γ ELISPOT assay to examine whether this dose induced detectable T-cell responses, which generally are not seen after a single plasmid DNA immunization (31). As shown in Fig. 1, substantial numbers of IFN-γ-producing cells in response to stimulation with a single pool of HIV peptides encompassing the entire Gag protein were observed repeatedly in PBMC samples of this macaque over time. This response was detected as early as 2 weeks p.i., peaked at 3 weeks p.i. to reach 120 spots/106, and declined but persisted to at least 26 weeks p.i. This is in striking contrast with the commonly weak or not detectable Gag responses that we and others have observed even following successive DNA immunizations of macaques in the absence of heterologous boost (35, 62). This result clearly demonstrated that the use of a single high dose of our DNA vaccine resulted in a substantial increase of the proportion of T-cell responses specific to HIV Gag antigens without any subsequent boost in an NHP model.
FIG. 1.
IFN-γ ELISPOT responses induced by a single high dose of Δ4SHIVKU2 DNA vaccine. Macaque AL49 was immunized with 30 mg Δ4SHIVKU2, and PBMCs were isolated at different weeks after immunization as indicated on the x axis. PBMCs were used for the ELISPOT assay to detect IFN-γ-producing cells in response to the pool of HIV Gag peptides as described in Materials and Methods. Numbers of spots were counted under a stereomiscroscope and are indicated on the y axis. The background response against the HIV peptides used was determined at week 0 before immunizing the animal.
Broad, polyfunctional, and persistent CD8+ T-cell responses induced by a single dose of Δ4SHIVKU2 DNA vaccine in macaques.
In mice, we previously found that a single dose of the Δ4SHIVKU2 DNA vaccine mostly induced HIV-specific CD8+ T cells that did not produce IFN-γ but proliferated vigorously in response to HIV antigen stimulation and produced the lytic molecule granzyme B (5). In the present study, we developed a novel and highly sensitive multiparametric flow-cytometry assay similar to the one we used with mice. We first used this test to examine the characteristics of T-cell responses found in the immunized animal (AL49) that showed substantial IFN-γ ELISPOT responses. Pools of peptides specific for Gag, Env, and the combination of Tat+ Rev+ Nef (TRN) were used as antigens for in vitro cell stimulation. The expansion of Ag-specific T cells using CFSE labeling, their phenotyping, and the assessment of their capacity to accumulate intracellular IFN-γ and/or granzyme B were performed as described in Materials and Methods.
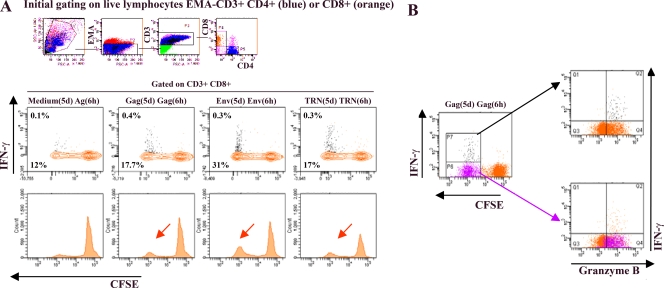
As shown in Fig. 2A, EMA− CD3+ CD8+ T cells exhibited vigorous proliferative capacity against the three pools of HIV-specific antigens. Interestingly, similarly to our previously reported data using the mouse model, the vast majority (more than 95%) of the proliferating T cells did not produce detectable IFN-γ upon restimulation. At 28 weeks p.i., 5% of total CD3+ CD8+ T cells proliferated, but only 0.2% produced IFN-γ in response to TRN peptides. A similar response was measured against Gag, showing that 5.7% of the CD3+ CD8+ T-cell population proliferated, while only 0.3% produced IFN-γ. The highest response was measured against Env, with 19% of total CD3+ CD8+ T cells proliferating but only 0.2% producing IFN-γ. Taken together, these data demonstrated that 7 months after the single high-dose plasmid DNA immunization and in the absence of any additional boost, macaque AL49 contained a pool of HIV-specific CD8+ T cells in the peripheral blood that rapidly expanded ex vivo in the presence of HIV antigens and represented up to 30% of total CD8+ T cells.
FIG. 2.
Polyfunctional HIV-specific CD8+ T cells in vaccinated macaque AL49 at 28 weeks p.i. PBMCs from macaque AL49 were labeled with CFSE and cultured in the presence of 2 μg/ml of specific pools of HIV peptides Gag, Env, TRN, or medium only for 5 days [designated Gag(5d), Env(5d), TRN(5d), and medium(5d)]. On day 5, cells were harvested and restimulated for 6 h with the same mix of peptides [Ag(6h)] in the presence of costimulatory antibodies and brefeldin A. Cells then were surface stained with anti-CD3, anti-CD8, and anti-CD4 MAbs in the presence of EMA (to allow the exclusion of dead cells) and subsequently permeabilized and stained with anti-IFN-γ and anti-granzyme B MAbs. For flow cytometry analysis, we gated on low-FSC/SSC, EMA−, CD3+, and large CD8+ T-cell populations (colored in orange). The proportions of cells producing IFN-γ (contour plot, upper number) and proliferating (CFSE dilution; contour plot, lower number; and histogram [arrow]) and dot plots (B) in response to specific antigens are presented. Frequencies for antigen-specific responses are reported (see Results) as the percent cytokine-secreting or proliferating CD8+ T cells after the subtraction of backgrounds obtained with cells cultured for 5 days with medium only and restimulated for 6 h with relevant mixes of peptides [medium(5d) Ag(6 h)]. (B) Proliferating-only (purple dots) as well as proliferating and IFN-γ-producing (black dots) Gag-specific CD8+ T cells are superposed to the total CD8+ T-cell population (in orange) for granzyme B expression detection (left panels). For simplicity, only the Gag response is shown, but Env and TRN antigens gave similar results.
To further characterize these HIV-specific proliferating T cells, we examined their content of lytic proteins by staining the cells for granzyme B. As shown in Fig. 2B, all Gag-specific proliferating CD3+ CD8+ T cells were found to express granzyme B. Similar results were obtained for Env and TRN antigens (data not shown). Similarly to our previously reported data with the mouse model, the present data confirmed in NHP that a high proportion of HIV-specific CD3+ CD8+ T cells induced by Δ4SHIVKU2 exert cytotoxic activity in the absence of IFN-γ. In the present study, we did not examine the cytotoxic activity of these cells; however, in our previous study we demonstrated that expanded HIV-specific CD8+ T cells from HIV-infected patients that express granzyme or perforin (Perf) exert cytotoxic activity (3).
To determine whether this type of vaccine-induced response was reproducible, we used the same protocol to immunize four additional rhesus macaques, two at the time, using the same single dose of 30 mg of Δ4SHIVKU2 DNA vaccine. All four immunized animals were monitored weekly for the first 4 weeks to detect and characterize the vaccine-induced T-cell responses using the multiparametric flow-cytometry analysis. In addition, similarly to the AL49 animal, two of these animals (AI99 and AJ07) also were examined at later time points (weeks 12 to 23 [W12 to W23]). Because of the limited blood samples that we can obtain weekly from immunized animals, we prioritized the responses specific to Gag.
Samples collected from each macaque before DNA injection were used to establish in our experimental conditions the threshold that helps to distinguish the random noise for PBMCs from each animal from the true proliferation values due to the specificity of the responses against HIV antigens. The threshold of unspecific proliferative response was determined based on the analysis of data from samples cultured in medium for 5 days and then stimulated with HIV Ag for 6 h as well as samples cultured in HIV Ag for 5 days and then restimulated with HIV Ag for an additional 6 h from unvaccinated animals. The threshold of unspecific proliferative response was found to be 0.25%, and that of unspecific IFN-γ secretion was 0.01%. In our conditions, Ag-specific proliferative responses above 0.25% were considered positive. In addition, we systematically included, at each time point, samples of cells from each vaccinated animal that were incubated with complete medium alone (for 5 days) and then HIV Ags (for 6 h) and were used as internal controls to determine the background responses. The values of these background responses then were subtracted from those of measured Ag-specific proliferative responses. The background values of spontaneous proliferation among these five animals ranged from 0.1 to 6.0% (median = 3.6%; n = 28). However, unlike rodents, which are raised in pathogen-free conditions and consequently generate minimal background in this type of experiment, macaques may have unapparent nonpathogenic infections that generate increased background responses. At one time point, we obtained a high proliferative background (12% CFSE-low IFN− CD8+ T cells) with samples from macaque AL49 at W28 p.i. (Fig. 2A) with medium for 5 days and then Gag for 6 h that might reflect this type of infection, in which the proliferation of CD8+ T cells is promoted by components in the medium. This cross-sensitivity overlapped the proliferative responses and may have resulted in the decrease of the true antigen-specific proliferative responses.
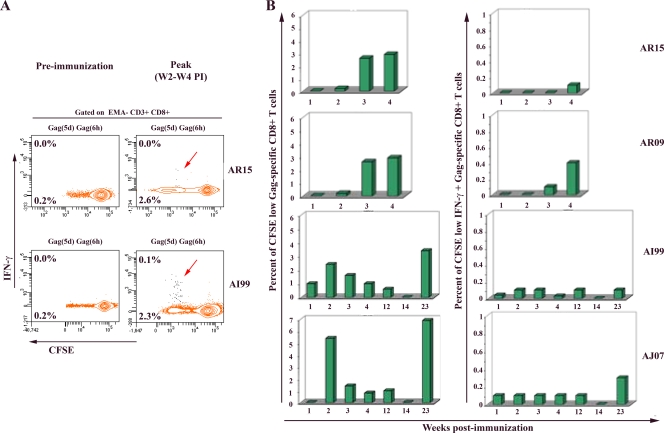
Characteristic results obtained at the initial peak of the IR are shown in Fig. 3A for animals AR15 and AI99. Results of sequential samples for all four macaques (AR15, AR09, AI99 and AJ07) are summarized in Fig. 3B. These results clearly show that each one of the immunized animals was able to mount Gag-specific CD8+ T-cell responses within the first 4 weeks following immunization. The proportion of Gag-specific proliferative CD8+ T cells was found to be between 2.0 and 5.7% of total CD8+ T cells at the initial peak of the IR. In addition, similarly to our previously reported data with the mouse model and those observed here with macaque AL49, only a minor fraction (0.1 to 0.4%) of these proliferative T cells were producing IFN-γ upon restimulation. However, all HIV-specific CD8+ T cells contained lytic granzyme B molecules, as shown in Fig. 2B. In addition, at two time points we collected larger volumes of blood from immunized animals, and we examined the expression of IL-2 and MIP-1β cytokines. While no IL-2 secretion was detected in any of these samples, all HIV-specific CD8+ T cells were found to express MIP-1β (data not shown). Interestingly, the examination of the profile of Gag T-cell responses up to 5 months p.i. for macaques AI99 and AJ07 clearly identified an initial expansion followed by contraction phases of HIV-specific T-cell responses, ending with a reproducible late reemergence in blood, as observed in macaque AL49 and seen in mice. The mechanisms underlying this observation are unclear at present but suggest an ability of the primary Gag-specific CD8+ T cells to constitute a pool of memory T cells that were able to generate secondary Gag-specific CD8+ circulating T-cell responses with features similar to those of the primary response (e.g., the coexistence of two populations of secondary IFN-γ-producer and non-IFN-γ-producer CD8+ T cells, both with vigorous proliferative capacities). Importantly, this occurred in the absence of any additional boost.
FIG. 3.
Longitudinal analysis of polyfunctional Gag-specific CD8+ T-cell responses in four additional vaccinated macaques. Macaques AR15, AR09, AI99, and AJ07 were immunized with 30 mg Δ4SHIVKU2. At the indicated preimmunization and postimmunization times, PBMCs were labeled with CFSE, cultured, restimulated, and stained using the same procedure as that described in the legend of Fig. 2. (A) The proportion of cells producing IFN-γ (contour plot, upper number) and proliferating (CFSE dilution; contour plot, lower number) in response to specific antigens are presented for two representative macaques at the preimmunization time point and at the peak of vaccine-induced IR (between 2 and 4 weeks p.i.). (B) Summary of the frequency of proliferating-only (CFSE low) as well as proliferating and IFN-γ-producing (CFSE low IFN-γ+) Gag-specific CD8+ T cells detected in each immunized animal following weeks of immunization.
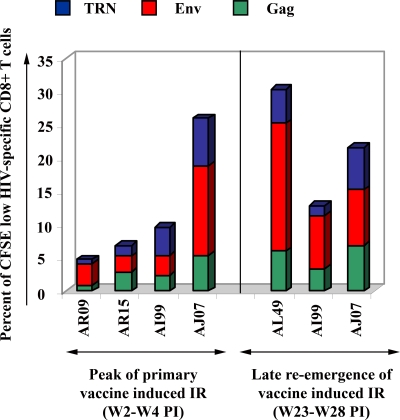
In addition, when the collection of larger samples of blood was possible at the time of the initial peak of the primary IR and the late reemergence, we examined the proliferative responses against the other HIV Ags expressed by our DNA vaccine using Env and a combination of TRN peptides. We found that EMA− CD3+ CD8+ T cells exhibit vigorous proliferative capacity against all HIV-specific antigens (Fig. 4). Among all five animals, the response measured against Env ranged from 2.4 to 13% at the time of the primary IR and 8.0 to 19% of total CD3+ CD8+ T cells at the time of reemergence. The responses against TRN antigens were between 0.7 and 7.0% during the primary phase and 1.5 and 6.3% at the reemergence phase. Taken together, these data demonstrate that 6 to 7 months after the single immunization, vaccinated macaques developed and maintained between 13 and 30% CD8+ T cells that react to multiple HIV antigens, with a vast majority of cells responding to Gag and Env.
FIG. 4.
Proliferative CD8+ T-cell responses in response to other HIV antigens. At the peak (between 2 and 4 weeks p.i.) and late reemergence (between 23 and 28 weeks p.i.) phases of induced IR, larger amounts of PBMCs were obtained for all five immunized macaques to examine the proliferative responses to all three pools of peptides. PBMCs were labeled with CFSE, cultured, restimulated, and stained using the procedure described in the legend to Fig. 2. Summaries of the frequencies of proliferating-only (CFSE low) Gag (green bar), Env (red), and TRN (blue)-specific CD8+ T cells detected in each immunized animal are displayed.
Overall, these results lead to the first demonstration that a single high dose of our HIV DNA vaccine induces broad, long-lasting, and polyfunctional CD8+ T cell responses against Gag and other HIV antigens in all five vaccinated rhesus macaques.
Functional recall memory to HIV Gag in response to IL-15 plasmid DNA injection.
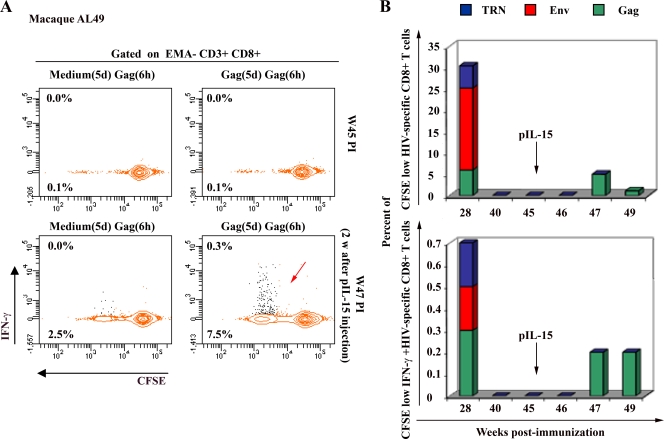
The expansion and contraction phases observed in our study classically are associated with the development of Ag-specific CD8+ T-cell responses. However, the late reemergence of HIV-specific CD8+ T cells observed after week 30 p.i. is intriguing. To examine the persistence of this secondary response, we extended our longitudinal study up to 45 weeks p.i. for the macaque AL49. As shown in Fig. 5A and B, Gag-specific CD8+ T-cell responses became undetectable in the peripheral blood of this animal at week 40 p.i. We hypothesized that Gag-specific memory CD8+ T cells hide in lymphatic reservoir organs after such an extensive absence of viral antigen expression. To test this hypothesis, we chose to inject this animal with 5 mg of IL-15 plasmid i.m., expecting that IL-15 would mobilize and expand resting memory T cells in the periphery. This is exactly what we observed. Two weeks following IL-15 plasmid injection, marked frequencies of detectable Gag-specific CD8+ T cells reemerged in the peripheral blood in a proportion similar to that detected at week 28 p.i. (Fig. 5A and B; other antigens were not tested), suggesting that these memory cells can be mobilized very rapidly given adequate homeostatic signals. Similarly to the present study, in an earlier mouse study we used the same HIV DNA vaccine for immunization, but a mouse-IL-15 expression plasmid (pmIL-15) was given to only one immunized group at the time the HIV-specific CD8+ T-cell responses became undetectable. We observed that the reemergence of immune responses occurred only in mice that received pmIL-15 DNA (our unpublished data). This result clearly indicated that the reemergence of IR was specific to IL-15 injection.
FIG. 5.
Functional recall memory of Gag-specific CD8+ T cells in response to IL-15 plasmid injection. A longitudinal study using the procedure described in the legend to Fig. 2 was performed to monitor macaque AL49 from week 28 to week 45 p.i. At this time, the Gag-specific CD8+ T-cell responses became repeatedly undetectable. We then injected 5 mg of IL-15 plasmid DNA intramuscularly and monitored the weekly IR monitoring up to 49 weeks p.i. (A) The proportion of cells producing IFN-γ (contour plot, upper number) and proliferating (CFSE dilution; contour plot, lower number) in response to Gag peptides is presented at the time of undetectable response (W45 p.i.) and 2 weeks after plasmid IL-15 injection (W47 p.i.). Backgrounds obtained with medium after 5 days [medium(5d)] and Gag after 6 h [Gag(6h)] were subtracted from the reported values shown in Results. (B) Summary of the frequency of proliferating-only (CFSE low; top) as well as proliferating and IFN-γ-producing (CFSE low IFN-γ+; bottom) Gag-specific CD8+ T cells detected from week 28 to week 49 p.i.
Taken together, these results indicated that an injection of a single dose of our HIV DNA vaccine allows the development of polyfunctional Gag-specific CD8+ T cells that demonstrated strong functional recall memory capacity in response to IL-15 cytokine.
Vaccine-induced CD4+ T-cell and B cell responses.
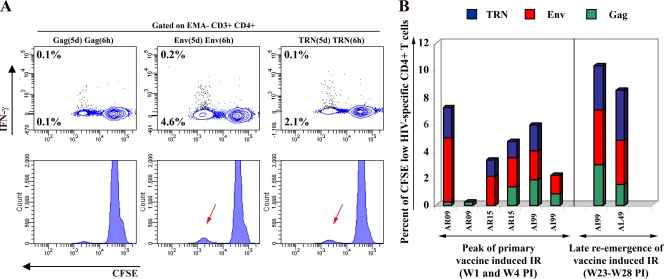
Since we included CD4 staining in our multiparametric flow cytometry analysis, we also examined vaccine-induced CD4+ T-cell responses. As summarized in Fig. 6, Ag-specific CD4+ T cells were detected against all HIV antigens expressed by the vaccine. Similarly to CD8+ T cells, CD4+ T cells also exhibited strong proliferative responses following stimulation with HIV-specific Ags, and only a minor fraction secreted IFN-γ (Fig. 6A). Interestingly, the magnitude of CD4+ T-cell proliferative responses paralleled the CD8+ T-cell findings, with an initial peak response followed by contraction and the late reemergence of antigen-specific T cells in blood, although the kinetics of antigen-specific CD4+ and CD8+ T-cell responses were not synchronized. The proportion of cells that responded to Env ranged from 2.1 to 4.8% during the primary IR and 3.3 to 4.0% of total CD3+ CD4+ T cells during the reemergence of the response. Smaller proportions of Gag-specific cells were detected ranging from 0.2 to 1.9% during the initial peak of the IR and from 1.5 to 3.0% of total CD3+ CD4+ T cells during the reemergence of the response. The proportion of cells specific to TRN antigens ranged from 1.2 to 2.2% during the initial phase and from 3.3 to 3.7% of total CD3+ CD4+ T cells at the reemergence phase of the response. Overall, these results demonstrated that macaques immunized with a single dose of our HIV DNA vaccine, in the absence of any additional boost, developed and maintained CD4+ T cells that may expand to reach a proportion of 8.5 to 10% in response to HIV antigens. The vast majority of responding cells are directed against Env and TRN antigens.
FIG. 6.
HIV-specific CD4+ T-cell responses in immunized macaques. For all five vaccinated macaques, PBMCs were processed and labeled at the peak (between 2 and 4 weeks following immunization) and late reemergence (between 23 and 28 weeks following immunization) phases of vaccine-induced IR using the procedure described in the legend to Fig. 2. (A) Flow cytometry data analyses were performed by gating on a low-FSC/SSC, EMA−, CD3+, and high CD4+ T-cell population (colored in blue). Representative results for macaque AR09 at the peak of the IR are displayed. The proportion of cells producing IFN-γ (contour plot, upper number) and proliferating (CFSE dilution; contour plot, lower number) in response to Gag, Env, and TRN peptides are indicated. (B) Summary of the frequency of proliferating-only (CFSE low) Gag (green bar)-, Env (red)-, and TRN (blue)-specific CD4+ T cells detected in samples from each immunized animal.
With respect to B cell responses induced by our DNA vaccine, we used preimmune and postimmune sera from all immunized macaques to examine the presence of total anti-HIV antibodies by a sensitive ELISA. While sera from SHIV-infected animals used as positive controls all exhibited strong positive ELISA values, none of the macaque sera collected between weeks 0 and 45 were found to contain any detectable anti-HIV antibodies (data not shown), suggesting that heterologous booster immunizations likely would be needed to generate such responses.
Safety issues: plasmid integration and anti-DNA Ab response.
Two main questions were addressed with respect to the safety of injecting a high dose of our DNA vaccine: (i) the integration of plasmid DNA in circulating mononuclear cells, and (ii) the induction of autoimmunity against injected DNA. To examine whether the injection of vaccine DNA was associated with systemic integration into the genome of host peripheral mononuclear cells, DNA samples were isolated from PBMCs of immunized animals at various time points from W0 to W45 p.i. and were used to detect the vaccine genome using the sensitive real-time PCR assay. Results (not shown) demonstrated that none of the animals contained integrated plasmid of the vaccine, while samples of SHIV-infected animals provided easily detectable provirus signals.
In addition, using a sensitive commercial anti-DNA antibody ELISA kit, we evaluated whether the injection of a high dose of DNA was associated with the development of anti-DNA antibodies that may induce SLE-like disease syndrome. All plasma samples collected from week 0 to 45 failed to exhibit any detectable anti-DNA antibody, while the positive control serum provided with the kit was strongly positive (data not shown).
Taken together, these results indicated that the single high dose of vaccine plasmid DNA was associated neither with systemic DNA integrations nor with autoantibody responses to DNA, suggesting that its use as a vaccine is safe.
DISCUSSION
Plasmid DNA vaccines against highly pathogenic agents are considered safe and highly versatile immunogens for humans and animals. However, HIV DNA vaccine candidates alone were found to be relatively poor immunogens in humans compared to viral vectors (49). Therefore, strategies based on DNA prime or multiprime and boost(s) either with protein or recombinant viruses have been developed extensively in animal models and humans. Nevertheless, the immune responses that were monitored mainly were those induced by the boost(s), and the tools used to make such assessments mostly were IFN-γ ELISPOT and intracellular cytokine analyses. While these tools have been informative, one has to keep in mind that the logistics of prime-boost immunization on a large scale will be daunting, and although the cost of manufacture for any vaccine may be a fundamental issue, the need for accessing a patient multiple times is likely a greater challenge. It therefore is imperative that a vaccine be developed with a single immunization modality and a minimal number of boosters, if any. However, to monitor such immunization, it is becoming evident that more comprehensive immune analyses are needed (1). Accordingly, we and others (5, 14, 23) have shown that the use of a more sensitive assay that simultaneously measures proliferative capacity, cytotoxic content, and other immune functions (IFN-γ, IL-2, tumor necrosis factor alpha, and MIP-1β) helps to better characterize the induced T-cell responses to better define the immunogenicity of vaccine candidates.
Using this approach, we revisited the immune responses induced by our HIV DNA vaccine, Δ4SHIVKU2, which has proven to be efficacious against SHIV challenge following repeated low-dose injections in cynomolgus macaques. Our objectives were 2-fold: first, to use a single high dose of DNA vaccine, like we previously did in rodents, and second, to better characterize the type of immune T cells induced by this DNA in NHP. HIV DNA immunogens commonly induce potent HIV-specific T-cell responses in rodents, but in NHP and humans, CD8+ T-cell responses remain consistently weak or undetectable, particularly after a primary immunization. Nevertheless, studies of the nature of protective responses against HIV and SIV indicated that the magnitude and breadth of CD8+ T-cell responses to Gag and other HIV antigens correlate with the control of virus replication (26, 28, 65). Using first the mouse model, we reported that a single-dose injection of Δ4SHIVKU2 induced long-lasting and polyfunctional CD8+ T-cell responses against all HIV antigens with substantial responses toward Gag (5). Recently, Liu et al. (33) also demonstrated that, compared to a lower dose, a single high dose of DNA showed a greater-than 10-fold increase in anti-Gag-specific CD8+ T-cell responses in mice. In humans, the administration of a high dose of DNA alone (7.2 mg) given intramuscularly increased the magnitude and quality of T-cell responses (8). In this report, we also demonstrated that, similarly to the mouse model, a single high dose of our vaccine injected intramuscularly in macaques induced strong, long-lasting, and polyfunctional CD8+ T cell responses to all HIV antigens expressed by the construct. Taken together, these results strongly suggest that the previous failure of the detection of T-cell responses in NHP and humans resulted from insufficient doses of DNA vaccines leading to inefficient antigen expression. In addition, our data reemphasized the importance of the dose in the kinetics of primary T-cell IR as well as their memorization in the absence of any additional boost with HIV antigens. However, even though this high dose has provided a proof of principle in NHP, it might be difficult to transpose it to humans. So far, investigators have focused on alternative approaches to keep the amount of DNA small but increase the DNA uptake. Interestingly, in vivo electroporation (EP) has been shown to augment the immunogenicity of plasmid DNA and allowed a more-than 10-fold reduction in vaccine dose (34). Indeed, EP has been used commonly in basic research for many years to introduce DNA into recipient cells in vitro. This technology recently has been adapted for the in vivo gene transfer of DNA vaccines and therapeutic plasmids to the skin, muscle, tumors, and other tissues and has resulted in high levels of protein expression in many species, including humans (6, 13, 32, 50). It will be interesting to evaluate whether such a route of immunization will help to reduce the amount of DNA given without altering the quantitative and qualitative characteristics of the induced IR. As an additional mechanism, using our HIV DNA construct we emphasized in a recent report the importance of induced cell death in the generation of Gag-specific CD8+ T-cell responses (4). Such an event commonly is associated with electroporation methods (7, 22, 46, 56, 60), likely contributing to the cross-presentation of apoptotic bodies by DCs. In a recent study, Yan et al. evaluated the immunogenicity of three separate DNA plasmids coding for consensus Gag, Env, and Pol in mice and cynomolgus macaques. After three repeated low-dose immunizations, they detected IFN-γ ELISPOT as well as proliferative responses. These responses were dominant toward Env and Pol, but Gag-specific CD8+ T-cell responses remained poor (62). These data are in contrast to those we obtained using a single dose and a single DNA expressing all genes showing an efficient priming of Gag responses.
Our comprehensive analysis of the immune responses induced by this single-dose approach suggests that the priming event plays a crucial role in imprinting long-lasting properties to the immune T-cell responses. As previously observed with the mouse model, the characteristics of vaccine-induced T-cell responses that reemerged several weeks later after the contraction phase remained very close to those detected in the primary responses. Moreover, spontaneous reemerging cells share characteristics with cells reemerging following induction by IL-15 expression. Whether T cells of these early and late responses have a similar efficacy to control the virus replication and to protect from pathogenic virus infection remain to be determined. Indeed, the heterologous challenge of immunized animals at early (8 weeks), middle (20 weeks), and late (40 weeks) time points p.i. with pathogenic SIVmac239 certainly will indicate which population of T cells is associated with protection.
Single-dose vaccination with our Δ4SHIVKU2 vaccine both in mice and in macaques induced sustained CD8+ T-cell responses predominantly composed of proliferating T cells with lytic granzyme B content, suggesting that the nature of T-cell IR induced by our DNA vaccine is not species specific. Importantly, similarly to this data, proliferative HIV-specific CD8+ T cells expressing the lytic molecule perforin (Perf) with only a fraction expressing IFN-γ were observed in the peripheral blood of long-term nonprogressor (LTNP) patients (40, 41). In our hands, using the same protocol, we examined blood cells from two LTNP patients and also confirmed the presence of Perf+ IFN-γ− CD8+ T cells (data not shown). These results found in both LTNP and DNA-vaccinated animals suggest that this type of cell is induced in the absence of persistent antigen expression and is in accordance with the recent study reported by Hansen et al. (20). The authors have shown that immunization with live recombinant vector allowing the persistent expression of Ags was associated with persistent effector memory T-cell responses responsible for the protection of rhesus macaques from mucosal SIV challenge. This is in contrast to nonpersistent vectors, which produce Ag for a limited time and elicit secondary lymphoid tissue-based or central memory T cells. These Ag-specific T cells require expansion, differentiation, and migration out of the lymphoid tissue to produce the peak of effector responses and prevent extensive replication at viral replication sites. In our mouse study, most of the vaccine-elicited CD8+ T cells exhibited a Tcm phenotype from the peak of the IR up to their late reemergence (5). One cannot exclude that Tcm-like cells also have been generated in our immunized macaques. Notably, at the time the T-cell IR became undetectable, we induced the reemergence of HIV-specific CD8+ T-cell responses in the periphery shortly after IL-15 DNA injection and in the absence of antigenic boost. This also may indicate that our protocol of a single Δ4SHIVKU2 DNA injection favors the development of secondary lymphoid tissue-based HIV-specific T cells. Whether the injection of recombinant IL-15 accelerates this process of reactivation and recirculation without altering the qualitative and/or quantitative responses remains unknown. The spread of HIV-specific T cells at mucosal sites is critical. Nevertheless, macaques vaccinated with live-attenuated SIV that develop poor mucosal IR still controlled SIVmac239 virus inoculated intravenously (39).
Consistently with our previously reported data (5, 21, 35, 54) and unlike other reports (18, 19, 37, 51, 63), immunization with our vaccine failed to induce any detectable antibody response to any HIV antigens, likely due to the absence of a booster immunization. If both neutralizing antibodies and T-cell responses are indispensable to prevent HIV infection, as is believed currently (12, 16, 17, 25, 52), this immunization protocol alone will not be sufficient for protection.
The major concerns with this type of vaccine in regard to safety are predominantly the potential integration of the plasmid DNA into the host genome and adverse immunopathological effects generated by the formation of anti-DNA antibodies that may lead to autoimmune disease. Since this is the first description of the use of a high dose of HIV DNA vaccine in NHP, we examined these two issues. In our recently published data, we clearly demonstrated that the great majority of primary cells transfected by our Δ4SHIVKU2 DNA vaccine underwent apoptosis following the expression of viral proteins (4). Assuming that this mechanism also occurs in cells expressing our construct in vivo, we believe that this results in the physical elimination of recipient cells, thereby decreasing dramatically the proportion of cells that could have integrated the vaccine genome. In addition, our highly sensitive real-time PCR technique failed to detect any proviral genome in PBMCs from all five immunized animals. However, we cannot totally exclude that DNA has persisted in the muscle injection site (2, 44). In a recent DNA vaccine study of humans, the authors found a delayed-type hypersensitivity (DTH) response at the site of DNA injection following a protein boost (27). Taken together, persistent DNA and/or DTH may participate in the persistence and/or late reemergence of IR observed in the absence of any antigen boost in our study.
Finally, the absence of anti-DNA antibodies in the sera of all immunized animals has demonstrated that our immunization protocol was not associated with the development of autoimmunity that could be responsible for SLE-like disease. Thus, we can conclude that increasing the dose of our DNA vaccine given in one shot did not result in an increase of risks linked to safety. Taken together, our comprehensive analysis demonstrated for the first time that a single high dose of HIV DNA vaccine alone is safe to induce potent, long-lasting, and polyfunctional T-cell responses in the NHP model. This approach brings new insights for the design of future HIV vaccines.
Acknowledgments
This work was supported by grants 2P20 RR016443-07 and R01 AI062340-04 from the National Institutes of Health and funding from INRA, Department of Animal Health.
We thank the National Institutes of Health AIDS Research and Reference Reagent Program for providing HIV overlapping Gag, Env, Tat, Rev, and Nef peptides (catalog no. 8117, 6451, 5138, 6445, and 5189, respectively) and the Resource for NHP Immune Reagents (http://pathology.emory.edu/Villinger/index.htm) for the IL-15 expression construct.
Footnotes
Published ahead of print on 18 November 2009.
In memory of Opendra Narayan.
REFERENCES
- 1.Appay, V., D. C. Douek, and D. A. Price. 2008. CD8+ T-cell efficacy in vaccination and disease. Nat. Med. 14:623-628. [DOI] [PubMed] [Google Scholar]
- 2.Armengol, G., L. M. Ruiz, and S. Orduz. 2004. The injection of plasmid DNA in mouse muscle results in lifelong persistence of DNA, gene expression, and humoral response. Mol. Biotechnol. 27:109-118. [DOI] [PubMed] [Google Scholar]
- 3.Arrode, G., J. S. Finke, H. Zebroski, F. P. Siegal, and R. M. Steinman. 2005. CD8+ T cells from most HIV-1-infected patients, even when challenged with mature dendritic cells, lack functional recall memory to HIV gag but not other viruses. Eur. J. Immunol. 35:159-170. [DOI] [PubMed] [Google Scholar]
- 4.Arrode, G., R. Hegde, Y. Jin, D. K. Singh, O. Narayan, and Y. Chebloune. 2008. Nef modulates the immunogenicity of Gag encoded in a non-infectious HIV DNA vaccine. Vaccine 26:3795-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arrode, G., R. Hegde, A. Mani, Y. Jin, Y. Chebloune, and O. Narayan. 2007. Phenotypic and functional analysis of immune CD8+ T-cell responses induced by a single injection of a HIV DNA vaccine in mice. J. Immunol. 178:2318-2327. [DOI] [PubMed] [Google Scholar]
- 6.Aung, W., S. Hasegawa, M. Koshikawa-Yano, T. Obata, H. Ikehira, T. Furukawa, I. Aoki, and T. Saga. 2009. Visualization of in vivo electroporation-mediated transgene expression in experimental tumors by optical and magnetic resonance imaging. Gene Ther. 16:830-839. [DOI] [PubMed] [Google Scholar]
- 7.Babiuk, S., M. E. Baca-Estrada, M. Foldvari, D. M. Middleton, D. Rabussay, G. Widera, and L. A. Babiuk. 2004. Increased gene expression and inflammatory cell infiltration caused by electroporation are both important for improving the efficacy of DNA vaccines. J. Biotechnol. 110:1-10. [DOI] [PubMed] [Google Scholar]
- 8.Bansal, A., B. Jackson, K. West, S. Wang, S. Lu, J. S. Kennedy, and P. A. Goepfert. 2008. Multifunctional T-cell characteristics induced by a polyvalent DNA prime/protein boost human immunodeficiency virus type 1 vaccine regimen given to healthy adults are dependent on the route and dose of administration. J. Virol. 82:6458-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbour, J. D., L. C. Ndhlovu, Q. Xuan Tan, T. Ho, L. Epling, B. M. Bredt, J. A. Levy, F. M. Hecht, and E. Sinclair. 2009. High CD8+ T cell activation marks a less differentiated HIV-1 specific CD8+ T cell response that is not altered by suppression of viral replication. PLoS One 4:e4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barouch, D. H. 2008. Challenges in the development of an HIV-1 vaccine. Nature 455:613-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Betts, M. R., M. C. Nason, S. M. West, S. C. De Rosa, S. A. Migueles, J. Abraham, M. M. Lederman, J. M. Benito, P. A. Goepfert, M. Connors, M. Roederer, and R. A. Koup. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, H., A. Piechocka-Trocha, T. Miura, M. A. Brockman, B. D. Julg, B. M. Baker, A. C. Rothchild, B. L. Block, A. Schneidewind, T. Koibuchi, F. Pereyra, T. M. Allen, and B. D. Walker. 2009. Differential neutralization of human immunodeficiency virus (HIV) replication in autologous CD4 T cells by HIV-specific cytotoxic T lymphocytes. J. Virol. 83:3138-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daud, A. I., R. C. DeConti, S. Andrews, P. Urbas, A. I. Riker, V. K. Sondak, P. N. Munster, D. M. Sullivan, K. E. Ugen, J. L. Messina, and R. Heller. 2008. Phase I trial of interleukin-12 plasmid electroporation in patients with metastatic melanoma. J. Clin. Oncol. 26:5896-5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Rosa, S. D., and M. J. McElrath. 2008. T cell responses generated by HIV vaccines in clinical trials. Curr. Opin. HIV AIDS 3:375-379. [DOI] [PubMed] [Google Scholar]
- 15.Dubie, R. A., S. Maksaereekul, B. L. Shacklett, D. Lemongello, K. S. Cole, F. Villinger, S. A. Blozis, P. A. Luciw, and E. E. Sparger. 2009. Co-immunization with IL-15 enhances cellular immune responses induced by a vif-deleted simian immunodeficiency virus proviral DNA vaccine and confers partial protection against vaginal challenge with SIVmac251. Virology 386:109-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fauci, A. S., M. I. Johnston, C. W. Dieffenbach, D. R. Burton, S. M. Hammer, J. A. Hoxie, M. Martin, J. Overbaugh, D. I. Watkins, A. Mahmoud, and W. C. Greene. 2008. HIV vaccine research: the way forward. Science 321:530-532. [DOI] [PubMed] [Google Scholar]
- 17.Friedrich, T. C., and D. I. Watkins. 2008. Wanted: correlates of vaccine-induced protection against simian immunodeficiency virus. Curr. Opin. HIV AIDS 3:393-398. [DOI] [PubMed] [Google Scholar]
- 18.Graham, B. S., R. A. Koup, M. Roederer, R. T. Bailer, M. E. Enama, Z. Moodie, J. E. Martin, M. M. McCluskey, B. K. Chakrabarti, L. Lamoreaux, C. A. Andrews, P. L. Gomez, J. R. Mascola, and G. J. Nabel. 2006. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 DNA candidate vaccine. J. Infect. Dis. 194:1650-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamajima, K., S. Sasaki, J. Fukushima, T. Kaneko, K. Q. Xin, I. Kudoh, and K. Okuda. 1998. Intranasal administration of HIV-DNA vaccine formulated with a polymer, carboxymethylcellulose, augments mucosal antibody production and cell-mediated immune response. Clin. Immunol. Immunopathol. 88:205-210. [DOI] [PubMed] [Google Scholar]
- 20.Hansen, S. G., C. Vieville, N. Whizin, L. Coyne-Johnson, D. C. Siess, D. D. Drummond, A. W. Legasse, M. K. Axthelm, K. Oswald, C. M. Trubey, M. Piatak, Jr., J. D. Lifson, J. A. Nelson, M. A. Jarvis, and L. J. Picker. 2009. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 15:293-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hegde, R., Z. Liu, G. Mackay, M. Smith, Y. Chebloune, O. Narayan, and D. K. Singh. 2005. Antigen expression kinetics and immune responses of mice immunized with noninfectious simian-human immunodeficiency virus DNA. J. Virol. 79:14688-14697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofmann, F., H. Ohnimus, C. Scheller, W. Strupp, U. Zimmermann, and C. Jassoy. 1999. Electric field pulses can induce apoptosis. J. Membr. Biol. 169:103-109. [DOI] [PubMed] [Google Scholar]
- 23.Horton, H., E. P. Thomas, J. A. Stucky, I. Frank, Z. Moodie, Y. Huang, Y. L. Chiu, M. J. McElrath, and S. C. De Rosa. 2007. Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J. Immunol. Methods 323:39-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, R. P., R. L. Glickman, J. Q. Yang, A. Kaur, J. T. Dion, M. J. Mulligan, and R. C. Desrosiers. 1997. Induction of vigorous cytotoxic T-lymphocyte responses by live attenuated simian immunodeficiency virus. J. Virol. 71:7711-7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnston, M. I., and A. S. Fauci. 2008. An HIV vaccine—challenges and prospects. N. Engl. J. Med. 359:888-890. [DOI] [PubMed] [Google Scholar]
- 26.Kawada, M., T. Tsukamoto, H. Yamamoto, N. Iwamoto, K. Kurihara, A. Takeda, C. Moriya, H. Takeuchi, H. Akari, and T. Matano. 2008. Gag-specific cytotoxic T-lymphocyte-based control of primary simian immunodeficiency virus replication in a vaccine trial. J. Virol. 82:10199-10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kennedy, J. S., M. Co, S. Green, K. Longtine, J. Longtine, M. A. O'Neill, J. P. Adams, A. L. Rothman, Q. Yu, R. Johnson-Leva, R. Pal, S. Wang, S. Lu, and P. Markham. 2008. The safety and tolerability of an HIV-1 DNA prime-protein boost vaccine (DP6-001) in healthy adult volunteers. Vaccine 26:4420-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, K. Bishop, M. van der Stok, K. Nair, N. Khan, H. Crawford, R. Payne, A. Leslie, J. Prado, A. Prendergast, J. Frater, N. McCarthy, C. Brander, G. H. Learn, D. Nickle, C. Rousseau, H. Coovadia, J. I. Mullins, D. Heckerman, B. D. Walker, and P. Goulder. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46-53. [DOI] [PubMed] [Google Scholar]
- 29.Korber, B. T., N. L. Letvin, and B. F. Haynes. 2009. T-cell vaccine strategies for human immunodeficiency virus, the virus with a thousand faces. J. Virol. 83:8300-8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kutzler, M. A., and D. B. Weiner. 2008. DNA vaccines: ready for prime time? Nat. Rev. Genet. 9:776-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwissa, M., R. R. Amara, H. L. Robinson, B. Moss, S. Alkan, A. Jabbar, F. Villinger, and B. Pulendran. 2007. Adjuvanting a DNA vaccine with a TLR9 ligand plus Flt3 ligand results in enhanced cellular immunity against the simian immunodeficiency virus. J. Exp. Med. 204:2733-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laddy, D. J., J. Yan, A. S. Khan, H. Andersen, A. Cohn, J. Greenhouse, M. Lewis, J. Manischewitz, L. R. King, H. Golding, R. Draghia-Akli, and D. B. Weiner. 2009. Electroporation of synthetic DNA antigens offers protection in nonhuman primates challenged with highly pathogenic avian influenza virus. J. Virol. 83:4624-4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, J., M. Hellerstein, M. McDonnel, R. R. Amara, L. S. Wyatt, B. Moss, and H. L. Robinson. 2007. Dose-response studies for the elicitation of CD8 T cells by a DNA vaccine, used alone or as the prime for a modified vaccinia Ankara boost. Vaccine 25:2951-2958. [DOI] [PubMed] [Google Scholar]
- 34.Liu, J., R. Kjeken, I. Mathiesen, and D. H. Barouch. 2008. Recruitment of antigen-presenting cells to the site of inoculation and augmentation of human immunodeficiency virus type 1 DNA vaccine immunogenicity by in vivo electroporation. J. Virol. 82:5643-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, Z., D. K. Singh, D. Sheffer, M. S. Smith, S. Dhillon, Y. Chebloune, R. Hegde, S. Buch, and O. Narayan. 2006. Immunoprophylaxis against AIDS in macaques with a lentiviral DNA vaccine. Virology 351:444-454. [DOI] [PubMed] [Google Scholar]
- 36.Liu, Z. Q., S. Muhkerjee, M. Sahni, C. McCormick-Davis, K. Leung, Z. Li, V. H. Gattone, Jr., C. Tian, R. W. Doms, T. L. Hoffman, R. Raghavan, O. Narayan, and E. B. Stephens. 1999. Derivation and biological characterization of a molecular clone of SHIV(KU-2) that causes AIDS, neurological disease, and renal disease in rhesus macaques. Virology 260:295-307. [DOI] [PubMed] [Google Scholar]
- 37.Luckay, A., M. K. Sidhu, R. Kjeken, S. Megati, S. Y. Chong, V. Roopchand, D. Garcia-Hand, R. Abdullah, R. Braun, D. C. Montefiori, M. Rosati, B. K. Felber, G. N. Pavlakis, I. Mathiesen, Z. R. Israel, J. H. Eldridge, and M. A. Egan. 2007. Effect of plasmid DNA vaccine design and in vivo electroporation on the resulting vaccine-specific immune responses in rhesus macaques. J. Virol. 81:5257-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Makaroff, L. E., D. W. Hendricks, R. E. Niec, and P. J. Fink. 2009. Postthymic maturation influences the CD8 T cell response to antigen. Proc. Natl. Acad. Sci. USA 106:4799-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mansfield, K., S. M. Lang, M. C. Gauduin, H. B. Sanford, J. D. Lifson, R. P. Johnson, and R. C. Desrosiers. 2008. Vaccine protection by live, attenuated simian immunodeficiency virus in the absence of high-titer antibody responses and high-frequency cellular immune responses measurable in the periphery. J. Virol. 82:4135-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Migueles, S. A., A. C. Laborico, W. L. Shupert, M. S. Sabbaghian, R. Rabin, C. W. Hallahan, D. Van Baarle, S. Kostense, F. Miedema, M. McLaughlin, L. Ehler, J. Metcalf, S. Liu, and M. Connors. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 3:1061-1068. [DOI] [PubMed] [Google Scholar]
- 41.Migueles, S. A., C. M. Osborne, C. Royce, A. A. Compton, R. P. Joshi, K. A. Weeks, J. E. Rood, A. M. Berkley, J. B. Sacha, N. A. Cogliano-Shutta, M. Lloyd, G. Roby, R. Kwan, M. McLaughlin, S. Stallings, C. Rehm, M. A. O'Shea, J. Mican, B. Z. Packard, A. Komoriya, S. Palmer, A. P. Wiegand, F. Maldarelli, J. M. Coffin, J. W. Mellors, C. W. Hallahan, D. A. Follman, and M. Connors. 2008. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity 29:1009-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulligan, M. J., N. D. Russell, C. Celum, J. Kahn, E. Noonan, D. C. Montefiori, G. Ferrari, K. J. Weinhold, J. M. Smith, R. R. Amara, and H. L. Robinson. 2006. Excellent safety and tolerability of the human immunodeficiency virus type 1 pGA2/JS2 plasmid DNA priming vector vaccine in HIV type 1 uninfected adults. AIDS Res. Hum. Retrovir. 22:678-683. [DOI] [PubMed] [Google Scholar]
- 43.Nchinda, G., J. Kuroiwa, M. Oks, C. Trumpfheller, C. G. Park, Y. Huang, D. Hannaman, S. J. Schlesinger, O. Mizenina, M. C. Nussenzweig, K. Uberla, and R. M. Steinman. 2008. The efficacy of DNA vaccination is enhanced in mice by targeting the encoded protein to dendritic cells. J. Clin. Investig. 118:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orság, P., V. Kvardova, M. Raska, A. D. Miller, M. Ledvina, and J. Turanek. 2008. Quantitative real-time PCR study on persistence of pDNA vaccine pVax-Hsp60 TM814 in beef muscles. Genet. Vaccines Ther. 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pantaleo, G., and R. A. Koup. 2004. Correlates of immune protection in HIV-1 infection: what we know, what we don't know, what we should know. Nat. Med. 10:806-810. [DOI] [PubMed] [Google Scholar]
- 46.Piñero, J., M. Lopez-Baena, T. Ortiz, and F. Cortes. 1997. Apoptotic and necrotic cell death are both induced by electroporation in HL60 human promyeloid leukaemia cells. Apoptosis 2:330-336. [DOI] [PubMed] [Google Scholar]
- 47.Reynolds, M. R., A. M. Weiler, K. L. Weisgrau, S. M. Piaskowski, J. R. Furlott, J. T. Weinfurter, M. Kaizu, T. Soma, E. J. Leon, C. MacNair, D. P. Leaman, M. B. Zwick, E. Gostick, S. K. Musani, D. A. Price, T. C. Friedrich, E. G. Rakasz, N. A. Wilson, A. B. McDermott, R. Boyle, D. B. Allison, D. R. Burton, W. C. Koff, and D. I. Watkins. 2008. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J. Exp. Med. 205:2537-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robb, M. L. 2008. Failure of the Merck HIV vaccine: an uncertain step forward. Lancet 372:1857-1858. [DOI] [PubMed] [Google Scholar]
- 49.Robinson, H. L., and K. J. Weinhold. 2006. Phase 1 clinical trials of the National Institutes of Health Vaccine Research Center HIV/AIDS candidate vaccines. J. Infect. Dis. 194:1625-1627. [DOI] [PubMed] [Google Scholar]
- 50.Rosati, M., A. Valentin, R. Jalah, V. Patel, A. von Gegerfelt, C. Bergamaschi, C. Alicea, D. Weiss, J. Treece, R. Pal, P. D. Markham, E. T. Marques, J. T. August, A. Khan, R. Draghia-Akli, B. K. Felber, and G. N. Pavlakis. 2008. Increased immune responses in rhesus macaques by DNA vaccination combined with electroporation. Vaccine 26:5223-5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sasaki, S., K. Sumino, K. Hamajima, J. Fukushima, N. Ishii, S. Kawamoto, H. Mohri, C. R. Kensil, and K. Okuda. 1998. Induction of systemic and mucosal immune responses to human immunodeficiency virus type 1 by a DNA vaccine formulated with QS-21 saponin adjuvant via intramuscular and intranasal routes. J. Virol. 72:4931-4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sattentau, Q. 2008. Correlates of antibody-mediated protection against HIV infection. Curr. Opin. HIV AIDS 3:368-374. [DOI] [PubMed] [Google Scholar]
- 53.Sekaly, R. P. 2008. The failed HIV Merck vaccine study: a step back or a launching point for future vaccine development? J. Exp. Med. 205:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh, D. K., Z. Liu, D. Sheffer, G. A. Mackay, M. Smith, S. Dhillon, R. Hegde, F. Jia, I. Adany, and O. Narayan. 2005. A noninfectious simian/human immunodeficiency virus DNA vaccine that protects macaques against AIDS. J. Virol. 79:3419-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith, M. S., Y. Niu, Z. Li, I. Adany, D. M. Pinson, Z. Q. Liu, T. Berry, D. Sheffer, F. Jia, and O. Narayan. 2002. Systemic infection and limited replication of SHIV vaccine virus in brains of macaques inoculated intracerebrally with infectious viral DNA. Virology 301:130-135. [DOI] [PubMed] [Google Scholar]
- 56.Stacey, K. J., I. L. Ross, and D. A. Hume. 1993. Electroporation and DNA-dependent cell death in murine macrophages. Immunol. Cell Biol. 71:75-85. [DOI] [PubMed] [Google Scholar]
- 57.Villinger, F., S. S. Brar, A. Mayne, N. Chikkala, and A. A. Ansari. 1995. Comparative sequence analysis of cytokine genes from human and nonhuman primates. J. Immunol. 155:3946-3954. [PubMed] [Google Scholar]
- 58.Wahid, R., R. Salerno-Goncalves, C. O. Tacket, M. M. Levine, and M. B. Sztein. 2008. Generation of specific effector and memory T cells with gut- and secondary lymphoid tissue-homing potential by oral attenuated CVD 909 typhoid vaccine in humans. Mucosal Immunol. 1:389-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watkins, D. I., D. R. Burton, E. G. Kallas, J. P. Moore, and W. C. Koff. 2008. Nonhuman primate models and the failure of the Merck HIV-1 vaccine in humans. Nat. Med. 14:617-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weaver, J. C. 1993. Electroporation: a general phenomenon for manipulating cells and tissues. J. Cell Biochem. 51:426-435. [DOI] [PubMed] [Google Scholar]
- 61.Wyand, M. S., K. Manson, D. C. Montefiori, J. D. Lifson, R. P. Johnson, and R. C. Desrosiers. 1999. Protection by live, attenuated simian immunodeficiency virus against heterologous challenge. J. Virol. 73:8356-8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan, J., D. A. Hokey, M. P. Morrow, N. Corbitt, K. Harris, D. Harris, and D. B. Weiner. 2009. Novel SIVmac DNA vaccines encoding Env, Pol and Gag consensus proteins elicit strong cellular immune responses in cynomolgus macaques. Vaccine 27:3260-3266. [DOI] [PubMed] [Google Scholar]
- 63.Yan, J., H. Yoon, S. Kumar, M. P. Ramanathan, N. Corbitt, M. Kutzler, A. Dai, J. D. Boyer, and D. B. Weiner. 2007. Enhanced cellular immune responses elicited by an engineered HIV-1 subtype B consensus-based envelope DNA vaccine. Mol. Ther. 15:411-421. [DOI] [PubMed] [Google Scholar]
- 64.Yankee, T. M., D. Sheffer, Z. Liu, S. Dhillon, F. Jia, Y. Chebloune, E. B. Stephens, and O. Narayan. 2009. Longitudinal study to assess the safety and efficacy of a live-attenuated SHIV vaccine in long term immunized rhesus macaques. Virology 383:103-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zuñiga, R., A. Lucchetti, P. Galvan, S. Sanchez, C. Sanchez, A. Hernandez, H. Sanchez, N. Frahm, C. H. Linde, H. S. Hewitt, W. Hildebrand, M. Altfeld, T. M. Allen, B. D. Walker, B. T. Korber, T. Leitner, J. Sanchez, and C. Brander. 2006. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J. Virol. 80:3122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]